Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer
Abstract
1. Introduction
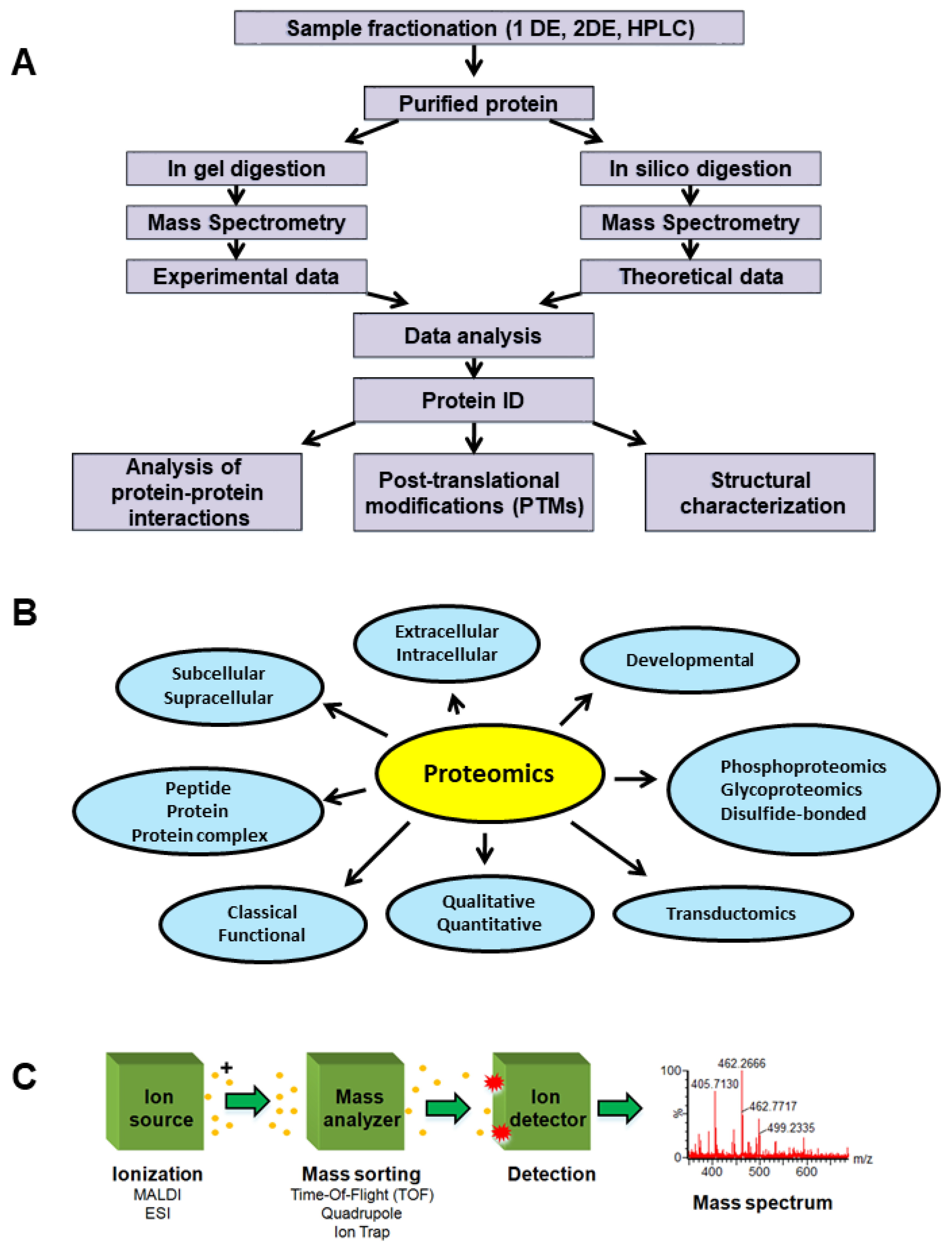
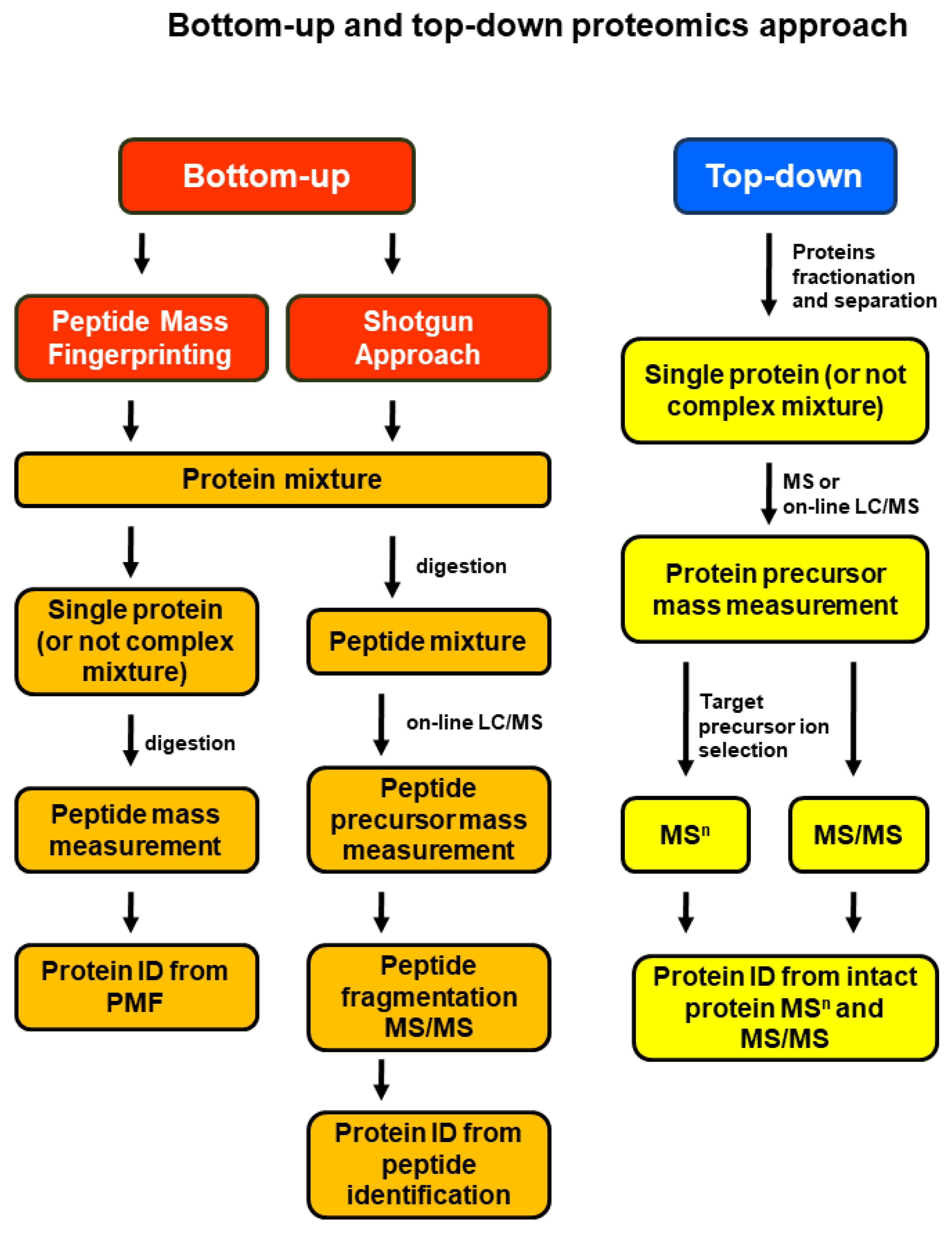
2. Proteomics-Based Investigation of Dysregulated Steroid Receptors and HER2
3. Proteomics-Based Investigation of Transcriptional and Translational Dysregulation in BC
4. Proteomics-Based Identification of Dysregulated Proteins Involved in BC EMT, Invasion and Metastasis
5. Proteomics-Based Identification of Dysregulated Proteins Involved in Intermediary Metabolism Reprogramming in BC Cells
| Protein | Gene Name | Biological and Pathological Role in BC | Methods of Identification | Status in BC | Potential Clinical Use |
|---|---|---|---|---|---|
| Steroid receptors and HER2 | |||||
| Estrogen receptors | ER isoforms: ERα & ERβ | Nuclear receptors/TFs that regulates transcription of estrogen target genes [67]; ERα is a promoter of cell proliferation/tumorigenesis in BC, and ERβ suppresses cell proliferation [68] | IHC [116], MALDI-TOF MS [67], LC-SRM MS [27]; multiplex IHC-MALDI-MSI (MALDI-IHC) [17] | More than 70% of all BC are ERα [117] | Diagnostic biomarkers, classification of BC subtypes [67] |
| nLC/ESI-MS/MS; MALDI-MS/MS (MSn) [70] | PTMs and PPI modulate activity: ubiquitination [117]; phosphorylation [70] | Tamoxifen resistance [72] | |||
| Progesterone receptors | PR isoforms: PRA & PRB | TFs that modulate ERα action in BC [118]; exhibits both activatory and repressive effect on gene transcription [119] | IHC [116]; LC-SRM MS [27]; multiplex IHC-MALDI-MSI (MALDI-IHC) [17] | Association between ERα/PR induces cell proliferation and tumor growth [120] | Predictive biomarker [121], prognostic and predictive biomarker of response to endocrine therapy [118] |
| Androgen receptor | AR | Nuclear TF that mediates the biological effects of androgens; tumor suppressor in ER+ BC and inducer of tumor progression in ER- BC, including HER2+ and TNBC [68], it has a crucial role in BC pathology and progression [122] | IHC [123], PRM targeted proteomic [124] | Expressed in 70–90% of the BCs [122]; upregulated in luminal A & B subtypes of BC and a subset of TNBC; positive immunostaining was associated with smaller tumor size [123] | Possible prognostic biomarker [123]; potential therapeutic target in AR+ BC patients [122] |
| Human epidermal growth factor receptor 2 | HER2/neu, c-erbB2 | Membrane tyrosine kinase and oncogene [76]; regulates cell growth, survival, differentiation and proliferation [74] | IHC, FISH, CISH, SISH [76]; MALDI-MSI [77], LC-MS/MS+SRM assay+FISH+IHC [125]; LC-SRM MS [27]; multiplex IHC-MALDI-MSI (MALDI-IHC) [17] | Overexpressed in 20–30% of BC [76] | Predictive and prognostic biomarker; treatment target [76]; poor prognosis and increased likehood of metastasis especially in node-positive BC [126] |
| Transcription and translation regulation | |||||
| Core binding factor subunit beta | CBFB | Translation regulation in cytoplasm and transcription regulation in the nucleus [78] | IHC, IF, immunoblotting, MS [78] | Highly mutated in solid tumors, including BC [78], mutations mainly occur in HR+/HER2- BC [127] | Putative prognostic biomarker in HR+/HER2- BC [127] |
| Catenin beta 1 | CTNNB1 | Transcriptional regulation in the Wnt signaling pathway and cell adhesion molecule by linking cadherins to the actin cytoskeleton [92]; downregulation inhibited cell proliferation, migration, and invasion and induced apoptosis in RCC [128] | LC-MS/MS [56]; IHC [129] | Key role in most cancers as an oncogene [128]; β-catenin/Wnt pathway activation is preferentially found in TN-BL breast carcinomas [130] | Prognostic biomarker [131]; poor clinical outcome in BC [130] |
| Histone H1 | H1 (seven somatic proteoforms [132]) | Chromatin organization and transcriptional regulation; knock-down in BC results in altered gene expression, proliferation, and IFN response [133] | Immunoblotting, IHC, LC-MS, LC-MS/MS [132] | H1 showed PTMs in BC cells [133] | Putative biomarker of proliferation BC cells [132] |
| EMT, cytoskeleton reorganization, cell adhesion, ECM, invasion and metastasis | |||||
| Vimentin | VIM | EMT; intermediate filament family protein; in IDC is associated with low ER, low PR, increased basement membrane invasiveness, and resistance to BC chemotherapy [134] | IHC [134], IF [135]; LC-MS for detection of phosphorylated isoform that increases mobility in cancer cells [87]; MALDI-TOF MS/MS for detection of methylated isoform [89] and interaction VIM-garlic phytochemical with anti-metastatic activity [91] | Overexpressed in BC, especially in BLBC [89] | Mesenchymal marker, poor prognostic factor of BC [134] |
| Epithelial (E)-cadherin | CDH1 | EMT; adhesion molecule of the epithelial adherens junction; dual role in BC: putative tumor suppressor [136] or promotor of metastasis and invasiveness [137] | IHC [137,138], IF [135]; 2D-DIGE and MS [139] | Downregulated in BC [140] | Phenotypic marker; biomarker of tumor subtypes [136]; prognostic biomarker for patients with lymph node metastasis and TNBC [141] |
| Filamin A | FLNA | EMT; actin cross-linking protein, involved in regulation of BRCA1 expression in BC [142] | IHC [142]; LC-MS/MS [56] | Upregulated in BC, especially in myoepithelial cells [142] | Putative prognostic biomarker [142] |
| Pleckstrin homology domain-containing family G member 2 | PLEKHG2 | Actin cytoskeleton reorganization and transcriptional regulation, regulation of cell morphology [28] | MALDI-MSI, LC-MALDI-MS/MS [28] | Phosphorylated in TNBC [28] | Prognostic biomarker [28] |
| SRY-related high-mobility-group (HMG) box 11 | SOX11 | Transcription factor and embryonic mammary epithelial marker associated with mesenchymal state and embryonic phenotype of BC cells [135]; involved in BC growth, migration, and invasion, regulating the BLBCs phenotype [28] | WB, IF [135]; IHC [143], MALDI-MSI, LC-MS/MS [28] | Upregulated in BLBC [144] | Prognostic biomarker [28] for BC with elevated risk of distant metastases and poor outcome [135], therapeutic target [28]; ER negative DCIS SOX11+ tumor cells metastasize to brain and bone at greater frequency than in lungs [135] |
| Collagen type I alpha 1 chain | COL1A1 | EMT; promotes BC metastasis [98]; upregulation is a risk factor for radiation-associated secondary diseases in BC [145] | IHC [98], MALDI-MSI, LC-MALDI-MS/MS [28] | Upregulated in invasive BC (IDC) [[28,97] | Prognostic biomarker [28], poor survival in ER+ BC, potential therapeutic target [98] |
| Collagen type I alpha 2 chain | COL1A2 | EMT; ECM assembly; upregulation is a risk factor for radiation-associated secondary diseases in BC [145] | MALDI-MSI, LC-MALDI-MS/MS [28] | Upregulated in invasive BC [28] | Prognostic biomarker [28] |
| Cytokeratins | CKs | IFs [146]; CK+ cells are enriched in cancer stem cell proprieties [92] | IHC [146]; multiplex IHC-MALDI-MSI (MALDI-IHC) [17] | CK 5/6 upregulated in ER+ BC and BLBC [147] | Adjuncts in diagnosis, classification and prognostication of BC [146] |
| Intermediary metabolism reprogramming | |||||
| Fatty acid synthase | FASN | FAM; enhances malignant progression [148], migration, metastasis [149], proliferation, drug resistance, and apoptosis [150]; inhibition reduces cell proliferation, suppresses migration and invasion and induces apoptosis [151] | LC-MS/MS [56], MALDI-TOF/TOF MS/MS [126]; IHC [150] | Overexpressed in cancer cells [148]; highly expressed in different sex hormone-related malignant tumors, positive expression in TNBC correlated with lymph node metastasis and stage [150] | Prognostic biomarker in TNBC [150] |
| Triose-phosphate isomerase | TPI1 | Glycolysis; promotes tumor development and progression of BC in tissue and cell lines, proliferation, metastasis, activates PI3K/Akt/mTOR, regulates EMT [152] | WB, IHC, IF [152]; MALDI-TOF/TOF MS/MS [126] | Upregulated in multiple cancers [152] | Therapeutic target for BC [152] |
| Alpha-enolase | ENO1 | Cell growth, hypoxia tolerance, autoimmune activities, glycolysis pathway [153] | WB [154], IHC [155], LC-MS/MS [156]; MALDI-TOF/TOF MS/MS [126] | Upregulated in BC [153,154] | Prognostic biomarker [155,157] |
| Phosphoglycerate kinase 1 | PGK1 | Glycolysis, hypoxia; cancer progression, metastases; invasion promoter, regulates HIF-1α-mediated EMT [158] | MALDI-TOF/TOF MS/MS [126] | Overexpressed in BC [158] | Poor prognosis, potential survival biomarker in BC [158] |
| Cell cycle, cellular division, mitotic spindle, cell proliferation | |||||
| Jumping translocation breakpoint protein/prostate androgen regulated protein | JTB/PAR | Dual role: tumor suppressor or oncogene; involved in cell proliferation, tumorigenesis, genomic instability [159] | WB, immunoprecipitation, IF [159] | Overexpressed in many cancers, including BC [159] | Putative target for therapeutic intervention [159] |
| Beta-tubulin | TUBB | Carcinogenesis, metastasis [160] | LC-MS/MS [56] | Upregulated in BC tissue [160] | Potential prognostic biomarker for worse prognosis in ERα+ and better prognosis in ERα- BC [160] |
| Proliferation marker protein Ki-67 | MKI67 | Proliferation-associated nuclear antigen involved in cell proliferation and growth, migration, invasion, tumor progression, maintenance of stem cell characteristics [161] | IHC [162,163]; LC-MS/MS [56] | Overexpressed in cancer cells [164] | Marker of cell proliferation, prognostic and predictive biomarker in invasive BC [165,166] |
| Aminoimidazole-4- carboxamide ribonucleotide | ATIC | Cell proliferation [28] | MALDI MSI [28] | Upregulated in TNBC [28] | Putative prognostic biomarker [28] and therapeutic target in BC resistant to tamoxifen [28,167] |
| Mutant tumor suppressor p53 protein | TP53/mtp53 | Driver oncogene [168], transcription factor involved in cell cycle; mtp53-related proteome targets cholesterol biosynthesis, DNA replication and repair pathways [168] | IHC [169,170], SILAC coupled to MS/MS [168] | The most frequently mutated gene in invasive BC; mutated in 30–35% of all BCs, and 80% in TNBC [171] | Potential biomarker and therapeutic target for BC patients, especially for TNBC [171] |
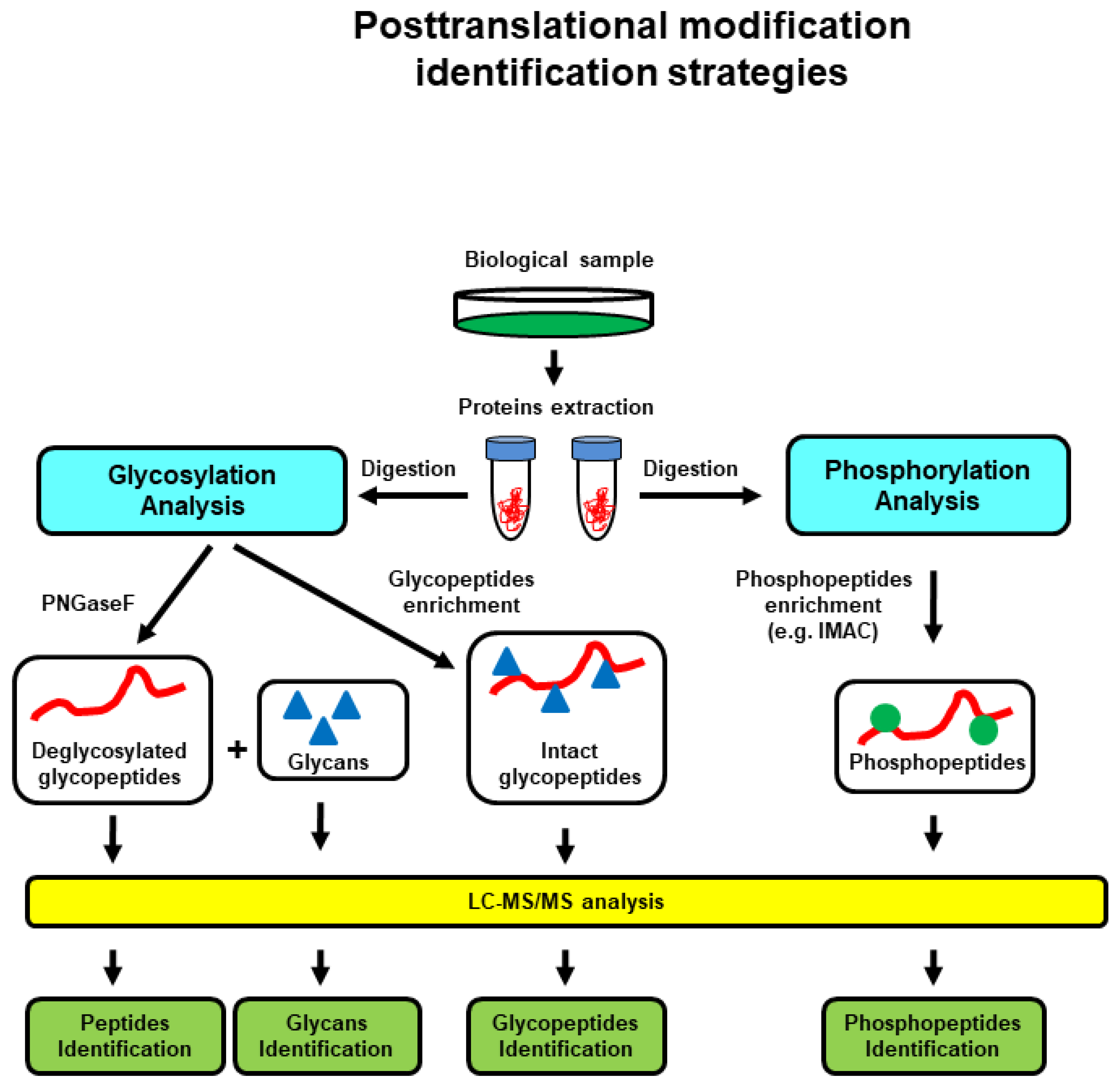
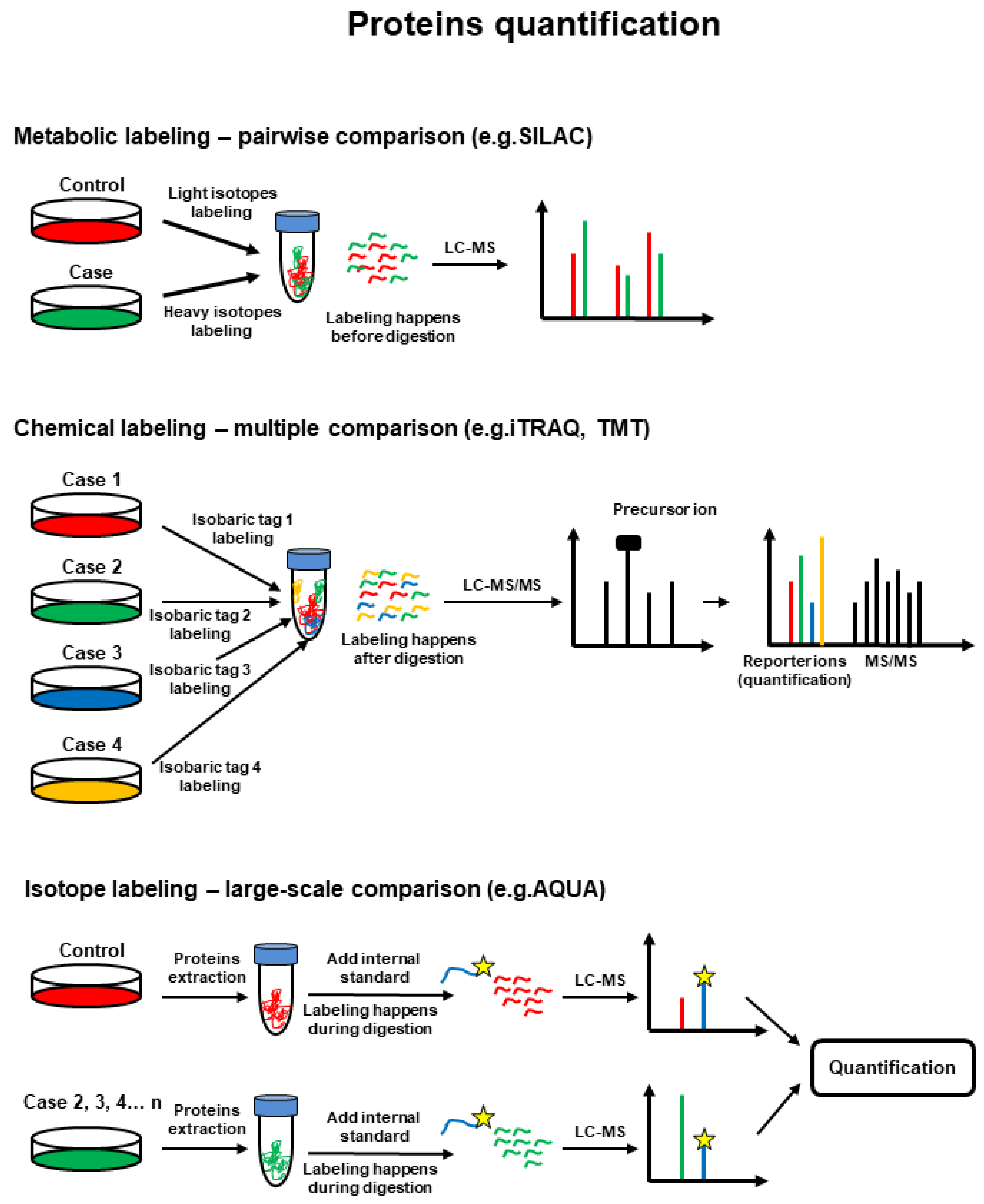
6. Proteomics-Based Investigation of PTMs and PPIs in BC
7. Proteomics-Based Investigation of Dysregulated Proteins in Diverse Liquid Biopsies/Body Fluids
| Body Fluids | Proteomics-Based Techniques | Applications in BC |
|---|---|---|
| Blood/plasma/serum | SELDI-TOF MS, MALDI-TOF/TOF MS | Identification of panels of serum biomarkers for BC [49] |
| 2DE, MALDI-TOF MS | Serum proteomic differences between patients with MBC and healthy controls [204] | |
| LC-MS/MS | BC grading and subtyping, identification of biomarkers for cell growth, ECM and cell-to-cell communication, energy metabolism and gene transcription, cell death and cancer development, transcription regulation, tumorigenesis and invasion, redox balance, and EMT [205]; secretome of BC CAFs [207]; exosomal BC proteome [211]; exosomal phosphoproteome [213] | |
| Proximal fluid proteomics/nipple aspirate fluid (NAF)/dried NAF spots on Guthrie cards | MALDI-TOF MS [214], LC-MS/MS [66], SELDI-TOF MS [215] | Early BC detection; biomarkers discovery in BC [216] |
| nLC-ESI-Q-TOF MS | Early BC screening and subtype classification [219] | |
| Milk | LC-MS/MS | Differential protein pattern between breastfeeding mothers with BC compared with healthy women; identification of putative biomarkers for BC [42]; detection of αS1-casein [221]; EVs proteome identification [224] |
| Urine | LC-MS/MS | Detection of overexpressed proteins in DCIS samples, early invasive and metastatic BC [32]; progressive changes during BC development in rat model [34] |
| MALDI-TOF/TOF, LC-MS/MS | Detection of urinary proteome alterations in HER2 enriched BC [33] | |
| Tears | SELDI-TOF MS [40], MALDI-TOF/TOF [38], LC-MS/MS [39] | Identification of differential biomarker profiles for BC patients compared to healthy controls; identification of dysregulated proteins involved in ECM remodeling [39], host immune system pathways, metabolic regulation [38] |
| Saliva | LC-MS/MS | Identification of biomarkers for DCIS or HER2/neu positive or negative BC [36] |
| nLC-Q-TOF MS | Differential immune landscape, molecular transport and signaling pathways between FA and IDC [239] | |
| MALDI-TOF MS, MALDI-TOF/TOF MS [240] | Identification of new BC biomarkers [240] | |
| SELDI-TOF MS [231], ESI-TOF MS, MALDI-TOF MS [233], ESI-Orbitrap MS, ESI-Q-TOF MS [234] | Panels of biomarkers for accurate discrimination between BC stages [235] or between BC patients and healthy controls [236] |
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rossi, C.; Cicalini, I.; Cufaro, M.C.; Consalvo, A.; Upadhyaya, P.; Sala, G.; Antonucci, I.; Del Boccio, P.; Stuppia, L.; De Laurenzi, V. Breast cancer in the era of integrating “Omics” approaches. Oncogenesis 2022, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Glusman, G.; Brogaard, K.; Price, N.D.; Hood, L. P4 medicine: How systems medicine will transform the healthcare sector and society. Per. Med. 2013, 10, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, G.; Phillips, K. Precision Medicine: From Science To Value. Health Affairs 2018, 37, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Pires, I.M.; Denysyuk, H.V.; Villasana, M.V.; Sá, J.; Lameski, P.; Chorbev, I.; Zdravevski, E.; Trajkovik, V.; Morgado, J.F.; Garcia, N.M. Mobile 5P-Medicine Approach for Cardiovascular Patients. Sensors 2021, 21, 6986. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Li, J.; Guo, Y.; Golubnitschaja, O. Mass spectrometry analysis of human tear fluid biomarkers specific for ocular and systemic diseases in the context of 3P medicine. EPMA J. 2021, 12, 449–475. [Google Scholar] [CrossRef]
- Hadi, N.I. “OMIC” tumor markers for breast cancer: A review. Pak. J. Med. Sci. 2015, 31, 1256–1262. [Google Scholar] [CrossRef]
- Stenemo, M.; Teleman, J.; Sjöström, M.; Grubb, G.; Malmström, E.; Malmström, J.; Niméus, E. Cancer associated proteins in blood plasma: Determining normal variation. Proteomics 2016, 16, 1928–1937. [Google Scholar] [CrossRef]
- Zhou, Y.; Lih, T.M.; Pan, J.; Höti, N.; Dong, M.; Cao, L.; Hu, Y.; Cho, K.-C.; Chen, S.-Y.; Eguez, R.V.; et al. Proteomic signatures of 16 major types of human cancer reveal universal and cancer-type-specific proteins for the identification of potential therapeutic targets. J. Hematol. Oncol. 2020, 13, 170. [Google Scholar] [CrossRef]
- Sajood, S.; Prasad, T.S.K.; Bhat, B.; Bhat, Z.F.; Shah, R.A.; Bhat, H.F. Comparison of co-immunoprecipitation techniques for effective identification of SNTA1 interacting proteins in breast cancer cells. bioRxiv 2022. [Google Scholar] [CrossRef]
- He, Z.; Chen, Z.; Tan, M.; Elingarami, S.; Liu, Y.; Li, T.; Deng, Y.; He, N.; Li, S.; Fu, J.; et al. A review on methods for diagnosis of breast cancer cells and tissues. Cell Prolif. 2020, 53, e12822. [Google Scholar] [CrossRef]
- Tian, F.; Zhang, S.; Liu, C.; Han, Z.; Liu, Y.; Deng, J.; Li, Y.; Wu, X.; Cai, L.; Qin, L.; et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 2021, 12, 2536. [Google Scholar] [CrossRef] [PubMed]
- Hudelist, G.; Pacher-zavisin, M.; Singer, C.; Holper, T.; Kubista, E.; Schreiber, M.; Manavi, M.; Bilban, M.; Czerwenka, K. Use of High-Throughput Protein Array for Profiling of Differentially Expressed Proteins in Normal and Malignant Breast Tissue. Breast Cancer Res. Treat. 2004, 86, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Boellner, S.; Becker, K.-F. Reverse Phase Protein Arrays-Quantitative Assessment of Multiple Biomarkers in Biopsies for Clinical Use. Microarrays 2015, 4, 98–114. [Google Scholar] [CrossRef]
- Masuda, H.; Qi, Y.; Liu, S.; Hayashi, N.; Kogawa, T.; Hortobagyi, G.N.; Tripathy, D.; Ueno, N.T. Reverse phase protein array identification of triple-negative breast cancer subtypes and comparison with mRNA molecular subtypes. Oncotarget 2017, 8, 70481–70495. [Google Scholar] [CrossRef] [PubMed]
- Ősz, Á.; Lánczky, A.; Győrffy, B. Survival analysis in breast cancer using proteomic data from four independent datasets. Sci. Rep. 2021, 11, 16787. [Google Scholar] [CrossRef] [PubMed]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of immunohistochemistry. J. Pharm. Bioallied Sci. 2012, 4, S307–S309. [Google Scholar] [CrossRef]
- Yagnik, G.; Liu, Z.; Rothschild, K.J.; Lim, M.J. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J. Am. Soc. Mass Spectrom. 2021, 32, 977–988. [Google Scholar] [CrossRef]
- Yang, T.; Xu, F.; Fang, D.; Chen, Y. Targeted Proteomics Enables Simultaneous Quantification of Folate Receptor Isoforms and Potential Isoform-based Diagnosis in Breast Cancer. Sci. Rep. 2015, 5, 16733. [Google Scholar] [CrossRef] [PubMed]
- Im, K.; Mareninov, S.; Diaz, M.F.P.; Yong, W.H. An Introduction to Performing Immunofluorescence Staining. Methods Mol. Biol. 2019, 1897, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Neagu, A.-N.; Jayathirtha, M.; Baxter, E.; Donnelly, M.; Petre, B.A.; Darie, C.C. Applications of Tandem Mass Spectrometry (MS/MS) in Protein Analysis for Biomedical Research. Molecules 2022, 27, 2411. [Google Scholar] [CrossRef] [PubMed]
- Darie-Ion, L.; Whitham, D.; Jayathirtha, M.; Rai, Y.; Neagu, A.-N.; Darie, C.C.; Petre, B.A. Applications of MALDI-MS/MS-Based Proteomics in Biomedical Research. Molecules 2022, 27, 6196. [Google Scholar] [CrossRef] [PubMed]
- Chantalat, E.; Boudou, F.; Laurell, H.; Palierne, G.; Houtman, R.; Melchers, D.; Rochaix, P.; Filleron, T.; Stella, A.; Burlet-Schiltz, O.; et al. The AF-1-deficient estrogen receptor ERα46 isoform is frequently expressed in human breast tumors. Breast Cancer Res. BCR 2016, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Neagu, A.-N.; Whitham, D.; Buonanno, E.; Jenkins, A.; Alexa-Stratulat, T.; Tamba, B.I.; Darie, C.C. Proteomics and its applications in breast cancer. Am. J. Cancer Res. 2021, 11, 4006–4049. [Google Scholar] [PubMed]
- Zhou, M.; Uwugiaren, N.; Williams, S.M.; Moore, R.J.; Zhao, R.; Goodlett, D.; Dapic, I.; Paša-Tolić, L.; Zhu, Y. Sensitive Top-Down Proteomics Analysis of a Low Number of Mammalian Cells Using a Nanodroplet Sample Processing Platform. Anal. Chem. 2020, 92, 7087–7095. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, I.; Woods, A.G.; Wagner, J.; Dorler, J.; Wormwood, K.; Thome, J.; Darie, C.C. Mass spectrometry for proteomics-based investigation of oxidative stress and heat shock proteins. In Oxidative Stress: Diagnostics, Prevention, and Therapy; Andreescu, S., Hepel, M., Eds.; American Chemical Society: Washington, DC, USA, 2011. [Google Scholar]
- Woods, A.G.; Sokolowska, I.; Ngounou Wetie, A.G.; Channaveerappa, D.; Dupree, E.J.; Jayathirtha, M.; Aslebagh, R.; Wormwood, K.L.; Darie, C.C. Mass Spectrometry for Proteomics-Based Investigation. Adv. Exp. Med. Biol. 2019, 1140, 1–26. [Google Scholar] [CrossRef]
- Chen, Y.; Britton, D.; Wood, E.; Brantley, S.; Fournier, M.; Wloch, M.; Williams, V.; Johnson, J.; Magliocco, A.; Pike, I.; et al. Quantification of Breast Cancer Protein Biomarkers at Different Expression Levels in Human Tumors. Methods Mol. Biol. 2018, 1788, 251–268. [Google Scholar] [PubMed]
- Phillips, L.; Gill, A.J.; Baxter, R.C. Novel Prognostic Markers in Triple-Negative Breast Cancer Discovered by MALDI-Mass Spectrometry Imaging. Front. Oncol. 2019, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Gawin, M.; Kurczyk, A.; Niemiec, J.; Stanek-Widera, A.; Grela-Wojewoda, A.; Adamczyk, A.; Biskup-Frużyńska, M.; Polanska, J.; Widlak, P. Intra-Tumor Heterogeneity Revealed by Mass Spectrometry Imaging Is Associated with the Prognosis of Breast Cancer. Cancers 2021, 13, 4349. [Google Scholar] [CrossRef]
- Rujchanarong, D.; Lefler, J.; Saunders, J.E.; Pippin, S.; Spruill, L.; Bethard, J.R.; Ball, L.E.; Mehta, A.S.; Drake, R.R.; Ostrowski, M.C.; et al. Defining the Tumor Microenvironment by Integration of Immunohistochemistry and Extracellular Matrix Targeted Imaging Mass Spectrometry. Cancers 2021, 13, 4419. [Google Scholar] [CrossRef]
- Swiatly, A.; Horala, A.; Hajduk, J.; Matysiak, J.; Nowak-Markwitz, E.; Kokot, Z.J. MALDI-TOF-MS analysis in discovery and identification of serum proteomic patterns of ovarian cancer. BMC Cancer 2017, 17, 472. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Millar, E.K.A.; Schwartz, P.; Graham, P.H.; Li, Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE 2015, 10, e0141876. [Google Scholar] [CrossRef]
- Wu, J.; Guo, Z.; Gao, Y. Early biomarker discovery in urine of Walker 256 subcutaneous rat model. bioRxiv 2017. [Google Scholar] [CrossRef]
- Gajbhiye, A.; Dhabhi, R.; Taunk, K.; Garikapati, V.; Roy Choudhury, S.; Adhav, R.; Seal, S.; Mane, A.; Santhakumari, B.; Santra, M.; et al. Urinary proteome alterations in HER2 enriched breast cancer revealed by multipronged quantitative proteomics. Proteomics 2016, 16, 2403–2418. [Google Scholar] [CrossRef] [PubMed]
- Paweletz, C.; Trock, B.; Pennanen, M.; Tsangaris, T.; Magnant, C.; Liotta, L.; Petricoin, E. Proteomic Patterns of Nipple Aspirate Fluids Obtained by SELDI-TOF: Potential for New Biomarkers to Aid in the Diagnosis of Breast Cancer. Dis. Markers 2001, 17, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Streckfus, C.F.; Mayorga-Wark, O.; Arreola, D.; Edwards, C.; Bigler, L.; Dubinsky, W.P. Breast Cancer Related Proteins Are Present in Saliva and Are Modulated Secondary to Ductal Carcinoma In Situ of the Breast. Cancer Investig. 2008, 26, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Streckfus, C.F.; Arreola, D.; Edwards, C.; Bigler, L. Salivary Protein Profiles among HER2/neu-Receptor-Positive and -Negative Breast Cancer Patients: Support for Using Salivary Protein Profiles for Modeling Breast Cancer Progression. J. Oncol. 2012, 2012, 413256. [Google Scholar] [CrossRef]
- Böhm, D.; Keller, K.; Pieter, J.; Boehm, N.; Wolters, D.; Siggelkow, W.; Lebrecht, A.; Schmidt, M.; Kölbl, H.; Pfeiffer, N.; et al. Comparison of tear protein levels in breast cancer patients and healthy controls using a de novo proteomic approach. Oncol. Rep. 2012, 28, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Daily, A.; Ravishankar, P.; Harms, S.; Klimberg, V.S. Using tears as a non-invasive source for early detection of breast cancer. PLoS ONE 2022, 17, e0267676. [Google Scholar] [CrossRef] [PubMed]
- Lebrecht, A.; Boehm, D.; Schmidt, M.; Koelbl, H.; Schwirz, R.L.; Grus, F.H. Diagnosis of Breast Cancer by Tear Proteomic Pattern. Cancer Genom. Proteom. 2009, 6, 177–182. [Google Scholar]
- Aslebagh, R.; Channaveerappa, D.; Arcaro, K.F.; Darie, C.C. Proteomics analysis of human breast milk to assess breast cancer risk. Electrophoresis 2018, 39, 653–665. [Google Scholar] [CrossRef]
- Aslebagh, R.; Channaveerappa, D.; Pentecost, B.T.; Arcaro, K.F.; Darie, C.C. Combinatorial Electrophoresis and Mass Spectrometry-Based Proteomics in Breast Milk for Breast Cancer Biomarker Discovery. In Advancements of Mass Spectrometry in Biomedical Research; Woods, A.G., Darie, C.C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 451–467. [Google Scholar] [CrossRef]
- Ahmed, H.; Ajat, M.; Mahmood, R.; Mansor, R.; Razak, I.; Al-Obaidi, J.; Razali, N.; Jaji, Z.; Danmaigoro, A.; Abu Bakar, M.Z. LC-MS/MS Proteomic Study of MCF-7 Cell Treated with Dox and Dox-Loaded Calcium Carbonate Nanoparticles Revealed Changes in Proteins Related to Glycolysis, Actin Signalling, and Energy Metabolism. Biology 2021, 10, 909. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Lemieux, G.; Hassis, M.; Olshen, A.; Fisher, S.; Werb, Z. Quantitative proteomic analyses of mammary organoids reveals distinct signatures after exposure to environmental chemicals. Proc. Natl. Acad. Sci. USA 2016, 113, 201600645. [Google Scholar] [CrossRef] [PubMed]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Donato, C.; Buczak, K.; Schmidt, A.; Aceto, N. Mass spectrometry analysis of circulating breast cancer cells from a Xenograft mouse model. STAR Protocols 2021, 2, 100480. [Google Scholar] [CrossRef]
- Lawrence, R.; Perez, L.; Hernández, D.; Miller, C.P.; Haas, K.M.; Irie, H.Y.; Lee, S.-I.; Blau, C.; Villén, J. The Proteomic Landscape of Triple-Negative Breast Cancer. Cell Rep. 2015, 11, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Pralea, I.-E.; Moldovan, R.-C.; Țigu, A.-B.; Ionescu, C.; Iuga, C.-A. Mass Spectrometry-Based Omics for the Characterization of Triple-Negative Breast Cancer Bio-Signature. J. Pers. Med. 2020, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.; Moore, K.; Phillips, L.; Boyle, F.M.; Marsh, D.J.; Baxter, R.C. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 2014, 16, R63. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, S.C.; Park, Y.S.; Jeon, Y.; Lee, J.; Jung, S.-Y.; Park, I.; Jang, S.; Park, H.; Yoo, C.; et al. Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer 2011, 11, 465. [Google Scholar] [CrossRef]
- Sanders, M.E.; Dias, E.C.; Xu, B.J.; Mobley, J.A.; Billheimer, D.; Roder, H.; Grigorieva, J.; Dowsett, M.; Arteaga, C.L.; Caprioli, R.M. Differentiating Proteomic Biomarkers in Breast Cancer by Laser Capture Microdissection and MALDI MS. J. Proteome Res. 2008, 7, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Al-Wajeeh, A.S.; Salhimi, S.M.; Al-Mansoub, M.A.; Khalid, I.A.; Harvey, T.M.; Latiff, A.A.; Ismail, M.N. Comparative proteomic analysis of different stages of breast cancer tissues using ultra high performance liquid chromatography tandem mass spectrometer. PLoS ONE 2020, 15, e0227404. [Google Scholar] [CrossRef]
- Pozniak, Y.; Balint Lahat, N.; Rudolph, J.D.; Lindskog, C.; Katzir, R.; Avivi, C.; Pontén, F.; Ruppin, E.; Barshack, I.; Geiger, T. System-wide Clinical Proteomics of Breast Cancer Reveals Global Remodeling of Tissue Homeostasis. Cell Syst. 2016, 2, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Goto, R.; Nakamura, Y.; Takami, T.; Sanke, T.; Tozuka, Z. Quantitative LC-MS/MS Analysis of Proteins Involved in Metastasis of Breast Cancer. PLoS ONE 2015, 10, e0130760. [Google Scholar] [CrossRef]
- Gámez-Pozo, A.; Trilla-Fuertes, L.; Berges-Soria, J.; Selevsek, N.; López-Vacas, R.; Díaz-Almirón, M.; Nanni, P.; Arevalillo, J.; Navarro, H.; Grossmann, J.; et al. Functional proteomics outlines the complexity of breast cancer molecular subtypes. Sci. Rep. 2017, 7, 10100. [Google Scholar] [CrossRef] [PubMed]
- Jayathirtha, M.; Neagu, A.-N.; Whitham, D.; Alwine, S.; Darie, C. Investigation of the effects of overexpression of jumping translocation breakpoint (JTB) protein in MCF7 cells for potential use as a biomarker in breast cancer. Am. J. Cancer Res. 2022, 12, 1784–1823. [Google Scholar]
- Jayathirtha, M.; Neagu, A.-N.; Whitham, D.; Alwine, S.; Darieet, C.C. Investigation of the effects of downregulation of jumping translocation breakpoint (JTB) protein expression in MCF7 cells for potential use as a biomarker in breast cancer. Am. J. Cancer Res. 2022, 12, 4373–4398. [Google Scholar]
- Hebert, J.D.; Myers, S.A.; Naba, A.; Abbruzzese, G.; Lamar, J.; Carr, S.A.; Hynes, R.O. Proteomic profiling of the ECM of xenograft breast cancer metastases in different organs reveals distinct metastatic niches. Cancer Res. 2020, 80, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Umbreit Feder, C.; Krüger, T.; Pelzel, D.; Ernst, G.; Kniemeyer, O.; Guntinas-Lichius, O.; Berndt, A.; von Eggeling, F. Identification of proteomic markers in head and neck cancer using MALDI-MS Imaging, LC-MS/MS and Immunohistochemistry. PROTEOMICS Clin. Appl. 2018, 13, 1700173. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.D.; Keller, A.; Zhou, B.; Tian, R.; Bruce, J.E. Cellular Interactome Dynamics during Paclitaxel Treatment. Cell Rep. 2019, 29, 2371–2383.e2375. [Google Scholar] [CrossRef]
- Chavez, J.D.; Schweppe, D.K.; Eng, J.K.; Zheng, C.; Taipale, A.; Zhang, Y.; Takara, K.; Bruce, J.E. Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun. 2015, 6, 7928. [Google Scholar] [CrossRef]
- Garza, K.Y.; Zhang, J.; Lin, J.Q.; Carter, S.; Suliburk, J.; Nagi, C.; Eberlin, L.S. Abstract P1-20-04: Advanced development of the MasSpec Pen technology to aid in breast cancer surgical margin evaluation and diagnosis during surgery. Cancer Res. 2020, 80, P1–P20. [Google Scholar] [CrossRef]
- Zhang, J.; Sans, M.; DeHoog, R.J.; Garza, K.Y.; King, M.E.; Feider, C.L.; Bensussan, A.; Keating, M.F.; Lin, J.Q.; Povilaitis, S.; et al. Direct Molecular Analysis of In Vivo and Freshly Excised Tissues in Human Surgeries with the MasSpec Pen Technology. medRxiv 2020. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, M.; Pestell, R.; Curran, E.M.; Welshons, W.V.; Fuqua, S.A.W. Phosphorylation of Estrogen Receptor α Blocks Its Acetylation and Regulates Estrogen Sensitivity. Cancer Res. 2004, 64, 9199–9208. [Google Scholar] [CrossRef] [PubMed]
- Holm, J.; Yu, N.Y.-L.; Johansson, A.; Ploner, A.; Hall, P.; Lindström, L.S.; Czene, K. Concordance of Immunohistochemistry-Based and Gene Expression-Based Subtyping in Breast Cancer. JNCI Cancer Spectr. 2020, 5, pkaa087. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.A.; He, J.; Lu, M.; Souda, P.; Saxton, R.E.; Faull, K.F.; Whitelegge, J.P.; Chang, H.R. Mass spectrometry (LC-MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid. J. Proteome Res. 2012, 11, 5034–5045. [Google Scholar] [CrossRef] [PubMed]
- Heger, Z.; Rodrigo, M.A.M.; Krizkova, S.; Zitka, O.; Beklova, M.; Kizek, R.; Adam, V. Identification of estrogen receptor proteins in breast cancer cells using matrix-assisted laser desorption/ionization time of flight mass spectrometry (Review). Oncol. Lett. 2014, 7, 1341–1344. [Google Scholar] [CrossRef] [PubMed][Green Version]
- You, C.-P.; Leung, M.-H.; Tsang, W.-C.; Khoo, U.-S.; Tsoi, H. Androgen Receptor as an Emerging Feasible Biomarker for Breast Cancer. Biomolecules 2022, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Paterni, I.; Granchi, C.; Katzenellenbogen, J.; Minutolo, F. Estrogen Receptors Alpha (ERα) and Beta (ERβ): Subtype-Selective Ligands and Clinical Potential. Steroids 2014, 90C, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Atsriku, C.; Britton, D.J.; Held, J.M.; Schilling, B.; Scott, G.K.; Gibson, B.W.; Benz, C.C.; Baldwin, M.A. Systematic Mapping of Posttranslational Modifications in Human Estrogen Receptor-α with Emphasis on Novel Phosphorylation Sites. Mol. Cell. Proteom. 2009, 8, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Scabia, V.; Ayyanan, A.; De Martino, F.; Agnoletto, A.; Battista, L.; Laszlo, C.; Treboux, A.; Zaman, K.; Stravodimou, A.; Jallut, D.; et al. Estrogen receptor positive breast cancers have patient specific hormone sensitivities and rely on progesterone receptor. Nat. Commun. 2022, 13, 3127. [Google Scholar] [CrossRef] [PubMed]
- Leeuw, R.; Neefjes, J.; Michalides, R. A Role for Estrogen Receptor Phosphorylation in the Resistance to Tamoxifen. Int. J. Breast Cancer 2011, 2011, 232435. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Bouchekioua-Bouzaghou, K.; Sentis, S.; Corbo, L.; Romancer, M. Post-translational modifications modulate estrogen receptor alpha activity in breast tumors. Méd. Sci. 2011, 26, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Iqbal, N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar] [CrossRef] [PubMed]
- Furrer, D.; Paquet, C.; Jacob, S.; Diorio, C. The Human Epidermal Growth Factor Receptor 2 (HER2) as a Prognostic and Predictive Biomarker: Molecular Insights into HER2 Activation and Diagnostic Implications. In Cancer Prognosis; Lemamy, G., Ed.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Gutierrez, C.; Schiff, R. HER2: Biology, Detection, and Clinical Implications. Arch. Pathol. Lab. Med. 2011, 135, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Rauser, S.; Marquardt, C.; Balluff, B.; Deininger, S.-O.; Albers, C.; Belau, E.; Hartmer, R.; Suckau, D.; Specht, K.; Ebert, M.P.; et al. Classification of HER2 Receptor Status in Breast Cancer Tissues by MALDI Imaging Mass Spectrometry. J. Proteome Res. 2010, 9, 1854–1863. [Google Scholar] [CrossRef]
- Malik, N.; Yan, H.; Moshkovich, N.; Palangat, M.; Yang, H.; Sanchez, V.; Cai, Z.; Peat, T.J.; Jiang, S.; Liu, C.; et al. The transcription factor CBFB suppresses breast cancer through orchestrating translation and transcription. Nat. Commun. 2019, 10, 2071. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Shyr, Y. Dysregulated transcription across diverse cancer types reveals the importance of RNA-binding protein in carcinogenesis. BMC Genom. 2015, 16, S5. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, L.; Fu, L.; Yuan, Y.; Dai, H.; Zhu, T.; Zhou, Y.; Yuan, F. Integrated Bioinformatics Analysis the Function of RNA Binding Proteins (RBPs) and Their Prognostic Value in Breast Cancer. Front. Pharmacol. 2019, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Shan, M.; Gregory, B. Using RNA Affinity Purification Followed by Mass Spectrometry to Identify RNA-Binding Proteins (RBPs). Methods Mol Biol. 2020, 2166, 241–253. [Google Scholar] [PubMed]
- Zamanian, M.; Qader Hamadneh, L.A.; Veerakumarasivam, A.; Abdul Rahman, S.; Shohaimi, S.; Rosli, R. Calreticulin mediates an invasive breast cancer phenotype through the transcriptional dysregulation of p53 and MAPK pathways. Cancer Cell Int. 2016, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Vaklavas, C.; Blume, S.; Grizzle, W. Translational Dysregulation in Cancer: Molecular Insights and Potential Clinical Applications in Biomarker Development. Front. Oncol. 2017, 7, 158. [Google Scholar] [CrossRef]
- Stuelten, C.H.; Parent, C.A.; Montell, D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 2018, 18, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Palma, C.; Grassi, M.; Thomé, C.; Ferreira, G.; Albuquerque, D.; Pinto, M.; Melo, F.; Kashima, S.; Covas, D.; Pitteri, S.; et al. Proteomic analysis of epithelial to mesenchymal transition reveals crosstalk between SNAIL and HDAC1 in breast cancer cells. Mol. Cell. Proteom. 2016, 15, 906–917. [Google Scholar] [CrossRef]
- Leggett, S.E.; Hruska, A.M.; Guo, M.; Wong, I.Y. The epithelial-mesenchymal transition and the cytoskeleton in bioengineered systems. Cell Commun. Signal. 2021, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Kraxner, J.; Lorenz, C.; Menzel, J.; Parfentev, I.; Silbern, I.; Denz, M.; Urlaub, H.; Schwappach, B.; Köster, S. Post-Translational Modifications Soften Vimentin Intermediate Filaments. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sharma, P.; Alsharif, S.; Fallatah, A.; Chung, B.M. Intermediate Filaments as Effectors of Cancer Development and Metastasis: A Focus on Keratins, Vimentin, and Nestin. Cells 2019, 8, 497. [Google Scholar] [CrossRef] [PubMed]
- Ulirsch, J.; Fan, C.; Knafl, G.; Wu, M.; Coleman, B.; Perou, C.; Swift-Scanlan, T. Vimentin DNA methylation predicts survival in breast cancer. Breast Cancer Res. Treat. 2012, 137, 383–396. [Google Scholar] [CrossRef]
- Raja, M. p62/SQSTM1 interacts with vimentin to enhance breast cancer metastasis. Carcinogenesis 2019, 38, 1092–1103. [Google Scholar]
- Kaschula, C.H.; Tuveri, R.; Ngarande, E.; Dzobo, K.; Barnett, C.; Kusza, D.A.; Graham, L.M.; Katz, A.A.; Rafudeen, M.S.; Parker, M.I.; et al. The garlic compound ajoene covalently binds vimentin, disrupts the vimentin network and exerts anti-metastatic activity in cancer cells. BMC Cancer 2019, 19, 248. [Google Scholar] [CrossRef]
- McGinn, O.; Ward, A.; Fettig, L.; Riley, D.; Ivie, J.; Paul, K.; Kabos, P.; Finlay-Schultz, J.; Sartorius, C. Cytokeratin 5 alters β-catenin dynamics in breast cancer cells. Oncogene 2020, 39, 2478–2492. [Google Scholar] [CrossRef]
- Mohd Sobri, S.N.; Abdul Sani, S.F.; Sabtu, S.N.; Looi, L.M.; Chiew, S.F.; Pathmanathan, D.; Chio-Srichan, S.; Bradley, D.A. Structural Studies of Epithelial Mesenchymal Transition Breast Tissues. Sci. Rep. 2020, 10, 1997. [Google Scholar] [CrossRef] [PubMed]
- Farndale, R.; Lisman, T.; Bihan, D.; Hamaia, S.; Smerling, C.; Pugh, N.; Konitsiotis, A.; Leitinger, B.; Groot, P.; Jarvis, G.; et al. Cell-collagen interactions: The use of peptide Toolkits to investigate collagen-receptor interactions. Biochem. Soc. Trans. 2008, 36, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Wołczynski, S.; Surażyński, A.; Swiatecka, J.; Pałka, J. Estrogenic and antiestrogenic effects of raloxifene on collagen metabolism in breast cancer MCF-7 cells. Gynecol. Endocrinol. 2001, 15, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Bourgot, I.; Primac, I.; Louis, T.; Noël, A.; Maquoi, E. Reciprocal Interplay Between Fibrillar Collagens and Collagen-Binding Integrins: Implications in Cancer Progression and Metastasis. Front. Oncol. 2020, 10, 1488. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, H.; Rustogi, N.; Hadjisavvas, A.; Tanaka, K.; Kyriacou, K.; Sutton, C.W. Proteomic Profiling of Breast Tissue Collagens and Site-specific Characterization of Hydroxyproline Residues of Collagen Alpha-1-(I). J. Proteome Res. 2012, 11, 5890–5902. [Google Scholar] [CrossRef]
- Liu, J.; Shen, J.-X.; Wu, H.-T.; Li, X.-L.; Wen, X.-F.; Du, C.-W.; Zhang, G.-J. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov. Med. 2018, 25, 211–223. [Google Scholar] [PubMed]
- Danen, E. Integrins: An Overview of Structural and Functional Aspects. In Madame Curie Bioscience Database; 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6259/ (accessed on 15 August 2022).
- Jena, M.K.; Janjanam, J. Role of extracellular matrix in breast cancer development: A brief update. F1000Research 2018, 7, 274. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, P.; Sesé, M.; Guijarro, P.J.; Emperador, M.; Sánchez-Redondo, S.; Peinado, H.; Hümmer, S.; Ramón y Cajal, S. ITGB3-mediated uptake of small extracellular vesicles facilitates intercellular communication in breast cancer cells. Nat. Commun. 2020, 11, 4261. [Google Scholar] [CrossRef]
- Yuan, L.; Shu, B.; Chen, L.; Qian, K.; Wang, Y.; Qian, G.; Zhu, Y.; Cao, X.; Xie, C.; Xiao, Y.; et al. Overexpression of COL3A1 confers a poor prognosis in human bladder cancer identified by co-expression analysis. Oncotarget 2017, 8, 70508–70520. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, C.; Li, Y.; Xing, C.; Wang, S.; Yu, Z.; Chen, L.; Li, P.; Dai, M. Data mining-based study of collagen type III alpha 1 (COL3A1) prognostic value and immune exploration in pan-cancer. Bioengineered 2021, 12, 3634–3646. [Google Scholar] [CrossRef]
- Rizwan, A.; Bulte, C.; Kalaichelvan, A.; Cheng, M.; Krishnamachary, B.; Bhujwalla, Z.M.; Jiang, L.; Glunde, K. Metastatic breast cancer cells in lymph nodes increase nodal collagen density. Sci. Rep. 2015, 5, 10002. [Google Scholar] [CrossRef]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A.; et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Angel, P.M.; Schwamborn, K.; Comte-Walters, S.; Clift, C.L.; Ball, L.E.; Mehta, A.S.; Drake, R.R. Extracellular Matrix Imaging of Breast Tissue Pathologies by MALDI–Imaging Mass Spectrometry. PROTEOMICS Clin. Appl. 2019, 13, e1700152. [Google Scholar] [CrossRef] [PubMed]
- Dekker, T.; Balluff, B.; Jones, E.; Schöne, C.; Schmitt, M.; Aubele, M.; Kroep, J.; Smit, V.; Tollenaar, R.; Mesker, W.; et al. Multicenter Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI MSI) Identifies Proteomic Differences in Breast-Cancer-Associated Stroma. J. Proteome Res. 2014, 13, 4730–4738. [Google Scholar] [CrossRef]
- Boyle, S.T.; Mittal, P.; Kaur, G.; Hoffmann, P.; Samuel, M.S.; Klingler-Hoffmann, M. Uncovering Tumor–Stroma Inter-relationships Using MALDI Mass Spectrometry Imaging. J. Proteome Res. 2020, 19, 4093–4103. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.; Das, G.M. Metabolic Reprogramming in Breast Cancer and Its Therapeutic Implications. Cells 2019, 8, 89. [Google Scholar] [CrossRef]
- Lai, X.; Li, Q.; Wu, F.; Lin, J.; Chen, J.; Zheng, H.; Guo, L. Epithelial-Mesenchymal Transition and Metabolic Switching in Cancer: Lessons From Somatic Cell Reprogramming. Front. Cell Dev. Biol. 2020, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.; Koo, J.S. Glucose Metabolism and Glucose Transporters in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 728759. [Google Scholar] [CrossRef]
- Liang, J.; Cao, R.; Wang, X.; Zhang, Y.; Wang, P.; Gao, H.; Li, C.; Yang, F.; Zeng, R.; Wei, P.; et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res. 2017, 27, 329–351. [Google Scholar] [CrossRef]
- Arnold, J.; Gu, F.; Ambati, C.S.R.; Rasaily, U.; Ramirez-Pena, E.; Joseph, R.; Manikkam, M.; San Martin, R.; Charles, C.; Pan, Y.; et al. UDP-glucose 6-dehydrogenase regulates hyaluronic acid production and promotes breast cancer progression. Oncogene 2020, 39, 1. [Google Scholar] [CrossRef]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 2019, 120, 1090–1098. [Google Scholar] [CrossRef]
- Yoneten, K.K.; Kasap, M.; Akpinar, G.; Gunes, A.; Gurel, B.; Utkan, N.Z. Comparative Proteome Analysis of Breast Cancer Tissues Highlights the Importance of Glycerol-3-phosphate Dehydrogenase 1 and Monoacylglycerol Lipase in Breast Cancer Metabolism. Cancer Genom. Proteom. 2019, 16, 377–397. [Google Scholar] [CrossRef] [PubMed]
- Hicks, D.; Dell’Orto, P.; Falzon, M.; Hoff, K.; Levy, Y.Y.; McMahon, L.; Miller, K.; Russo, L.; Viale, G. Immunohistochemical Performance of Estrogen and Progesterone Receptor Antibodies on the Dako Omnis Staining Platform. Appl. Immunohistochem. Mol. Morphol. 2015, 25, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Tecalco-Cruz, A.; Ramírez-Jarquín, J.; Cruz-Ramos, E. Estrogen Receptor Alpha and its Ubiquitination in Breast Cancer Cells. Curr. Drug Targets 2018, 19, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, K.; Sartorius, C. Progesterone and Progesterone Receptors in Breast Cancer: Past, Present, Future. J. Mol. Endocrinol. 2020, 65, T49–T63. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, W.; Waliszczak, G.; Jach, R.; Dulińska-Litewka, J. Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development. Cancers 2021, 13, 4779. [Google Scholar] [CrossRef] [PubMed]
- Giulianelli, S.; Vaqué, J.; Wargon, V.; Soldati, R.; Vanzulli, S.; Martins, R.; Zeitlin, E.; Helguero, L.; Lamb, C.; Molinolo, A.; et al. The role of estrogen receptor alpha in breast cancer cell proliferation mediated by progestins. Medicina 2012, 72, 315–320. [Google Scholar] [PubMed]
- Badowska-Kozakiewicz, A.; Patera, J.; Sobol, M.; Przybylski, J. The role of oestrogen and progesterone receptors in breast cancer—Immunohistochemical evaluation of oestrogen and progesterone receptor expression in invasive breast cancer in women. Wspolczesna Onkol. 2015, 19, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Anestis, A.; Zoi, I.; Papavassiliou, A.G.; Karamouzis, M.V. Androgen Receptor in Breast Cancer-Clinical and Preclinical Research Insights. Molecules 2020, 25, 358. [Google Scholar] [CrossRef]
- Ismael, N.; Khairy, R.; Talaat, S.; El-Fattah, F. Immunohistochemical Expression of Androgen Receptors (AR) in Various Breast Cancer Subtypes. Open Access Maced. J. Med. Sci. 2019, 7, 1259–1265. [Google Scholar] [CrossRef]
- Vasiliou, S.K.; Filippou, P.S.; Clotet-Freixas, S.; Soosaipillai, A.; Batruch, I.; Viktor Tsianos, F.; Konvalinka, A.; Diamandis, E.P. Transcriptome profiling and proteomic validation reveals targets of the androgen receptor signaling in the BT-474 breast cancer cell line. Clin. Proteom. 2022, 19, 14. [Google Scholar] [CrossRef]
- Steiner, C.; Tille, J.-C.; Lamerz, J.; Geijtenbeek, S.; McKee, T.; Venturi, M.; Rubbia-Brandt, L.; Hochstrasser, D.; Cutler, P.; Lescuyer, P.; et al. Quantification Of HER2 By Targeted Mass Spectrometry in Formalin-Fixed Paraffin-Embedded Breast Cancer Tissues. Mol. Cell. Proteom. MCP 2015, 14, 2786–2799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tai, L.K.; Wong, L.L.; Chiu, L.-L.; Sethi, S.K.; Koay, E.S.C. Proteomic Study Reveals That Proteins Involved in Metabolic and Detoxification Pathways Are Highly Expressed in HER-2/neu-positive Breast Cancer*. Mol. Cell. Proteom. 2005, 4, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, B.; Zhang, G.; Wang, Y.; Cao, L.; Ren, C.; Wen, L.; Lin, J.; Wei, G.; Liao, N. The transcription factor CBFB mutations indicate an improved survival in HR+/HER2- breast cancer. Gene 2020, 759, 144970. [Google Scholar] [CrossRef]
- Yang, C.-M.; Ji, S.; Li, Y.; Fu, L.-Y.; Jiang, T.; Meng, F.-D. β-Catenin promotes cell proliferation, migration, and invasion but induces apoptosis in renal cell carcinoma. Onco Targets Ther. 2017, 10, 711–724. [Google Scholar] [CrossRef]
- Sefidbakht, S.; Saeedipour, H.; Saffar, H.; Mirzaian, E. Determination of β-catenin Expression in Breast Cancer and Its Relationship with Clinicopathologic Parameters. Asian Pac. J. Cancer Prev. 2021, 22, 3493–3498. [Google Scholar] [CrossRef] [PubMed]
- Geyer, F.C.; Lacroix-Triki, M.; Savage, K.; Arnedos, M.; Lambros, M.B.; MacKay, A.; Natrajan, R.; Reis-Filho, J.S. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol. 2011, 24, 209–231. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.-C. β-Catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef]
- Harshman, S.W.; Hoover, M.E.; Huang, C.; Branson, O.E.; Chaney, S.B.; Cheney, C.M.; Rosol, T.J.; Shapiro, C.L.; Wysocki, V.H.; Huebner, K.; et al. Histone H1 phosphorylation in breast cancer. J. Proteome Res. 2014, 13, 2453–2467. [Google Scholar] [CrossRef]
- Izquierdo-Bouldstridge, A.; Bustillos, A.; Bonet-Costa, C.; Aribau-Miralbés, P.; García-Gomis, D.; Dabad, M.; Esteve-Codina, A.; Pascual-Reguant, L.; Peiró, S.; Esteller, M.; et al. Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Res. 2017, 45, 11622–11642. [Google Scholar] [CrossRef]
- Khillare, C.; Sinai Khandeparkar, S.; Joshi, A.; Kulkarni, M.; Gogate, B.; Battin, S. Immunohistochemical Expression of Vimentin in Invasive Breast Carcinoma and Its Correlation with Clinicopathological Parameters. Niger. Med. J. 2019, 60, 17. [Google Scholar] [CrossRef]
- Oliemuller, E.; Newman, R.; Tsang, S.M.; Foo, S.; Muirhead, G.; Noor, F.; Haider, S.; Aurrekoetxea-Rodríguez, I.; Vivanco, M.d.; Howard, B.A. SOX11 promotes epithelial/mesenchymal hybrid state and alters tropism of invasive breast cancer cells. eLife 2020, 9, e58374. [Google Scholar] [CrossRef]
- Horne, H.; Oh, H.; Sherman, M.; Palakal, M.; Hewitt, S.; Schmidt, M.; Milne, R.; Hardisson, D.; Benítez, J.; Blomqvist, C.; et al. E-cadherin breast tumor expression, risk factors and survival: Pooled analysis of 5,933 cases from 12 studies in the Breast Cancer Association Consortium. Sci Rep. 2018, 8, 6574. [Google Scholar] [CrossRef]
- Karsten, N.; Kolben, T.; Mahner, S.; Beyer, S.; Meister, S.; Kuhn, C.; Schmoeckel, E.; Wuerstlein, R.; Harbeck, N.; Ditsch, N.; et al. The role of E-Cadherin expression in primary site of breast cancer. Arch. Gynecol. Obstet. 2022, 305, 913–920. [Google Scholar] [CrossRef]
- Grabenstetter, A.; Mohanty, A.S.; Rana, S.; Zehir, A.; Brannon, A.R.; D’Alfonso, T.M.; DeLair, D.F.; Tan, L.K.; Ross, D.S. E-cadherin immunohistochemical expression in invasive lobular carcinoma of the breast: Correlation with morphology and CDH1 somatic alterations. Hum. Pathol. 2020, 102, 44–53. [Google Scholar] [CrossRef]
- Rosso, M.; Lapyckyj, L.; Besso, M.J.; Monge, M.; Reventós, J.; Canals, F.; Quevedo Cuenca, J.O.; Matos, M.L.; Vazquez-Levin, M.H. Characterization of the molecular changes associated with the overexpression of a novel epithelial cadherin splice variant mRNA in a breast cancer model using proteomics and bioinformatics approaches: Identification of changes in cell metabolism and an increased expression of lactate dehydrogenase B. Cancer Metab. 2019, 7, 5. [Google Scholar] [CrossRef]
- Vergara, D.; Simeone, P.; Latorre, D.; Cascione, F.; Leporatti, S.; Trerotola, M.; Giudetti, A.M.; Capobianco, L.; Lunetti, P.; Rizzello, A.; et al. Proteomics analysis of E-cadherin knockdown in epithelial breast cancer cells. J. Biotechnol. 2015, 202, 3–11. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.W.; Zhu, L.P.; Wang, H.L.; Wang, B.; Zhao, Q.; Wang, X.Y. Significance and prognosis of epithelial-cadherin expression in invasive breast carcinoma. Oncol. Lett. 2018, 16, 1659–1665. [Google Scholar] [CrossRef]
- Guo, Y.; Li, M.; Bai, G.; Li, X.; Sun, Z.; Yang, J.; Wang, L.; Sun, J. Filamin A inhibits tumor progression through regulating BRCA1 expression in human breast cancer. Oncol. Lett. 2018, 16, 6261–6266. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-T.; Peng, Z.; Han, J.-Y.; Lin, F.-Z.; Bu, X.-M.; Xu, Q.-X. Clinical and Prognostic Significance of SOX11 in Breast Cancer. Asian Pac J Cancer Prev 2014, 15, 5483–5486. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shepherd, J.; Uray, I.; Mazumdar, A.; Tsimelzon, A.; Savage, M.; Hilsenbeck, S.; Brown, P. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget 2016, 7, 13106–13121. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Zhao, K.; Bao, K.; Li, J. Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med. 2022, 17, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.-M.; Chan, S.K.; Yu, A.M.C.; Lam, C.C.F.; Tsang, J.Y.S.; Lui, P.C.W.; Law, B.K.B.; Tan, P.-H.; Tse, G.M. Keratin expression in breast cancers. Virchows Archiv. 2012, 461, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Manjari, M.; Kahlon, S.; Kumar, P.; Kalra, N. Cytokeratin 5/6 expression in benign and malignant breast lesions. Indian J. Pathol. Microbiol. 2010, 53, 676–680. [Google Scholar] [CrossRef]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty acid synthase promotes breast cancer metastasis by mediating changes in fatty acid metabolism. Oncol. Lett. 2020, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Xing, X.-L.; Zhang, C.; Yi, L.; Xu, W.; Ou, J.; Zhu, N. MET and FASN as Prognostic Biomarkers of Triple Negative Breast Cancer: A Systematic Evidence Landscape of Clinical Study. Front. Oncol. 2021, 11, 604801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huang, J.; Tang, Y.; Yang, Y.; Huaidong, H. Inhibition of Fatty Acid Synthase (FASN) Affects the Proliferation and Apoptosis of HepG2 Hepatoma Carcinoma Cells via the β-catenin/C-myc Signaling Pathway. Ann. Hepatol. 2020, 19, 411–416. [Google Scholar] [CrossRef]
- Jin, X.; Wang, D.; Lei, M.; Guo, Y.; Cui, Y.; Chen, F.; Sun, W.; Chen, X. TPI1 activates the PI3K/AKT/mTOR signaling pathway to induce breast cancer progression by stabilizing CDCA5. J. Transl. Med. 2022, 20, 191. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Buttacavoli, M.; Roz, E.; Feo, S. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int. J. Mol. Sci. 2019, 20, 3952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, H.; Miao, L.; Ding, J. Silencing of ENO1 inhibits the proliferation, migration and invasion of human breast cancer cells. J. B.U.ON. Off. J. Balk. Union Oncol. 2020, 25, 696–701. [Google Scholar]
- Chu, P.-Y.; Hsu, N.C.; Liao, A.T.; Shih, N.-Y.; Hou, M.-F.; Liu, C.-H. Overexpression of α-enolase correlates with poor survival in canine mammary carcinoma. BMC Vet. Res. 2011, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Mittal, L.; Camarillo, I.G.; Varadarajan, G.S.; Srinivasan, H.; Aryal, U.K.; Sundararajan, R. High-throughput, Label-Free Quantitative Proteomic Studies of the Anticancer Effects of Electrical Pulses with Turmeric Silver Nanoparticles: An in vitro Model Study. Sci. Rep. 2020, 10, 7258. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.K.; Sun, Y.; Lv, L.; Ping, Y. ENO1 and Cancer. Mol. Ther. Oncolytics 2022, 24, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; He, C.; Wei, J.; Zhang, Z.; Luo, Y.; Tan, H.; Ren, C. PGK1 is a Potential Survival Biomarker and Invasion Promoter by Regulating the HIF-1α–Mediated Epithelial-Mesenchymal Transition Process in Breast Cancer. Cell. Physiol. Biochem. 2018, 51, 2434–2444. [Google Scholar] [CrossRef]
- Platica, M.; Ionescu, A.; Ivan, E.; Holland, J.; Mandeli, J.; Platica, O. PAR, a protein involved in the cell cycle, is functionally related to chromosomal passenger proteins. Int. J. Oncol. 2011, 38, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Alhammad, R. Bioinformatics Identification of TUBB as Potential Prognostic Biomarker for Worse Prognosis in ERα-Positive and Better Prognosis in ERα-Negative Breast Cancer. Diagnostics 2022, 12, 2067. [Google Scholar] [CrossRef] [PubMed]
- Mrouj, K.; Andrés-Sánchez, N.; Dubra, G.; Singh, P.; Sobecki, M.; Chahar, D.; Al Ghoul, E.; Aznar, A.B.; Prieto, S.; Pirot, N.; et al. Ki-67 regulates global gene expression and promotes sequential stages of carcinogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2026507118. [Google Scholar] [CrossRef]
- Aman, N.A.; Doukoure, B.; Koffi, K.D.; Koui, B.S.; Traore, Z.C.; Kouyate, M.; Toure, I.; Effi, A.B. Immunohistochemical Evaluation of Ki-67 and Comparison with Clinicopathologic Factors in Breast Carcinomas. As. Pac. J. Cancer Prev. 2019, 20, 73–79. [Google Scholar] [CrossRef]
- Tan, P.-H.; Bay, B.-H.; Yip, G.; Selvarajan, S.; Tan, P.; Wu, J.; Lee, C.-H.; Li, K.-B. Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod. Pathol. 2005, 18, 374–381. [Google Scholar] [CrossRef]
- Wu, S.-y.; Liao, P.; Yan, L.-y.; Zhao, Q.-y.; Xie, Z.-y.; Dong, J.; Sun, H.-t. Correlation of MKI67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 2021, 21, 416. [Google Scholar] [CrossRef]
- Davey, M.G.; Hynes, S.O.; Kerin, M.J.; Miller, N.; Lowery, A.J. Ki-67 as a Prognostic Biomarker in Invasive Breast Cancer. Cancers 2021, 13, 4455. [Google Scholar] [CrossRef] [PubMed]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (Review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Jiang, K.; Hong, R.; Xu, F.; Xia, W.; Qin, G.; Lee, K.; Zheng, Q.; Lu, Q.; Zhai, Q.; et al. Identification and characterization of critical genes associated with tamoxifen resistance in breast cancer. PeerJ 2020, 8, e10468. [Google Scholar] [CrossRef] [PubMed]
- Polotskaia, A.; Xiao, G.; Reynoso, K.; Martin, C.; Qiu, W.-G.; Hendrickson, R.C.; Bargonetti, J. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc. Natl. Acad. Sci. USA 2015, 112, E1220–E1229. [Google Scholar] [CrossRef] [PubMed]
- Bartley, A.; Ross, D. Validation of p53 Immunohistochemistry as a Prognostic Factor in Breast Cancer in Clinical Practice. Arch. Pathol. Lab. Med. 2002, 126, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-p.; Zhang, X.-m.; Zhang, Z.; Zheng, L.-h.; Jindal, S.; Liu, Y.-j. Association of p53 expression with poor prognosis in patients with triple-negative breast invasive ductal carcinoma. Medicine 2019, 98, e15449. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 in breast cancer: Potential as a therapeutic target and biomarker. Breast Cancer Res. Treat 2018, 170, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zangar, R.C. Protein modifications as potential biomarkers in breast cancer. Biomark Insights 2009, 4, 191–200. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Q.; Kang, Y.; Xu, S.; Pang, D. Unconventional protein post-translational modifications: The helmsmen in breast cancer. Cell Biosci. 2022, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Ko, P.-J.; Dixon, S.J. Protein palmitoylation and cancer. EMBO Rep. 2018, 19, e46666. [Google Scholar] [CrossRef]
- Huang, K.-L.; Wu, Y.; Primeau, T.; Wang, Y.-T.; Gao, Y.; McMichael, J.; Scott, A.; Cao, S.; Wendl, M.; Johnson, K.; et al. Regulated Phosphosignaling Associated with Breast Cancer Subtypes and Druggability. Mol. Cell. Proteom. 2019, 18, mcp.RA118.001243. [Google Scholar] [CrossRef] [PubMed]
- Villamar-Cruz, O.; Arias-Romero, L.E. Phosphoproteomics for the Mapping of Altered Cell Signaling Networks in Breast Cancer. In Oncogenomics and Cancer Proteomics—Novel Approaches in Biomarkers Discovery and Therapeutic Targets in Cancer; López-Camarillo, C., Aréchaga-Ocampo, E., Eds.; IntechOpen: London, UK, 2013. [Google Scholar]
- Scott, D.A.; Drake, R.R. Glycosylation and its implications in breast cancer. Expert Rev. Proteom. 2019, 16, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, J.; Smith, J.C.; Ethier, M.; Figeys, D. Proteomic Analysis of Ubiquitinated Proteins from Human MCF-7 Breast Cancer Cells by Immunoaffinity Purification and Mass Spectrometry. J. Proteome Res. 2005, 4, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Yoon, H.-S.; Yoo, S.-A.; Yun, S.H.; Park, J.-H.; Han, E.H.; Chi, S.-G.; Chung, Y.-H. Co-relation with novel phosphorylation sites of IκBα and necroptosis in breast cancer cells. BMC Cancer 2021, 21, 596. [Google Scholar] [CrossRef]
- Ko, H.; Kim, S.; Yang, K.; Kim, K. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis 2018, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, J.; Kang, Y.-L.; Woo, J.; Kim, Y.; Huh, J.; Park, J.-W. Ketohexokinase-A acts as a nuclear protein kinase that mediates fructose-induced metastasis in breast cancer. Nat. Commun. 2020, 11, 5436. [Google Scholar] [CrossRef]
- Chan, Y.-T.; Lai, A.C.-Y.; Lin, R.-J.; Wang, Y.-H.; Wang, Y.-T.; Chang, W.-W.; Wu, H.-Y.; Lin, Y.-J.; Chang, W.-Y.; Wu, J.-C.; et al. GPER-induced signaling is essential for the survival of breast cancer stem cells. Int. J. Cancer 2020, 146, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Liu, W.; Yuan, X.; Jiang, J.; Wang, W.; Mullen, M.; Zhao, X.; Zhang, Y.; Liu, F.; Du, S.; et al. Acetylation of ACAP4 regulates CCL18-elicited breast cancer cell migration and invasion. J. Mol. Cell Biol. 2018, 10, 559–572. [Google Scholar] [CrossRef]
- Tiede, S.; Meyer-Schaller, N.; Kalathur, R.K.R.; Ivanek, R.; Fagiani, E.; Schmassmann, P.; Stillhard, P.; Häfliger, S.; Kraut, N.; Schweifer, N.; et al. The FAK inhibitor BI 853520 exerts anti-tumor effects in breast cancer. Oncogenesis 2018, 7, 73. [Google Scholar] [CrossRef]
- Lang, L.; Hou, Y.; Chen, Y.; Tu, G.; Tao, J.; Yang, D.; Xi, L.; Fu, L.; Sun, K.; Yin, J.; et al. ATM-Mediated Phosphorylation of Cortactin Involved in Actin Polymerization Promotes Breast Cancer Cells Migration and Invasion. Cell. Physiol. Biochem. 2018, 51, 2972–2988. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-F.; Dong, Q.; Bai, Y.; Gu, J.; Tao, Q.; Yue, J.; Zhou, R.; Niu, X.; Zhu, L.; Song, C.; et al. c-Abl kinase-mediated phosphorylation of γ-tubulin promotes γ-tubulin ring complexes assembly and microtubule nucleation. J. Biol. Chem. 2022, 298, 101778. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e1431. [Google Scholar] [CrossRef]
- Whelan, S.A.; Lu, M.; He, J.; Yan, W.; Saxton, R.E.; Faull, K.F.; Whitelegge, J.P.; Chang, H.R. Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J. Proteome Res. 2009, 8, 4151–4160. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-W.; Lim, S.-O.; Xia, W.; Lee, H.-H.; Chan, L.-C.; Kuo, C.-W.; Khoo, K.-H.; Chang, S.-S.; Cha, J.-H.; Kim, T.; et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 2016, 7, 12632. [Google Scholar] [CrossRef] [PubMed]
- Pane, K.; Quintavalle, C.; Nuzzo, S.; Ingenito, F.; Roscigno, G.; Affinito, A.; Scognamiglio, I.; Pattanayak, B.; Gallo, E.; Accardo, A.; et al. Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study. Int. J. Mol. Sci. 2022, 23, 3989. [Google Scholar] [CrossRef]
- Wu, X.; Zahari, M.S.; Renuse, S.; Nirujogi, R.S.; Kim, M.-S.; Manda, S.S.; Stearns, V.; Gabrielson, E.; Sukumar, S.; Pandey, A. Phosphoproteomic Analysis Identifies Focal Adhesion Kinase 2 (FAK2) as a Potential Therapeutic Target for Tamoxifen Resistance in Breast Cancer. Mol. Cell. Proteom. 2015, 14, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Deshmukh, A.P.; den Hollander, P.; Addanki, S.; Kuburich, N.A.; Kudaravalli, S.; Joseph, R.; Chang, J.T.; Soundararajan, R.; Mani, S.A. EMTome: A resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br. J. Cancer 2021, 124, 259–269. [Google Scholar] [CrossRef]
- Lapek, J.D., Jr.; Greninger, P.; Morris, R.; Amzallag, A.; Pruteanu-Malinici, I.; Benes, C.H.; Haas, W. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017, 35, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, L.V.; Millikin, R.J.; Miller, R.M.; Anderson, L.C.; Fellers, R.T.; Ge, Y.; Kelleher, N.L.; LeDuc, R.D.; Liu, X.; Payne, S.H.; et al. Identification and Quantification of Proteoforms by Mass Spectrometry. Proteomics 2019, 19, e1800361. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, L.V.; Millikin, R.J.; Shortreed, M.R.; Scalf, M.; Smith, L.M. Improving Proteoform Identifications in Complex Systems Through Integration of Bottom-Up and Top-Down Data. J. Proteome Res. 2020, 19, 3510–3517. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef]
- Naryzhny, S.; Klopov, N.; Ronzhina, N.; Zorina, E.; Zgoda, V.; Kleyst, O.; Belyakova, N.; Legina, O. A database for inventory of proteoform profiles: “2DE-pattern”. Electrophoresis 2020, 41, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Asano, S. Pathophysiological Roles of Actin-Binding Scaffold Protein, Ezrin. Int. J. Mol. Sci. 2022, 23, 3246. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, W.; Xu, G.; Kong, S.; Ding, L.; Xiao, J.; Cao, X.; Wang, F. ACAP4 interacts with CrkII to promote the recycling of integrin β1. Biochem. Biophys. Res. Commun. 2019, 516, 8–14. [Google Scholar] [CrossRef]
- Brix, D.M.; Bundgaard Clemmensen, K.K.; Kallunki, T. Zinc Finger Transcription Factor MZF1-A Specific Regulator of Cancer Invasion. Cells 2020, 9, 223. [Google Scholar] [CrossRef]
- Veyssière, H.; Bidet, Y.; Penault-Llorca, F.; Radosevic-Robin, N.; Durando, X. Circulating proteins as predictive and prognostic biomarkers in breast cancer. Clin. Proteom. 2022, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Terkelsen, T.; Pernemalm, M.; Gromov, P.; Børresen-Dale, A.-L.; Krogh, A.; Haakensen, V.D.; Lethiö, J.; Papaleo, E.; Gromova, I. High-throughput proteomics of breast cancer interstitial fluid: Identification of tumor subtype-specific serologically relevant biomarkers. Mol. Oncol. 2021, 15, 429–461. [Google Scholar] [CrossRef]
- Zografos, E.; Anagnostopoulos, A.K.; Papadopoulou, A.; Legaki, E.; Zagouri, F.; Marinos, E.; Tsangaris, G.T.; Gazouli, M. Serum Proteomic Signatures of Male Breast Cancer. Cancer Genom. Proteom. 2019, 16, 129–137. [Google Scholar] [CrossRef]
- Yao, F.; Yan, C.; Zhang, Y.; Shen, L.; Zhou, D.; Ni, J. Identification of blood protein biomarkers for breast cancer staging by integrative transcriptome and proteome analyses. J. Proteom. 2021, 230, 103991. [Google Scholar] [CrossRef] [PubMed]
- Celis, J.E.; Moreira, J.M.A.; Cabezón, T.; Gromov, P.; Friis, E.; Rank, F.; Gromova, I. Identification of Extracellular and Intracellular Signaling Components of the Mammary Adipose Tissue and Its Interstitial Fluid in High Risk Breast Cancer Patients: Toward Dissecting The Molecular Circuitry of Epithelial-Adipocyte Stromal Cell Interactions. Mol. Cell. Proteom. 2005, 4, 492–522. [Google Scholar]
- Fu, Z.; Song, P.; Li, D.; Yi, C.; Chen, H.; Ruan, S.; Shi, Z.; Xu, W.; Fu, X.; Zheng, S. Cancer-associated fibroblasts from invasive breast cancer have an attenuated capacity to secrete collagens. Int. J. Oncol. 2014, 45, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Wen, H.; Mao, J.; Ren, Y.; Yang, H. Exosomes: A rising star in breast cancer (Review). Oncol. Rep. 2020, 44, 407–423. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Yang, S.; Pi, H.; Li, Z.; Yao, P.; Zhang, Q.; Wang, Q.; Shen, P.; Li, X.; et al. Proteomic Landscape of Exosomes Reveals the Functional Contributions of CD151 in Triple-Negative Breast Cancer. Mol. Cell. Proteom. 2021, 20, 100121. [Google Scholar] [CrossRef]
- Clark, D.J.; Fondrie, W.E.; Liao, Z.; Hanson, P.I.; Fulton, A.; Mao, L.; Yang, A.J. Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis. Anal. Chem. 2015, 87, 10462–10469. [Google Scholar] [CrossRef]
- Chen, I.H.; Xue, L.; Hsu, C.-C.; Paez, J.S.P.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.-K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef]
- Alexander, H.; Stegner, A.; Wagner-Mann, C.; Bois, G.; Alexander, S.; Sauter, E. Proteomic Analysis to Identify Breast Cancer Biomarkers in Nipple Aspirate Fluid. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 7500–7510. [Google Scholar] [CrossRef]
- Sauter, E.R.; Zhu, W.; Fan, X.J.; Wassell, R.P.; Chervoneva, I.; Du Bois, G.C. Proteomic analysis of nipple aspirate fluid to detect biologic markers of breast cancer. Br. J. Cancer 2002, 86, 1440–1443. [Google Scholar] [CrossRef]
- Teng, P.-n.; Bateman, N.W.; Hood, B.L.; Conrads, T.P. Advances in proximal fluid proteomics for disease biomarker discovery. J. Proteome Res. 2010, 12, 6091–6100. [Google Scholar] [CrossRef] [PubMed]
- Brunoro, G.; Carvalho, P.; Barbosa, V.; Pagnoncelli, D.; Gallo, C.; Perales, J.; Zahedi, R.; Valente, R.; Neves-Ferreira, A. Differential proteomic comparison of breast cancer secretome using a quantitative paired analysis workflow. BMC Cancer 2019, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Shaheed, S.-u.; Tait, C.; Kyriacou, K.; Mullarkey, J.; Burrill, W.; Patterson, L.H.; Linforth, R.; Salhab, M.; Sutton, C.W. Nipple aspirate fluid—A liquid biopsy for diagnosing breast health. Proteom. Clin. Appl. 2017, 11, 1700015. [Google Scholar] [CrossRef] [PubMed]
- Delmonico, L.; Areias, V.R.; Pinto, R.C.; Matos, C.D.S.; Rosa, M.F.F.; De Azevedo, C.M.; Alves, G. Protein identification from dried nipple aspirate fluid on Guthrie cards using mass spectrometry. Mol. Med. Rep. 2015, 12, 159–164. [Google Scholar] [CrossRef][Green Version]
- Schneider, S.; Aslebagh, R.; Wetie, A.; Sturgeon, S.; Darie, C.; Arcaro, K. Using Breast Milk to Assess Breast Cancer Risk: The Role of Mass Spectrometry-Based Proteomics. Adv. Exp. Med. Biol. 2014, 806, 399–408. [Google Scholar] [CrossRef]
- Altendorfer, I.; Koenig, S.; Braukmann, A.; Saenger, T.; Bleck, E.; Vordenbäumen, S.; Kubiak, A.; Schneider, M.; Jose, J. Quantification of αS1-casein in breast milk using a targeted mass spectrometry-based approach. J. Pharm. Biomed. Anal. 2014, 103C, 52–58. [Google Scholar] [CrossRef]
- Bonuccelli, G.; Castello-Cros, R.; Capozza, F.; Martinez-Outschoorn, U.E.; Lin, Z.; Tsirigos, A.; Xuanmao, J.; Whitaker-Menezes, D.; Howell, A.; Lisanti, M.P.; et al. The milk protein α-casein functions as a tumor suppressor via activation of STAT1 signaling, effectively preventing breast cancer tumor growth and metastasis. Cell Cycle 2012, 11, 3972–3982. [Google Scholar] [CrossRef]
- Samuel, M.; Fonseka, P.; Sanwlani, R.; Gangoda, L.; Chee, S.; Keerthikumar, S.; Spurling, A.; Chitti, S.V.P.; Zanker, D.; Ang, C.-S.; et al. Oral administration of bovine milk-derived extracellular vesicles induces senescence in the primary tumor but accelerates cancer metastasis. Nat. Commun. 2021, 12, 3950. [Google Scholar] [CrossRef]
- Galley, J.D.; Besner, G.E. The Therapeutic Potential of Breast Milk-Derived Extracellular Vesicles. Nutrients 2020, 12, 745. [Google Scholar] [CrossRef]
- van Herwijnen, M.J.C.; Zonneveld, M.I.; Goerdayal, S.; Nolte’t Hoen, E.N.M.; Garssen, J.; Stahl, B.; Maarten Altelaar, A.F.; Redegeld, F.A.; Wauben, M.H.M. Comprehensive Proteomic Analysis of Human Milk-derived Extracellular Vesicles Unveils a Novel Functional Proteome Distinct from Other Milk Components*. Mol. Cell. Proteom. 2016, 15, 3412–3423. [Google Scholar] [CrossRef]
- Lewis, K.M.; Harford-Wright, E.; Vink, R.; Ghabriel, M.N. Characterisation of Walker 256 breast carcinoma cells from two tumour cell banks as assessed using two models of secondary brain tumours. Cancer Cell Int. 2013, 13, 5. [Google Scholar] [CrossRef]
- Tkacikova, S.; Talian, I.; Sabo, J. Optimisation of urine sample preparation for shotgun proteomics. Open Chem. 2020, 18, 850–856. [Google Scholar] [CrossRef]
- Keller, K.; Pieter, J.; Boehm, D.; Boehm, N.; Wolters, D.; Koelbl, H.; Pfeiffer, N.; Grus, F. Proteomic Analysis Of Tear Fluid Of Breast Cancer Patients And Healthy Subjects Shows Differences In Protein Expression Levels. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3724. [Google Scholar]
- Jasim, H.; Olausson, P.; Hedenberg-Magnusson, B.; Ernberg, M.; Ghafouri, B. The proteomic profile of whole and glandular saliva in healthy pain-free subjects. Sci. Rep. 2016, 6, 39073. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.A.; Yan, W.; Ramachandran, P.; Wong, D.T. Comparative human salivary and plasma proteomes. J. Dent. Res. 2010, 89, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Perrone, D.; Cocchi, R.; Lo Russo, L.; De Lillo, A.; Giannatempo, G.; Lo Muzio, L. Novel possibilities in the study of the salivary proteomic profile using seldi-TOF/MS technology (Review). Oncol. Lett. 2016, 11, 1967–1972. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Streckfus, C.; Bigler, L.; Zwick, M. The use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to detect putative breast cancer markers in saliva: A feasibility study. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2006, 35, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Zambonin, C.; Aresta, A. MALDI-TOF/MS Analysis of Non-Invasive Human Urine and Saliva Samples for the Identification of New Cancer Biomarkers. Molecules 2022, 27, 1925. [Google Scholar] [CrossRef]
- Mitulović, G. Proteomics of the Salivary Fluid; 2019; Available online: https://www.intechopen.com/chapters/58474 (accessed on 14 August 2022).
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N.S. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol./Hematol. 2017, 110, 62–73. [Google Scholar] [CrossRef]
- Koopaie, M.; Kolahdooz, S.; Fatahzadeh, M.; Manifar, S. Salivary biomarkers in breast cancer diagnosis: A systematic review and diagnostic meta-analysis. Cancer Med. 2022, 11, 2644–2661. [Google Scholar] [CrossRef]
- Yan, W.; Apweiler, R.; Balgley, B.M.; Boontheung, P.; Bundy, J.L.; Cargile, B.J.; Cole, S.; Fang, X.; Gonzalez-Begne, M.; Griffin, T.J.; et al. Systematic comparison of the human saliva and plasma proteomes. Proteom. Clin. Appl. 2009, 3, 116–134. [Google Scholar] [CrossRef] [PubMed]
- Streckfus, C.F.; Bigler, L. A Catalogue of Altered Salivary Proteins Secondary to Invasive Ductal Carcinoma: A Novel In Vivo Paradigm to Assess Breast Cancer Progression. Sci. Rep. 2016, 6, 30800. [Google Scholar] [CrossRef] [PubMed]
- Delmonico, L.; Bravo, M.; Silvestre, R.T.; Ornellas, M.H.F.; De Azevedo, C.M.; Alves, G. Proteomic profile of saliva and plasma from women with impalpable breast lesions. Oncol. Lett. 2016, 12, 2145–2152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zambonin, C. MALDI-TOF/MS Analysis of Extracellular Vesicles Released by Cancer Cells. Appl. Sci. 2022, 12, 6149. [Google Scholar] [CrossRef]
| PTMs | References | Proteins | Function and Roles in BC |
|---|---|---|---|
| Phosphorylation | [175] | histone H1 isoforms [132] | Putative biomarker of proliferation BC cells [132] |
| YWHAH | BC cell migration [181] | ||
| PKA/BAD | Stemness and survival of BCSCs [182] | ||
| ACAP4 | Phosphorylated ezrin and phosphorylated ACAP4 interacts to induce membrane fusion of intracellular tubule-vesicles with the apical membrane; cancer progression and metastasis [199], cell migration, polarity, vesicle trafficking and tumorigenesis, regulation of cell adhesion [200] | ||
| ERα | Critical in development and progression of BC [70] | ||
| MZF1 [180] | Development of aggressive BC, control of genes involved in EMT, lysosome-mediated invasion/metastasis [201] | ||
| TUBG1 | Phosphorylation deficiency impairs centrosome construction and microtubules nucleation [186] | ||
| CTTN | Phosphorylated CTTN may play a critical role in promoting breast cancer cell mobility and invasion via actin polymerization [185] | ||
| IκBα | Phosphorylation of NF-κB inhibitor alpha is involved in NF-κB TF activity, regulating apoptosis and necroptosis in BC cells [179] | ||
| FAK autophosphorylation | Activation of FAK-SRC signaling complex that trigger pathways involved in cancer cell migration, invasion, proliferation, death and malignant tumor progression [184] | ||
| Glycosylation | [177] | membrane proteins [188], i.e., PD-L1 | Potential therapeutic strategies to increase cancer immune therapy efficacy [189] |
| Acetylation | [173] | nuclear proteins [187], ACAP4 [183] | BC cell migration and invasion [183] |
| Ubiquitination | [178] | PD-L1 | Potential therapeutic strategies to increase cancer immune therapy efficacy [189] |
| SUMOylation | [172] | MZF1 | Transcriptional activation or inactivation [201] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neagu, A.-N.; Jayathirtha, M.; Whitham, D.; Mutsengi, P.; Sullivan, I.; Petre, B.A.; Darie, C.C. Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer. Proteomes 2022, 10, 35. https://doi.org/10.3390/proteomes10040035
Neagu A-N, Jayathirtha M, Whitham D, Mutsengi P, Sullivan I, Petre BA, Darie CC. Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer. Proteomes. 2022; 10(4):35. https://doi.org/10.3390/proteomes10040035
Chicago/Turabian StyleNeagu, Anca-Narcisa, Madhuri Jayathirtha, Danielle Whitham, Panashe Mutsengi, Isabelle Sullivan, Brindusa Alina Petre, and Costel C. Darie. 2022. "Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer" Proteomes 10, no. 4: 35. https://doi.org/10.3390/proteomes10040035
APA StyleNeagu, A.-N., Jayathirtha, M., Whitham, D., Mutsengi, P., Sullivan, I., Petre, B. A., & Darie, C. C. (2022). Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer. Proteomes, 10(4), 35. https://doi.org/10.3390/proteomes10040035








