Algorithmically Reconstructed Molecular Pathways as the New Generation of Prognostic Molecular Biomarkers in Human Solid Cancers
Abstract
1. Introduction
2. Materials and Methods
2.1. Interactome Model and Gene-Centric Molecular Pathways
2.2. Classical Molecular Pathways
2.3. Gene Expression Data and Clinical Annotation
2.4. Calculation of Pathway Activation Levels
2.5. Statistical Analysis of Potential Cancer-Specific Biomarkers
2.6. Statistical Analysis of Survival Characteristics
2.7. Software
3. Results
3.1. Assessment of Potential Cancer-Type Biomarkers
3.2. Assessment of Potential Tumor Biomarkers
3.3. Assessment of Potential Survival Biomarkers
3.4. Prognostic Performance of Hazard Ratio for Overall Survival and Progression-Free Survival
4. Discussion
5. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Code Availability Statement
Conflicts of Interest
References
- Hong, L.; Yang, J.; Han, Y.; Lu, Q.; Cao, J.; Syed, L. High Expression of miR-210 Predicts Poor Survival in Patients with Breast Cancer: A Meta-Analysis. Gene 2012, 507, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Visser, E.; Franken, I.A.; Brosens, L.A.A.; Ruurda, J.P.; van Hillegersberg, R. Prognostic Gene Expression Profiling in Esophageal Cancer: A Systematic Review. Oncotarget 2017, 8, 5566–5577. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Wang, H.Y.; Liu, Y.; Zhao, M.C.; Zhang, H.J.; Lu, Z.Y.; Fang, Y.C.; Chen, X.F.; Liu, G.T. The Prognostic Value of PD-L1 Expression for Non-Small Cell Lung Cancer Patients: A Meta-Analysis. Eur. J. Surg. Oncol. (EJSO) 2015, 41, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Pirker, R.; Pereira, J.R.; von Pawel, J.; Krzakowski, M.; Ramlau, R.; Park, K.; de Marinis, F.; Eberhardt, W.E.E.; Paz-Ares, L.; Störkel, S.; et al. EGFR Expression as a Predictor of Survival for First-Line Chemotherapy plus Cetuximab in Patients with Advanced Non-Small-Cell Lung Cancer: Analysis of Data from the Phase 3 FLEX Study. Lancet Oncol. 2012, 13, 33–42. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Z.; Zhang, W.; Zhang, W.; Buzdin, A.; Mu, X.; Yan, Q.; Zhao, X.; Chang, H.-H.; Duhon, M.; et al. FDA-Approved and Emerging Next Generation Predictive Biomarkers for Immune Checkpoint Inhibitors in Cancer Patients. Front. Oncol. 2021, 11, 683419. [Google Scholar] [CrossRef] [PubMed]
- Buzdin, A.; Sorokin, M.; Garazha, A.; Glusker, A.; Aleshin, A.; Poddubskaya, E.; Sekacheva, M.; Kim, E.; Gaifullin, N.; Giese, A.; et al. RNA Sequencing for Research and Diagnostics in Clinical Oncology. Semin. Cancer Biol. 2020, 60, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Colomer, R.; Aranda-López, I.; Albanell, J.; García-Caballero, T.; Ciruelos, E.; López-García, M.Á.; Cortés, J.; Rojo, F.; Martín, M.; Palacios-Calvo, J. Biomarkers in Breast Cancer: A Consensus Statement by the Spanish Society of Medical Oncology and the Spanish Society of Pathology. Clin. Transl. Oncol. 2018, 20, 815–826. [Google Scholar] [CrossRef]
- Knyazeva, M.; Korobkina, E.; Karizky, A.; Sorokin, M.; Buzdin, A.; Vorobyev, S.; Malek, A. Reciprocal Dysregulation of MiR-146b and MiR-451 Contributes in Malignant Phenotype of Follicular Thyroid Tumor. Int. J. Mol. Sci. 2020, 21, 5950. [Google Scholar] [CrossRef]
- Lezhnina, K.; Kovalchuk, O.; Zhavoronkov, A.A.; Korzinkin, M.B.; Zabolotneva, A.A.; Shegay, P.V.; Sokov, D.G.; Gaifullin, N.M.; Rusakov, I.G.; Aliper, A.M.; et al. Novel Robust Biomarkers for Human Bladder Cancer Based on Activation of Intracellular Signaling Pathways. Oncotarget 2014, 5, 9022–9032. [Google Scholar] [CrossRef][Green Version]
- Hayashi, M.; Guida, E.; Inokawa, Y.; Goldberg, R.; Reis, L.O.; Ooki, A.; Pilli, M.; Sadhukhan, P.; Woo, J.; Choi, W.; et al. GULP1 Regulates the NRF2-KEAP1 Signaling Axis in Urothelial Carcinoma. Sci. Signal. 2020, 13, eaba0443. [Google Scholar] [CrossRef]
- Raevskiy, M.; Sorokin, M.; Vladimirova, U.; Suntsova, M.; Efimov, V.; Garazha, A.; Drobyshev, A.; Moisseev, A.; Rumiantsev, P.; Li, X.; et al. EGFR Pathway-Based Gene Signatures of Druggable Gene Mutations in Melanoma, Breast, Lung, and Thyroid Cancers. Biochemistry 2021, 86, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Ignatev, K.; Barbara, V.; Vladimirova, U.; Muraveva, A.; Suntsova, M.; Gaifullin, N.; Vorotnikov, I.; Kamashev, D.; Bondarenko, A.; et al. Molecular Pathway Activation Markers Are Associated with Efficacy of Trastuzumab Therapy in Metastatic HER2-Positive Breast Cancer Better than Individual Gene Expression Levels. Biochemistry 2020, 85, 758–772. [Google Scholar] [CrossRef]
- Zhu, Q.; Izumchenko, E.; Aliper, A.M.; Makarev, E.; Paz, K.; Buzdin, A.A.; Zhavoronkov, A.A.; Sidransky, D. Pathway Activation Strength Is a Novel Independent Prognostic Biomarker for Cetuximab Sensitivity in Colorectal Cancer Patients. Hum. Genome Var. 2015, 2, 15009. [Google Scholar] [CrossRef] [PubMed]
- Buzdin, A.; Sorokin, M.; Garazha, A.; Sekacheva, M.; Kim, E.; Zhukov, N.; Wang, Y.; Li, X.; Kar, S.; Hartmann, C.; et al. Molecular Pathway Activation—New Type of Biomarkers for Tumor Morphology and Personalized Selection of Target Drugs. Semin. Cancer Biol. 2018, 53, 110–124. [Google Scholar] [CrossRef]
- Buzdin, A.; Tkachev, V.; Zolotovskaia, M.; Garazha, A.; Moshkovskii, S.; Borisov, N.; Gaifullin, N.; Sorokin, M.; Suntsova, M. Using Proteomic and Transcriptomic Data to Assess Activation of Intracellular Molecular Pathways. Adv. Protein Chem. Struct. Biol. 2021, 127, 1–53. [Google Scholar] [CrossRef]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. PLoS Comput. Biol. 2012, 8, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Raevskiy, M.; Sorokin, M.; Zakharova, G.; Tkachev, V.; Borisov, N.; Kuzmin, D.; Kremenchutckaya, K.; Gudkov, A.; Kamashev, D.; Buzdin, A. Better Agreement of Human Transcriptomic and Proteomic Cancer Expression Data at the Molecular Pathway Activation Level. Int. J. Mol. Sci. 2022, 23, 2611. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Ozerov, I.V.; Lezhnina, K.V.; Izumchenko, E.; Artemov, A.V.; Medintsev, S.; Vanhaelen, Q.; Aliper, A.; Vijg, J.; Osipov, A.N.; Labat, I.; et al. In Silico Pathway Activation Network Decomposition Analysis (iPANDA) as a Method for Biomarker Development. Nat. Commun. 2016, 7, 13427. [Google Scholar] [CrossRef]
- Sorokin, M.; Borisov, N.; Kuzmin, D.; Gudkov, A.; Zolotovskaia, M.; Garazha, A.; Buzdin, A. Algorithmic Annotation of Functional Roles for Components of 3,044 Human Molecular Pathways. Front. Genet. 2021, 12, 617059. [Google Scholar] [CrossRef]
- Borisov, N.; Sorokin, M.; Garazha, A.; Buzdin, A. Quantitation of Molecular Pathway Activation Using RNA Sequencing Data. In Nucleic Acid Detection and Structural Investigations; Astakhova, K., Bukhari, S.A., Eds.; Springer: New York, NY, USA, 2020; Volume 2063, pp. 189–206. ISBN 978-1-07-160137-2. [Google Scholar]
- Poddubskaya, E.V.; Baranova, M.P.; Allina, D.O.; Sekacheva, M.I.; Makovskaia, L.A.; Kamashev, D.E.; Suntsova, M.V.; Barbara, V.S.; Kochergina-Nikitskaya, I.N.; Aleshin, A.A. Personalized Prescription of Imatinib in Recurrent Granulosa Cell Tumor of the Ovary: Case Report. Mol. Case Stud. 2019, 5, a003434. [Google Scholar] [CrossRef] [PubMed]
- Borisov, N.; Suntsova, M.; Sorokin, M.; Garazha, A.; Kovalchuk, O.; Aliper, A.; Ilnitskaya, E.; Lezhnina, K.; Korzinkin, M.; Tkachev, V.; et al. Data Aggregation at the Level of Molecular Pathways Improves Stability of Experimental Transcriptomic and Proteomic Data. Cell Cycle 2017, 16, 1810–1823. [Google Scholar] [CrossRef] [PubMed]
- Gudkov, A.; Shirokorad, V.; Kashintsev, K.; Sokov, D.; Nikitin, D.; Anisenko, A.; Borisov, N.; Sekacheva, M.; Gaifullin, N.; Garazha, A.; et al. Gene Expression-Based Signature Can Predict Sorafenib Response in Kidney Cancer. Front. Mol. Biosci. 2022, 9, 753318. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.L.; Sorokin, M.; Kantelhardt, S.R.; Kalasauskas, D.; Sprang, B.; Fauss, J.; Ringel, F.; Garazha, A.; Albert, E.; Gaifullin, N.; et al. Intratumoral Heterogeneity and Longitudinal Changes in Gene Expression Predict Differential Drug Sensitivity in Newly Diagnosed and Recurrent Glioblastoma. Cancers 2020, 12, 520. [Google Scholar] [CrossRef]
- Sorokin, M.; Zolotovskaia, M.; Nikitin, D.; Suntsova, M.; Poddubskaya, E.; Glusker, A.; Garazha, A.; Moisseev, A.; Li, X.; Sekacheva, M.; et al. Personalized Targeted Therapy Prescription in Colorectal Cancer Using Algorithmic Analysis of RNA Sequencing Data. BMC Cancer 2022, 22, 1113. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, M.; Poddubskaya, E.; Baranova, M.; Glusker, A.; Kogoniya, L.; Markarova, E.; Allina, D.; Suntsova, M.; Tkachev, V.; Garazha, A.; et al. RNA Sequencing Profiles and Diagnostic Signatures Linked with Response to Ramucirumab in Gastric Cancer. Cold Spring Harb. Mol. Case Stud. 2020, 6, a004945. [Google Scholar] [CrossRef]
- Venkova, L.; Aliper, A.; Suntsova, M.; Kholodenko, R.; Shepelin, D.; Borisov, N.; Malakhova, G.; Vasilov, R.; Roumiantsev, S.; Zhavoronkov, A.; et al. Combinatorial High-Throughput Experimental and Bioinformatic Approach Identifies Molecular Pathways Linked with the Sensitivity to Anticancer Target Drugs. Oncotarget 2015, 6, 27227–27238. [Google Scholar] [CrossRef]
- Buzdin, A.A.; Zhavoronkov, A.A.; Korzinkin, M.B.; Venkova, L.S.; Zenin, A.A.; Smirnov, P.Y.; Borisov, N.M. Oncofinder, a New Method for the Analysis of Intracellular Signaling Pathway Activation Using Transcriptomic Data. Front. Genet. 2014, 5, 55. [Google Scholar] [CrossRef]
- Zolotovskaia, M.A.; Tkachev, V.S.; Guryanova, A.A.; Simonov, A.M.; Raevskiy, M.M.; Efimov, V.V.; Wang, Y.; Sekacheva, M.I.; Garazha, A.V.; Borisov, N.M.; et al. OncoboxPD: Human 51 672 Molecular Pathways Database with Tools for Activity Calculating and Visualization. Comput. Struct. Biotechnol. J. 2022, 20, 2280–2291. [Google Scholar] [CrossRef]
- Zolotovskaia, M.; Tkachev, V.; Sorokin, M.; Garazha, A.; Kim, E.; Kantelhardt, S.R.; Bikar, S.-E.; Zottel, A.; Šamec, N.; Kuzmin, D.; et al. Algorithmically Deduced FREM2 Molecular Pathway Is a Potent Grade and Survival Biomarker of Human Gliomas. Cancers 2021, 13, 4117. [Google Scholar] [CrossRef]
- Zolotovskaia, M.A.; Kovalenko, M.A.; Tkachev, V.S.; Simonov, A.M.; Sorokin, M.I.; Kim, E.; Kuzmin, D.V.; Karademir-Yilmaz, B.; Buzdin, A.A. Next-Generation Grade and Survival Expression Biomarkers of Human Gliomas Based on Algorithmically Reconstructed Molecular Pathways. Int. J. Mol. Sci. 2022, 23, 7330. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, D. BioCarta; Biotech Software & Internet Report; Mary Ann Liebert: Larchmont, NY, USA, 2001; Volume 2, pp. 117–120. [Google Scholar] [CrossRef]
- Romero, P.; Wagg, J.; Green, M.L.; Kaiser, D.; Krummenacker, M.; Karp, P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004, 6, R2. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Li, C.; Marcu, A.; Badran, H.; Pon, A.; Budinski, Z.; Patron, J.; Lipton, D.; Cao, X.; Oler, E.; et al. PathBank: A Comprehensive Pathway Database for Model Organisms. Nucleic Acids Res. 2020, 48, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Wspolczesna Onkol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Haynes, W. Benjamini–Hochberg Method. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Therneau, T.; Lumley, T. Survival: Survival Analysis. 2016. Available online: https://cran.r-project.org/web/packages/survival/survival.pdf (accessed on 20 May 2023).
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Dietz, K., Gail, M., Krickeberg, K., Samet, J., Tsiatis, A., Eds.; Statistics for Biology and Health; Springer: New York, NY, USA, 2000; ISBN 978-1-4419-3161-0. [Google Scholar]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Survminer: Drawing Survival Curves Using “Ggplot2”. 2021. Available online: https://cran.r-project.org/web/packages/survminer/survminer.pdf (accessed on 20 May 2023).
- Haibe-Kains, B.; Desmedt, C.; Sotiriou, C.; Bontempi, G. A Comparative Study of Survival Models for Breast Cancer Prognostication Based on Microarray Data: Does a Single Gene Beat Them All? Bioinformatics 2008, 24, 2200–2208. [Google Scholar] [CrossRef]
- Schröder, M.S.; Culhane, A.C.; Quackenbush, J.; Haibe-Kains, B. Survcomp: An R/Bioconductor Package for Performance Assessment and Comparison of Survival Models. Bioinformatics 2011, 27, 3206–3208. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. 2016. Available online: https://ggplot2.tidyverse.org (accessed on 20 May 2023).
- Zolotovskaia, M.A.; Sorokin, M.I.; Petrov, I.V.; Poddubskaya, E.V.; Moiseev, A.A.; Sekacheva, M.I.; Borisov, N.M.; Tkachev, V.S.; Garazha, A.V.; Kaprin, A.D.; et al. Disparity between Inter-Patient Molecular Heterogeneity and Repertoires of Target Drugs Used for Different Types of Cancer in Clinical Oncology. Int. J. Mol. Sci. 2020, 21, 1580. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, P.; Aubry, M.-C.; Kosari, F.; Endo, C.; Molina, J.; Vasmatzis, G. Can Gene Expression Profiling Predict Survival for Patients with Squamous Cell Carcinoma of the Lung? Mol. Cancer 2004, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Y.; Hang, B.; Zou, X.; Mao, J.-H. A Novel Gene Expression-Based Prognostic Scoring System to Predict Survival in Gastric Cancer. Oncotarget 2016, 7, 55343–55351. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, W.; Liu, Z.-Q.; Zhou, H.-H. Associating Transcriptional Modules with Colon Cancer Survival through Weighted Gene Co-Expression Network Analysis. BMC Genom. 2017, 18, 361. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, H.; Sun, Z.; Bungum, A.O.; Edell, E.S.; Lingle, W.L.; Stoddard, S.M.; Zhang, M.; Jen, J.; Yang, P.; et al. Network-Based Approach Identified Cell Cycle Genes as Predictor of Overall Survival in Lung Adenocarcinoma Patients. Lung Cancer 2013, 80, 91–98. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, L.; Wu, L.; Chen, H.; Zhu, W.; He, Y. Identification of Prognostic Genes in Kidney Renal Clear Cell Carcinoma by RNA-Seq Data Analysis. Mol. Med. Rep. 2017, 15, 1661–1667. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, G.; Hossain, A.; Danilova, I.G.; Bolkov, M.A.; Liu, G.; Tuzankina, I.A.; Tan, W. A Co-Expression Network for Differentially Expressed Genes in Bladder Cancer and a Risk Score Model for Predicting Survival. Hereditas 2019, 156, 24. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Ahn, T.; Goo, T.; Lee, C.; Kim, S.; Han, K.; Park, S.; Park, T. Deep Learning-Based Identification of Cancer or Normal Tissue Using Gene Expression Data. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1748–1752. [Google Scholar]
- Xiao, Y.; Wu, J.; Lin, Z.; Zhao, X. A Deep Learning-Based Multi-Model Ensemble Method for Cancer Prediction. Comput. Methods Programs Biomed. 2018, 153, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Shi, Y.; Li, C.; Kim, J.; Cai, W.; Han, Z.; Feng, D.D. DeepGene: An Advanced Cancer Type Classifier Based on Deep Learning and Somatic Point Mutations. BMC Bioinform. 2016, 17, 476. [Google Scholar] [CrossRef] [PubMed]
- Mostavi, M.; Chiu, Y.-C.; Huang, Y.; Chen, Y. Convolutional Neural Network Models for Cancer Type Prediction Based on Gene Expression. BMC Med. Genom. 2020, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.; Chiu, Y.-C.; Hererra, A.; Mostavi, M.; Ramirez, J.; Chen, Y.; Huang, Y.; Jin, Y.-F. Classification of Cancer Types Using Graph Convolutional Neural Networks. Front. Phys. 2020, 8, 203. [Google Scholar] [CrossRef]
- Bludau, I.; Frank, M.; Dörig, C.; Cai, Y.; Heusel, M.; Rosenberger, G.; Picotti, P.; Collins, B.C.; Röst, H.; Aebersold, R. Systematic detection of functional proteoform groups from bottom-up proteomic datasets. Nat. Commun. 2021, 12, 3810. [Google Scholar] [CrossRef]



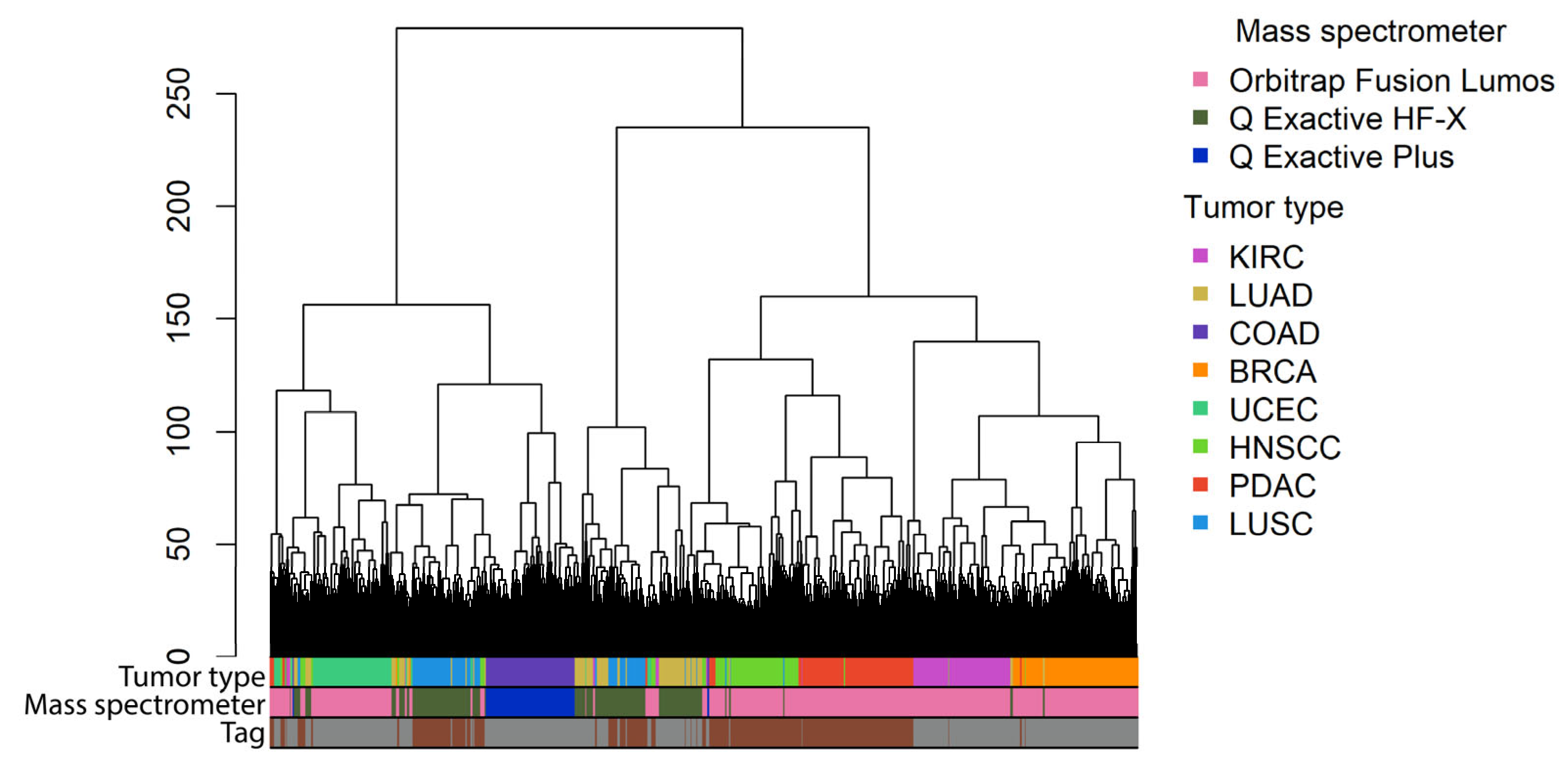






| Cancer Type | Abbreviation | RNAseq Data | Proteomic Data | ||||
|---|---|---|---|---|---|---|---|
| Number of Tumor Samples | Number of Samples with Survival Data | Number of Tumor-Matched Normal Samples | Number of Tumor Samples | Number of Samples with Survival Data | Number of Normal Samples | ||
| Urothelial carcinoma | BLCA * | 412 | 403 | 19 | - | - | - |
| Infiltrating ductal carcinoma of the breast | BRCA * | 1106 | 777 | 113 | 240 | 102 | 21 |
| Cervical squamous cell carcinoma | CESC * | 304 | 252 | 3 | - | - | - |
| Colon adenocarcinoma | COAD * | 478 | 452 | 41 | 95 | - | 100 |
| Esophageal carcinoma | ESCA | 162 | 162 | 11 | - | - | - |
| Head and neck squamous cell carcinoma | HNSC | 502 | 502 | 44 | 110 | - | 68 |
| Clear cell renal cell carcinoma | KIRC | 540 | 532 | 72 | 110 | 101 | 84 |
| Papillary renal cell carcinoma | KIRP | 290 | 290 | 32 | - | - | - |
| Hepatocellular carcinoma | LIHC * | 371 | 355 | 50 | - | - | - |
| Lung adenocarcinoma | LUAD | 537 | 516 | 59 | 113 | 105 | 102 |
| Lung squamous cell carcinoma | LUSC | 502 | 501 | 49 | 110 | 107 | 102 |
| Infiltrating ductal adenocarcinoma of the pancreas | PAAD * | 178 | 143 | 4 | 137 | 108 | 74 |
| Pheochromocytoma | PCPG * | 179 | 149 | 3 | - | - | - |
| Prostate adenocarcinoma | PRAD * | 500 | 484 | 52 | - | - | - |
| Rectal adenocarcinoma | READ | 166 | 165 | 10 | - | - | - |
| Sarcomas | SARC | 259 | 259 | 2 | - | - | - |
| Cutaneous melanoma | SKCM | 103 | 103 | 1 | - | - | - |
| Stomach adenocarcinoma | STAD | 375 | 375 | 32 | - | - | - |
| Thyroid carcinoma | THCA * | 504 | 496 | 59 | - | - | - |
| Thymomas | THYM * | 120 | 109 | 2 | - | - | - |
| Endometrioid adenocarcinoma | UCEC * | 553 | 401 | 35 | 103 | 88 | 30 |
| All cancer types | Total | 8141 | 7426 | 693 | 1018 | 611 | 581 |
| TCGA Cancer ID | Marker Genes | Marker Classical Pathways | Marker Gene-Centric Pathways | TCGA Cancer ID | Marker Genes | Marker Classical Pathways | Marker Gene-Centric Pathways |
|---|---|---|---|---|---|---|---|
| PCPG | 9601 (39%) | 1488 (49%) | 3691 (49%) | LUSC | 2610 (10%) | 974 (32%) | 2614 (35%) |
| BLCA | 1959 (8%) | 268 (9%) | 608 (8%) | PAAD | 3108 (13%) | 824 (27%) | 2085 (28%) |
| BRCA | 4590 (18%) | 350 (12%) | 709 (9%) | PRAD | 6412 (26%) | 451 (15%) | 1212 (16%) |
| CESC | 3743 (15%) | 1244 (41%) | 2961 (40%) | SKCM | 5969 (24%) | 1505 (50%) | 3652 (49%) |
| UCEC | 10,579 (43%) | 864 (29%) | 1904 (25%) | STAD | 6168 (25%) | 781 (26%) | 1595 (21%) |
| COAD | 9082 (37%) | 1334 (44%) | 3322 (44%) | THYM | 7846 (32%) | 1332 (44%) | 3164 (42%) |
| HNSC | 5263 (21%) | 636 (21%) | 2961 (40%) | THCA | 8343 (34%) | 727 (24%) | 1760 (24%) |
| KIRC | 6709 (27%) | 1198 (40%) | 2866 (38%) | SARC | 4042 (16%) | 1037 (34%) | 1954 (26%) |
| KIRP | 6186 (25%) | 989 (33%) | 2790 (37%) | ESCA | 7963 (32%) | 1898 (63%) | 4869 (65%) |
| LIHC | 13,180 (53%) | 760 (25%) | 2429 (33%) | READ | 7988 (32%) | 1304 (43%) | 3328 (45%) |
| LUAD | 1660 (7%) | 661 (22%) | 1780 (24%) | Total | 24,349 | 3020 | 7441 |
| CPTAC Project ID | Proteins | Classical Pathways | Gene-Centric Pathways | Label, TMT10/TMT11 | Mass Spectrometer |
|---|---|---|---|---|---|
| KIRC PDC000127 | 3585 (52%) | 4231 (57%) | 1691 (56%) | TMT10 | Orbitrap Fusion Lumos |
| LUAD PDC000153 | 1488 (22%) | 2000 (27%) | 793 (26%) | TMT10 | Q Exactive HF-X |
| COAD PDC000116 | 1554 (23%) | 1923 (26%) | 776 (26%) | TMT10 | Q Exactive Plus |
| BRCA PDC000120 | 3607 (53%) | 4131 (56%) | 1659 (55%) | TMT10 | Orbitrap Fusion Lumos |
| UCEC PDC000125 | 3220 (47%) | 3698 (50%) | 1620 (54%) | TMT10 | Orbitrap Fusion Lumos |
| HNSC PDC000221 | 1410 (21%) | 1631 (22%) | 659 (22%) | TMT11 | Orbitrap Fusion Lumos |
| LUSC PDC000234 | 2371 (35%) | 3888 (52%) | 1209 (40%) | TMT11 | Q Exactive HF-X |
| PDAC PDC000270 | 3977 (58%) | 4793 (65%) | 1983 (66%) | TMT11 | Orbitrap Fusion Lumos |
| Total | 6742 | 2950 | 7343 |
| Mass Spectrometer | Individual Proteins (%) | Classical Pathways (%) | Gene-Centric Pathways (%) |
|---|---|---|---|
| Orbitrap Fusion Lumos (5 datasets) | 46 | 50 | 51 |
| Q Exactive HF-X (2 datasets) | 29 | 39 | 33 |
| Q Exactive Plus (1 dataset) | 22 | 27 | 26 |
| TCGA Cancer ID | Marker Genes | Marker Classical Pathways | Marker Gene-Centric Pathways | TCGA Cancer ID | Marker Genes | Marker Classical Pathways | Marker Gene-Centric Pathways |
|---|---|---|---|---|---|---|---|
| PCPG | 0 (0%) | 0 (0%) | 0 (0%) | LUSC | 11,508 (46%) | 2013 (67%) | 4989 (67%) |
| BLCA | 7379 (30%) | 1294 (43%) | 2939 (39%) | PAAD | 0 (0%) | 0 (0%) | 0 (0%) |
| BRCA | 8328 (33%) | 1515 (50%) | 3427 (46%) | PRAD | 6031 (24%) | 1271 (42%) | 3005 (40%) |
| CESC | 0 (0%) | 537 (18%) | 784 (10%) | SKCM | 0 (0%) | 0 (0%) | 0 (0%) |
| UCEC | 9445 (38%) | 1568 (52%) | 3868 (52%) | STAD | 8032 (32%) | 813 (27%) | 1825 (24%) |
| COAD | 10,351 (42%) | 2048 (68%) | 4701 (63%) | THYM | 0 (0%) | 0 (0%) | 0 (0%) |
| HNSC | 6172 (25%) | 1011 (33%) | 2563 (34%) | THCA | 7481 (30%) | 845 (28%) | 1759 (23%) |
| KIRC | 10,559 (42%) | 1500 (50%) | 3471 (46%) | SARC | 0 (0%) | 0 (0%) | 0 (0%) |
| KIRP | 12,673 (51%) | 1738 (58%) | 4140 (55%) | ESCA | 10,671 (43%) | 1712 (57%) | 4537 (61%) |
| LIHC | 12,245 (49%) | 1276 (42%) | 3627 (48%) | READ | 10,510 (42%) | 2005 (66%) | 4640 (62%) |
| LUAD | 10,836 (44%) | 1271 (42%) | 3008 (40%) | Total | 24,548 | 3021 | 7466 |
| CPTAC Project ID | Proteins | Classical Pathways | Gene-Centric Pathways | Label, TMT10/TMT11 | Mass Spectrometer |
|---|---|---|---|---|---|
| KIRC PDC000127 | 5649 (57%) | 2078 (71%) | 5054 (69%) | TMT10 | Orbitrap Fusion Lumos |
| LUAD PDC000153 | 5622 (51%) | 1936 (66%) | 5419 (74%) | TMT10 | Q Exactive HF-X |
| COAD PDC000116 | 3657 (49%) | 1927 (67%) | 5116 (71%) | TMT10 | Q Exactive Plus |
| BRCA PDC000120 | 5417 (52%) | 2076 (71%) | 5299 (72%) | TMT10 | Orbitrap Fusion Lumos |
| UCEC PDC000125 | 6147 (57%) | 2047 (70%) | 4889 (67%) | TMT10 | Orbitrap Fusion Lumos |
| HNSC PDC000221 | 4650 (45%) | 1571 (53%) | 4607 (63%) | TMT11 | Orbitrap Fusion Lumos |
| PDAC PDC000270 | 4387 (43%) | 1844 (63%) | 4401 (60%) | TMT11 | Orbitrap Fusion Lumos |
| LUSC PDC000234 | 6673 (58%) | 2055 (70%) | 5386 (73%) | TMT11 | Q Exactive HF-X |
| Total |
| Reference | Disease | Input Data | Results | Gene Network Construction Method |
|---|---|---|---|---|
| [52] | Lung squamous cell carcinoma | RNA expression data for 15 patients | Seven out of 24 gene networks generated from differentially expressed genes were correlated with overall survival | http://www.ingenuity.com (accessed on 20 May 2023) |
| [53] | Gastric cancer | RNA expression data for 265 (TCGA) + 200 (GSE15459) patients | Gene correlation network of 249 genes significantly associated with overall survival. Four functional network components were highlighted | http://baderlab.org/Software/ExpressionCorrelation (accessed on 20 May 2023)) |
| [54] | Colon cancer | RNA expression data for 461 patients (GSE39582) | 11 gene networks associated with tumor grade and progression-free survival | WGCNA |
| [55] | Lung adenocarcinoma | RNA expression data for 82 patients | Gene network enriched with cell cycle-related genes correlated with tumor grade and overall survival | WGCNA |
| [56] | Renal clear cell carcinoma | RNA expression data for 533 patients (TCGA) | From 12 gene networks, two (“cell cycle” and “p53 signaling” pathways) associated with overall survival | WGCNA |
| [57] | Bladder cancer | RNA expression data for 414 patients (TCGA) | Protein interactions of 77 genes: 37 genes formed a network related to overall survival | WGCNA + STRING |
| This study; cancer type markers | Pan-cancer analysis: 21 cancer types | RNA expression data for 8141 patients (TCGA) | For 14 of 21 cancer types, both gene-centric and classical pathways were better cancer type-specific biomarkers than individual genes. In total, 3020 classical and 7441 genecentric pathways were identified as cancer type-specific biomarkers. | Classical and gene-centric molecular pathways |
| This study; cancer type markers | Pan-cancer analysis: 8 cancer types | Proteomic data for 1018 patients (CPTAC) | For all cancer types, both gene-centric and classical pathways had a higher percentage of significant biomarkers than single proteins. In total, 2950 classical and 7343 gene-centric pathways were identified as cancer-type-specific biomarkers. | Classical and gene-centric molecular pathways |
| This study; survival markers | Pan-cancer analysis: 21 cancer types | RNA expression data for 7426 patients (TCGA) | For overall survival, the highest percentage of biomarkers was observed in five and three cancer types for gene-centric and classical pathways, respectively. For progression-free survival, the advantage for gene-centric and classical pathways was shown for three and five cancer types, respectively. | Classical and gene-centric molecular pathways |
| This study; survival markers | Pan-cancer analysis: 6 cancer types | Proteomic data for 611 patients (CPTAC) | Statistically significant survival pathway-based biomarkers were found for pancreatic cancer (168 gene-centric and 45 classical pathways). Gene-centric pathways showed the highest percentage of biomarkers identified. | Classical and gene-centric molecular pathways |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zolotovskaia, M.; Kovalenko, M.; Pugacheva, P.; Tkachev, V.; Simonov, A.; Sorokin, M.; Seryakov, A.; Garazha, A.; Gaifullin, N.; Sekacheva, M.; et al. Algorithmically Reconstructed Molecular Pathways as the New Generation of Prognostic Molecular Biomarkers in Human Solid Cancers. Proteomes 2023, 11, 26. https://doi.org/10.3390/proteomes11030026
Zolotovskaia M, Kovalenko M, Pugacheva P, Tkachev V, Simonov A, Sorokin M, Seryakov A, Garazha A, Gaifullin N, Sekacheva M, et al. Algorithmically Reconstructed Molecular Pathways as the New Generation of Prognostic Molecular Biomarkers in Human Solid Cancers. Proteomes. 2023; 11(3):26. https://doi.org/10.3390/proteomes11030026
Chicago/Turabian StyleZolotovskaia, Marianna, Maks Kovalenko, Polina Pugacheva, Victor Tkachev, Alexander Simonov, Maxim Sorokin, Alexander Seryakov, Andrew Garazha, Nurshat Gaifullin, Marina Sekacheva, and et al. 2023. "Algorithmically Reconstructed Molecular Pathways as the New Generation of Prognostic Molecular Biomarkers in Human Solid Cancers" Proteomes 11, no. 3: 26. https://doi.org/10.3390/proteomes11030026
APA StyleZolotovskaia, M., Kovalenko, M., Pugacheva, P., Tkachev, V., Simonov, A., Sorokin, M., Seryakov, A., Garazha, A., Gaifullin, N., Sekacheva, M., Zakharova, G., & Buzdin, A. A. (2023). Algorithmically Reconstructed Molecular Pathways as the New Generation of Prognostic Molecular Biomarkers in Human Solid Cancers. Proteomes, 11(3), 26. https://doi.org/10.3390/proteomes11030026








