Proteomic Approach for Comparative Analysis of the Spike Protein of SARS-CoV-2 Omicron (B.1.1.529) Variant and Other Pango Lineages
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Determination of Physicochemical Properties
2.3. Prediction of Immunoproperties
2.4. Phylogenetic Tree Construction and Primary Amino Acid Sequence Alignment
2.5. Comparative Analysis of the Secondary and Tertiary Structure of Omicron
2.6. Protein-Protein Interactions
3. Results
3.1. Physical Parameters of Proteins
3.2. Prediction of Immune Properties
3.3. Comparative Sequences and Phylogenetic Analysis of Omicron Spike Protein
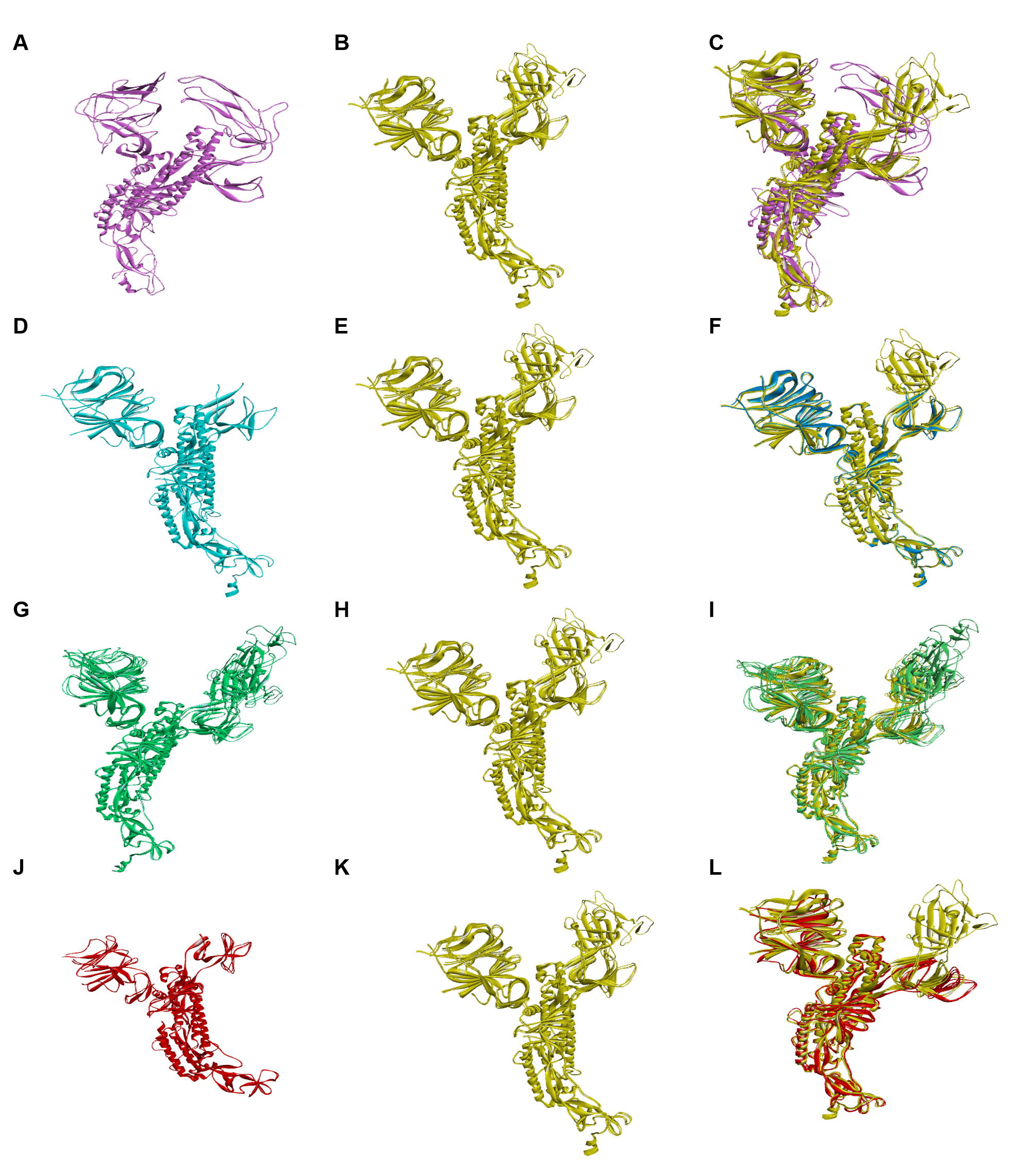
3.4. Secondary and Tertiary Structure Analysis
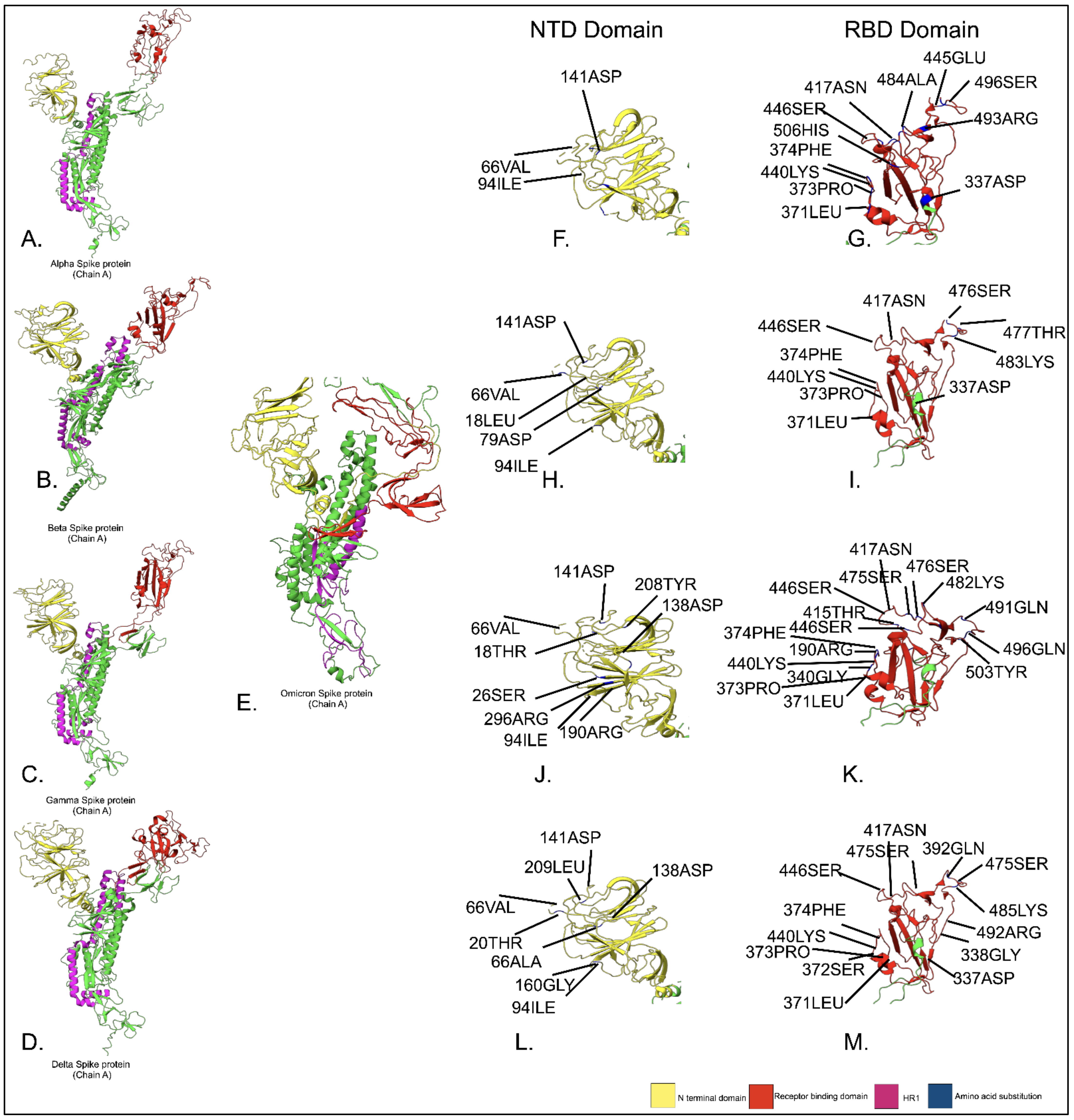
3.5. Proteome-Based Mutational Analysis of Spike Protein Domains
3.6. Protein-Protein Interaction Analysis: (Spike-SARS-CoV-2)-hACE2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, evaluation, and treatment of coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554776/ (accessed on 20 August 2022).
- Piret, J.; Boivin, G. Pandemics throughout history. Front. Microbiol. 2021, 11, 631736. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): An update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef] [PubMed]
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; Nahed, A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B. 1.1. 529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022, 209, 112816. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses 2021, 13, 109. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Natesh, R.; Schwager, S.L.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Shi, Y.; Ding, W.; Niu, T.; Sun, L.; Tan, Y.; Chen, Y.; Shi, J.; Xiong, Q.; Huang, X.; et al. Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes. Nat. Commun. 2021, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, C.; Wang, Y.; Hong, Q.; Zhang, C.; Li, Z.; Xu, S.; Zuo, Q.; Liu, C.; Huang, Z.; et al. Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with hACE2 receptor revealed by cryo-EM. Nat. Commun. 2021, 12, 7345. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; Veinotte, K.; et al. SARS-CoV-2 Spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain. BioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, X.; Yurkovetskiy, L.; Shen, K.; Luban, J.; Natalya, D.; Pascal, K.; Tomkins-Tinch, C.; Nyalile, T.; Wang, Y.; Baum, A.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Microsc. Microanal. 2021, 27 (Suppl. S1), 3260–3262. [Google Scholar] [CrossRef]
- Hong, Q.; Han, W.; Li, J.; Xu, S.; Wang, Y.; Xu, C.; Li, Z.; Wang, Y.; Zhang, C.; Huang, Z.; et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 2022, 604, 546–552. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Zhang, C.; Wang, Y.; Hong, Q.; Xu, S.; Li, Z.; Yang, Y.; Huang, Z.; Cong, Y. Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun. 2022, 13, 871. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.; Marquez, C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron Variant: hACE2 Binding, Cryo-EM Structure of Spike Protein-hACE2 Complex and Antibody Evasion. BioRxiv 2021. [Google Scholar] [CrossRef]
- Feurstein, C.; Meyer, V.; Jung, S. Structure-activity analysis using computational mining of protein databases to assist design of antimicrobial peptides. In Proceedings of the 17th Naples Workshop on Bioactive Peptides, Naples, Italy, 16–18 June 2022; EDIZIONI ZIINO—MASSMEDIA COMUNICAZIONE SRLS: Castellammare di Stabia, Italy, 2022; Volume 47. Available online: https://www.peptidesnaplesworkshop.it/images/Book%20of%20Abstracts%2017th%20Naples%20Workshop%20on%20Bioactive%20Peptides_compressed.pdf#page=47 (accessed on 21 August 2022).
- Al Saba, A.; Adiba, M.; Saha, P.; Hosen, M.I.; Chakraborty, S.; Nabi, A.N. An in-depth in silico and immunoinformatics approach for designing a potential multi-epitope construct for the effective development of vaccine to combat against SARS-CoV-2 encompassing variants of concern and interest. Comput. Biol. Med. 2021, 136, 104703. [Google Scholar] [CrossRef]
- Ayyagari, V.S.; TC, V.; Srirama, K. Design of a multi-epitope-based vaccine targeting M-protein of SARS-CoV2: An immunoinformatics approach. J. Biomol. Struct. Dyn. 2022, 40, 2963–2977. [Google Scholar] [CrossRef]
- Akter, S.; Shahab, M.; Sarkar, M.M.; Hayat, C.; Banu, T.A.; Goswami, B.; Jahan, I.; Osman, E.; Uzzaman, M.S.; Habib, M.A.; et al. Immunoinformatics approach to epitope-based vaccine design against the SARS-CoV-2 in Bangladeshi patients. J. Genet. Eng. Biotechnol. 2022, 20, 136. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021, 184, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Huang, X.; Miller, W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef]
- Sen, T.Z.; Jernigan, R.L.; Garnier, J.; Kloczkowski, A. GOR V server for protein secondary structure prediction. Bioinformatics 2005, 21, 2787–2788. [Google Scholar] [CrossRef]
- Sun, C.; Xie, C.; Bu, G.L.; Zhong, L.Y.; Zeng, M.S. Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2022, 7, 202. [Google Scholar] [CrossRef]
- Maiti, R.; Van Domselaar, G.H.; Zhang, H.; Wishart, D.S. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 2004, 32 (Suppl. S2), W590–W594. [Google Scholar] [CrossRef]
- Van Zundert, G.C.; Rodrigues, J.P.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.; van Dijk, M.; De Vries, S.J.; Bonvin, A.M. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Schwartz, O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022, 20, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Mohamed, M.E.; Abd El-Lateef, H.M.; Venugopala, K.N.; El-Beltagi, H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022, 94, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Zafar, I.; Iftikhar, R.; Ahmad, S.U.; Rather, M.A. Genome wide identification, phylogeny, and synteny analysis of sox gene family in common carp (Cyprinus carpio). Biotechnol. Rep. 2021, 30, e00607. [Google Scholar] [CrossRef]
- He, L.; Sun, S.; Zhang, Q.; Bao, X.; Li, P.K. Alignment-free sequence comparison for virus genomes based on location correlation coefficient. Infect. Genet. Evol. 2021, 96, 105106. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Fersht, A.R.; Serrano, L. Principles of protein stability derived from protein engineering experiments. Curr. Opin. Struct. Biol. 1993, 3, 75–83. [Google Scholar] [CrossRef]
- Matthews, B.W. Structural and genetic analysis of protein stability. Annu. Rev. Biochem. 1993, 62, 139–160. [Google Scholar] [CrossRef]
- Colson, P.; Lavagna, C.; Delerce, J.; Groshenry, G.; Yahi, N.; Fantini, J.; La Scola, B.; Althaus, T. First Detection of the SARS-CoV-2 Omicron BA. 5/22B in Monaco. Microorganisms 2022, 10, 1952. [Google Scholar] [CrossRef]
- Tekewe, A.; Connors, N.K.; Middelberg, A.P.; Lua, L.H. Design strategies to address the effect of hydrophobic epitope on stability and in vitro assembly of modular virus-like particle. Protein Sci. 2016, 25, 1507–1516. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, P.; Xiao, X.; Dimitrov, D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004, 314, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Mansbach, R.A.; Chakraborty, S.; Nguyen, K.; Montefiori, D.C.; Korber, B.; Gnanakaran, S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 2021, 7, eabf3671. [Google Scholar] [CrossRef]
- Hattori, T.; Koide, A.; Noval, M.G.; Panchenko, T.; Romero, L.A.; Teng, K.W.; Tada, T.; Landau, N.R.; Stapleford, K.A.; Koide, S. The ACE2-binding interface of SARS-CoV-2 Spike inherently deflects immune recognition. J. Mol. Biol. 2021, 433, 166748. [Google Scholar] [CrossRef]
- Yi, C.; Sun, X.; Ye, J.; Ding, L.; Liu, M.; Yang, Z.; Lu, X.; Zhang, Y.; Ma, L.; Gu, W.; et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020, 17, 621–630. [Google Scholar] [CrossRef]
- Francis, J.M.; Leistritz-Edwards, D.; Dunn, A.; Tarr, C.; Lehman, J.; Dempsey, C.; Hamel, A.; Rayon, V.; Liu, G.; Wang, Y.; et al. Allelic variation in class I HLA determines CD8+ T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci. Immunol. 2021, 7, eabk3070. [Google Scholar]
- Dzuvor, C.K.; Tettey, E.L.; Danquah, M.K. Aptamers as promising nanotheranostic tools in the COVID-19 pandemic era. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, 14, e1785. [Google Scholar] [CrossRef]
- Parums, D.V. First Approval of the Protein-Based Adjuvanted Nuvaxovid (NVX-CoV2373) Novavax Vaccine for SARS-CoV-2 Could Increase Vaccine Uptake and Provide Immune Protection from Viral Variants. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2022, 28, e936523-1–e936523-3. [Google Scholar] [CrossRef]
- Mosqueda, J.; Hernández-Silva, D.J.; Romero-Maldonado, A.; Mejia-López, S.; Mercado-Uriostegui, M.A. Innovative recombinant protein-based vaccines against SARS-CoV-2. In Biomedical Innovations to Combat COVID-19; Academic Press: Cambridge, MA, USA, 2022; Chapter 11; pp. 193–211. [Google Scholar] [CrossRef]

| Variants of SARS-CoV-2 | Exposed B Cell Epitopes | Predicted Probability of Antigenicity Score | Number of Epitopes Identified in CTLa | Protective Antigen Prediction Score | Number of Strong Binders in T-Cell | Immunogenicity Predication Score |
|---|---|---|---|---|---|---|
| α (7CYD) | 40 | 0.717053 | 37 | 0.4646 | 20 | 0.3019 |
| β (7VX1) | 40 | 0.643558 | 38 | 0.4542 | 09 | 1.23216 |
| δ (7W92) | 38 | 0.744007 | 35 | 0.4709 | 23 | 0.0304 |
| γ (6XS6) | 41 | 0.596261 | 34 | 0.4583 | 22 | 1.07515 |
| Omicron (7T9J) | 33 | 0.717053 | 35 | 0.4646 | 27 | 0.49637 |
| Parameters | α | β | δ | γ | Omicron |
|---|---|---|---|---|---|
| α helix (Hh) | 262 is 23.4% | 268 is 21.30% | 275 is 21.81% | 268 is 21.34% | 255 is 23.46% |
| 310 helix (Gg) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Pi helix (Ii) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| β bridge (Bb) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Extended strand (Ee) | 290 is 25.99% | 253 is 20.11% | 248 is 19.67% | 263 is 20.94% | 253 is 19.69% |
| β turn (Tt) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Bend region (Ss) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Random coil (Cc) | 564 is 50.54% | 737 is 58.59% | 738 is 58.52% | 725 is 57.72% | 777 is 60.47% |
| Ambiguous states (?) | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Other states | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% | 0 is 0.00% |
| Interaction | RMSD | α Carbon | Backbone | Heavy | All |
|---|---|---|---|---|---|
| α–Omicron (7CYD–7T9J) | Local | 2.785 | 2.783 | 2.903 | 2.903 |
| Global | 2.785 | 2.783 | 2.903 | 2.903 | |
| β–Omicron (7VX1–7T9J) | Local | 0.738 | 0.747 | 0.91 | 0.91 |
| Global | 0.738 | 0.747 | 0.91 | 0.91 | |
| γ–Omicron (6XS6–7T9J) | Local | 0.818 | 0.833 | 1.05 | 1.05 |
| Global | 0.818 | 0.833 | 1.05 | 1.05 | |
| δ–Omicron (7W92–7T9J) | Local | 1.437 | 1.438 | 1.673 | 1.673 |
| Global | 1.437 | 1.438 | 1.673 | 1.673 |
| Interaction | Chain | RMSD Value | α Carbon | Backbone | Heavy | All |
|---|---|---|---|---|---|---|
| α–Omicron (7CYD–7T9J) | A chain | Local | - | - | - | - |
| Global | - | - | - | - | ||
| B chain | Local | - | - | - | - | |
| Global | - | - | - | - | ||
| C chain | Local | 1.70 | 1.73 | 2.05 | 2.05 | |
| Global | 20.47 | 20.43 | 20.55 | 20.55 | ||
| β–Omicron (7VX1–7T9J) | A chain | Local | 43.60 | 43.59 | 43.48 | 43.48 |
| Global | 43.60 | 43.59 | 43.48 | 43.48 | ||
| B chain | Local | 44.04 | 44.03 | 43.89 | 43.89 | |
| Global | 44.04 | 44.03 | 43.89 | 43.89 | ||
| C chain | Local | 64.35 | 64.35 | 62.23 | 62.23 | |
| Global | 64.35 | 64.35 | 62.23 | 62.23 | ||
| γ–Omicron (6X6S–7T9J) | A chain | Local | 2.39 | 2.40 | 2.64 | 2.64 |
| Global | 2.39 | 2.40 | 2.64 | 2.64 | ||
| B chain | Local | 2.22 | 2.24 | 2.46 | 2.46 | |
| Global | 2.22 | 2.24 | 2.46 | 2.46 | ||
| C chain | Local | 2.41 | 2.43 | 2.63 | 2.63 | |
| Global | 2.41 | 2.43 | 2.63 | 2.63 | ||
| δ–Omicron (7W92–7T9J) | A chain | Local | 5.25 | 5.13 | 5.43 | 5.43 |
| Global | 4.57 | 4.57 | 4.75 | 4.75 | ||
| B chain | Local | 0.98 | 1.00 | 1.43 | 1.43 | |
| Global | 2.83 | 2.84 | 3.12 | 3.12 | ||
| C chain | Local | 1.28 | 1.32 | 1.63 | 1.63 | |
| Global | 15.10 | 15.09 | 15.20 | 15.20 |
| Interaction of SARS-CoV-2 Variant’s Spike Protein with hACE2 | Binding Affinity in kcal/mol |
|---|---|
| spike protein of α-hACE2 | −10.8 |
| spike protein of β-hACE2 | −10.5 |
| spike protein of δ-hACE2 | −8.3 |
| spike protein of γ-hACE2 | −9.5 |
| spike protein of omicron-hACE2 | −11.8 |
| Interacting Proteins | Variants | ||||
|---|---|---|---|---|---|
| Alpha | Beta | Gamma | Delta | Omicron | |
| Spike-RBD residues | R403, Y453, A475, G485, F486, N487, C488, Y489, Q493, Q498, T500, N501, Y505 | R408, T415,G416,N417,Y449, L452,Y453,L455,F456, A475, G476,T478,K484,F489,N487,Y489,Q493, G496, Q498,T500,Y501,G502,Y505 | E329, K353, D405, T417, L455, F456, K484, F486, Q498, T500,Y501, Y505 | R403, Y453, A475, G485, F486, N487, C488, Y489, Q493, Q498, T500, N501, Y505 | N417, Y449, Y453, L455, F456, F486, N487, Y489, F490, R493, S494, S496, Y501 |
| ACE2 residues | I21, Q24, K31, H34, D38, L39, Q42, M82, Y83, P84, E87 | S19, Q24, T27,F28,D30,K31,H34,E35,D38,Y41,Q42,L45,L79,M82, Y83 | Y41, D30, E35, E37, D38, L39, Q42, M82, Y83, P84, E87 | I21, Q24, K31, H34, D38, L39, Q42, M82, Y83, P84, E87 | T27, F28, D30, K31, H34, E35, D38, T78, L79, M82, K353 |
| SARS-CoV-2 Variant’s Spike Protein -hACE2 Interaction | Chain A (Spike-Variant) Residues | Chain B (hACE2) Residues | Salt Bridges | H-Bonding | Non-Bonded Contacts |
|---|---|---|---|---|---|
| α-hACE2 | 10 | 15 | 1 | 7 | 77 |
| β-hACE2 | 18 | 11 | 2 | 16 | 67 |
| δ-hACE2 | 18 | 12 | 1 | 8 | 73 |
| γ-hACE2 | 16 | 16 | 2 | 8 | 113 |
| Omicron-hACE2 | 12 | 17 | 3 | 32 | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, M.; Patil, N.; Gor, D.; Sharma, M.K.; Goel, N.; Kaushik, P. Proteomic Approach for Comparative Analysis of the Spike Protein of SARS-CoV-2 Omicron (B.1.1.529) Variant and Other Pango Lineages. Proteomes 2022, 10, 34. https://doi.org/10.3390/proteomes10040034
Jain M, Patil N, Gor D, Sharma MK, Goel N, Kaushik P. Proteomic Approach for Comparative Analysis of the Spike Protein of SARS-CoV-2 Omicron (B.1.1.529) Variant and Other Pango Lineages. Proteomes. 2022; 10(4):34. https://doi.org/10.3390/proteomes10040034
Chicago/Turabian StyleJain, Mukul, Nil Patil, Darshil Gor, Mohit Kumar Sharma, Neha Goel, and Prashant Kaushik. 2022. "Proteomic Approach for Comparative Analysis of the Spike Protein of SARS-CoV-2 Omicron (B.1.1.529) Variant and Other Pango Lineages" Proteomes 10, no. 4: 34. https://doi.org/10.3390/proteomes10040034
APA StyleJain, M., Patil, N., Gor, D., Sharma, M. K., Goel, N., & Kaushik, P. (2022). Proteomic Approach for Comparative Analysis of the Spike Protein of SARS-CoV-2 Omicron (B.1.1.529) Variant and Other Pango Lineages. Proteomes, 10(4), 34. https://doi.org/10.3390/proteomes10040034







