Molecular Dynamics Study of Citrullinated Proteins Associated with the Development of Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. List of Proteins with RA-Associated Post-Translational Modifications
| No. | UniProt ID * | Protein Name | Biological Process ** | PDB ID 3 * | Sequence with PTM Moiety | Mw, kDa | Number of a. a. | Aliphatic Index | Instability Index | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P06733-2 | α-enolase | An enzyme of glycolysis, growth control, hypoxia tolerance, and allergic responses | 7 | 266-DPS R/Cit YISPDQLADLYKSFIK-285 | 47.2 | 434 | 88.6 | 36.5 | [29] |
| 2 | P08670 | Vimentin | Class-III intermediate filaments | 2 | 136-EQLKGQGKS R/Cit LGDLYEEEMR-155 | 53.6 | 466 | 81.6 | 58.1 | [16] |
| 3 | 1 | 371-NMKEEMARHL R/Cit EYQDLLNVK-390 | [16] | |||||||
| 4 | 1 | 371-NMKEEMA R/Cit HLREYQDLLNVK-390 | [16] | |||||||
| 5 | Q9UM07 | protein-arginine deiminase type-4 | Citrullination/deimination of arginine residues of proteins such as histones | 18 | 204-A R/Cit SEMDKV R/Cit VFQAT R/Cit GK-220 | 74.1 | 663 | 82.3 | 39.2 | [16,30] |
| 6 | 18 | 375-GLKEFPIK R/Cit VMGPDFGYVTR-394 | [30] | |||||||
| 7 | 18 | 480-PAPDRKGFRLLLASP R/Cit SCYK-499 | [30] | |||||||
| 8 | P02778-2 | C-X-C chemokine 10 | Pro-inflammatory cytokine that is involved in apoptosis, chemotaxis, differentiation, regulation of cell growth | 4 | 24-LS R/Cit TVRCTCISISNQPVNPR-43 | 10.1 | 98 | 103.5 | 55.3 | [31] |
| 9 | P02671 | Fibrinogen α-chain | Hemostasis | 2 | 29-AEGGGV R/Cit GPRVVE R/Cit HQSACK-48 | 98 | 866 | 53.1 | 40.8 | [31] |
| 10 | 2 | 562-SHHPGIAEFPS R/Cit GKSSSYSK-581 | [31] | |||||||
| 11 | 2 | 583-FTSSTSYN R/Cit GDSTFESKSYK-602 | [31] | |||||||
| 12 | P02675 | Fibrinogen β-chain | Hemostasis | 28 | 266-Y R/Cit VYCDMNTENGGWTVIQNR-285 | 55.9 | 491 | 62.5 | 42.5 | [32] |
| 13 | 28 | 254-MYLIQPDSSVKPY R/Cit VYCDMR-273 | [32] | |||||||
| 14 | 4 | 55-EAPSL R/Cit PAPPPISGGGYRAR-74 | [32] | |||||||
| 15 | P60709 | Actin, cytoplasmic 1 | Production of filaments that form cross-linked networks in the cytoplasm of cells | 7 | 88-HTFYNEL R/Cit VAPEEHPVLLTEAPLNPK-113 | 41.7 | 375 | 82 | 35.3 | [33] |
| 16 | P01009 | α-1-antitrypsin | Inhibitor of serine proteases | 24 | 218-WE R/Cit PFEVKDTEEEDFHVDQVTTVK-241 | 46.7 | 418 | 91.2 | 31.6 | [33] |
| 17 | P02647 | Apolipoprotein A-I | Reverse transport of cholesterol from tissues to the liver | 17 | 231-AKPALEDL R/Cit QGLLPVLESFK-250 | 30.8 | 267 | 84.8 | 40.8 | [33] |
| 18 | P02656 | Apolipoprotein C-III | Triglyceride homeostasis | 1 | 47-LSSVQESQVAQQA R/Cit GWVTDGFSSLK-71 | 10.9 | 99 | 84.6 | 29.2 | [33] |
| 19 | P02649 | Apolipoprotein E | Core component of plasma lipoproteins | 7 | 186-EGAE R/Cit GLSAIR-198 | 10.9 | 99 | 84.5 | 29.2 | [33] |
| 20 | P02760 | Protein AMBP | Inhibition of trypsin, plasmin, and lysosomal granulocytic elastase | 2 | 294-GPC R/Cit AFIQLWAFDAVK-309 | 39 | 352 | 70.6 | 49.1 | [29] |
| 21 | P00734 | Prothrombin | Blood homeostasis, inflammation and wound healing | 27 | 453-YNW R/Cit ENLD Cit DIALMK-467 | 70 | 622 | 69.8 | 40.9 | [33] |
| 22 | 27 | 434-YE R/Cit NIEK-440 | [29] | |||||||
| 23 | P02768 | Serum albumin | Regulation of the colloidal osmotic pressure of blood | 125 | 97-LCTVATL R/Cit ETYGEMADCCAK-117 | 69.4 | 609 | 77.6 | 39.1 | [33] |
3.2. Structural Motifs of Protein Molecules Studied in This Work
3.2.1. β-β-Hairpin
3.2.2. α-α-Corner
3.2.3. β-α-β-Motif
3.2.4. 3ß-Corner
3.2.5. Right Superhelix
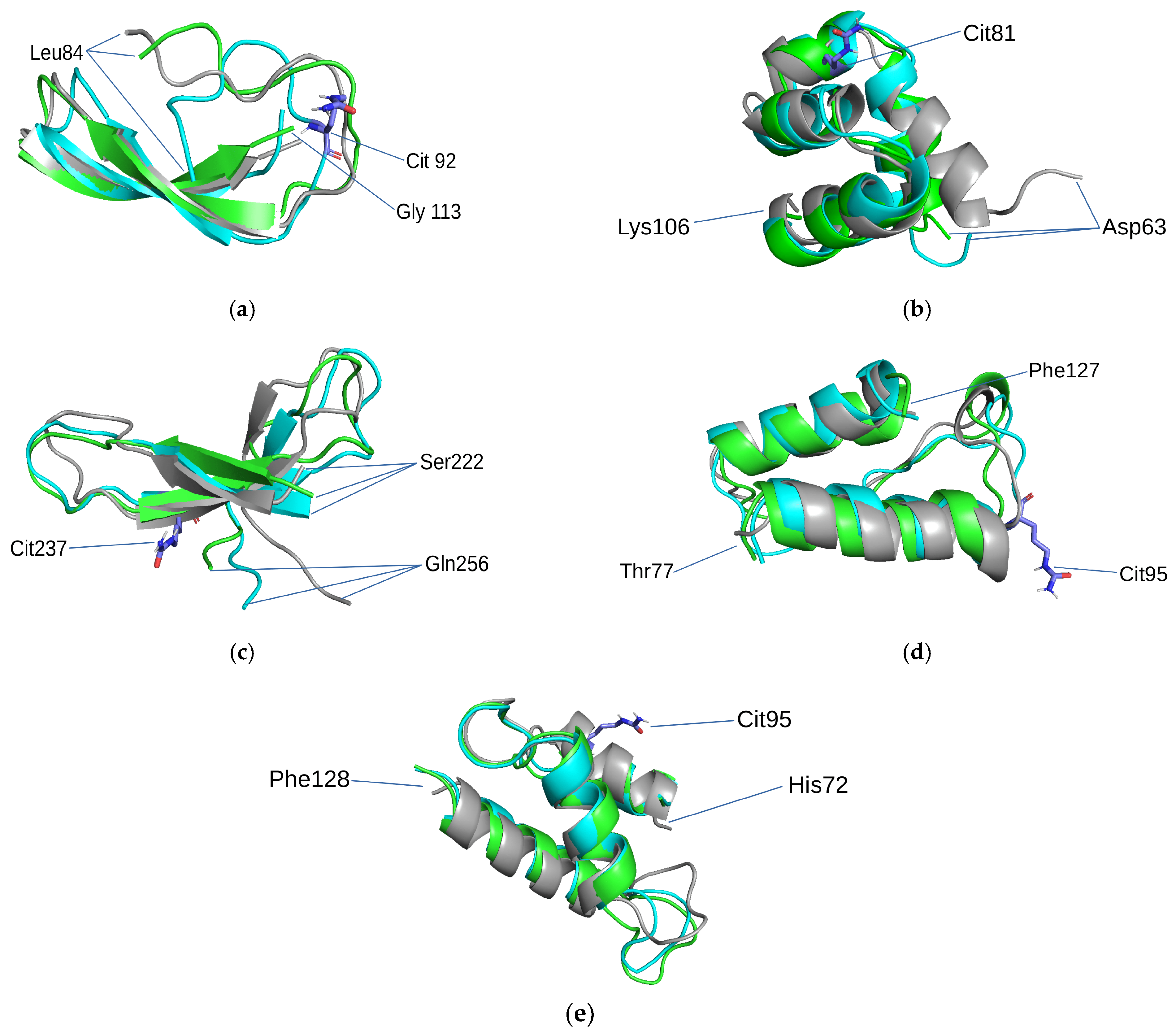
3.3. Results of MD Research
4. Discussion
4.1. Structural Motifs in Protein Structures—An Object of MD Research
4.2. The Role of Protein Citrullination in RA Development
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamada, R.; Suzuki, A.; Chang, X.; Yamamoto, K. Citrullinated Proteins in Rheumatoid Arthritis. Front. Biosci. J. Virtual Libr. 2005, 10, 54–64. [Google Scholar] [CrossRef]
- Jilani, A.A.; Mackworth-Young, C.G. The Role of Citrullinated Protein Antibodies in Predicting Erosive Disease in Rheumatoid Arthritis: A Systematic Literature Review and Meta-Analysis. Int. J. Rheumatol. 2015, 2015, e728610. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.L. Radiological Progression in Established Rheumatoid Arthritis. J. Rheumatol. Suppl. 2004, 69, 55–65. [Google Scholar] [PubMed]
- Trela, M.; Perera, S.; Sheeran, T.; Rylance, P.; Nelson, P.N.; Attridge, K. Citrullination Facilitates Cross-Reactivity of Rheumatoid Factor with Non-IgG1 Fc Epitopes in Rheumatoid Arthritis. Sci. Rep. 2019, 9, 12068. [Google Scholar] [CrossRef] [PubMed]
- Masson-Bessière, C.; Sebbag, M.; Girbal-Neuhauser, E.; Nogueira, L.; Vincent, C.; Senshu, T.; Serre, G. The Major Synovial Targets of the Rheumatoid Arthritis-Specific Antifilaggrin Autoantibodies Are Deiminated Forms of the Alpha- and Beta-Chains of Fibrin. J. Immunol. 2001, 166, 4177–4184. [Google Scholar] [CrossRef]
- Kinloch, A.; Tatzer, V.; Wait, R.; Peston, D.; Lundberg, K.; Donatien, P.; Moyes, D.; Taylor, P.C.; Venables, P.J. Identification of Citrullinated Alpha-Enolase as a Candidate Autoantigen in Rheumatoid Arthritis. Arthritis Res. Ther. 2005, 7, R1421–R1429. [Google Scholar] [CrossRef] [PubMed]
- Vossenaar, E.R.; Després, N.; Lapointe, E.; van der Heijden, A.; Lora, M.; Senshu, T.; van Venrooij, W.J.; Ménard, H.A. Rheumatoid Arthritis Specific Anti-Sa Antibodies Target Citrullinated Vimentin. Arthritis Res. Ther. 2004, 6, R142–R150. [Google Scholar] [CrossRef]
- Burkhardt, H.; Sehnert, B.; Bockermann, R.; Engström, A.; Kalden, J.R.; Holmdahl, R. Humoral Immune Response to Citrullinated Collagen Type II Determinants in Early Rheumatoid Arthritis. Eur. J. Immunol. 2005, 35, 1643–1652. [Google Scholar] [CrossRef]
- Rantapää-Dahlqvist, S.; de Jong, B.A.W.; Berglin, E.; Hallmans, G.; Wadell, G.; Stenlund, H.; Sundin, U.; van Venrooij, W.J. Antibodies against Cyclic Citrullinated Peptide and IgA Rheumatoid Factor Predict the Development of Rheumatoid Arthritis. Arthritis Rheum. 2003, 48, 2741–2749. [Google Scholar] [CrossRef]
- van der Helm-van Mil, A.H.M.; Verpoort, K.N.; Breedveld, F.C.; Toes, R.E.M.; Huizinga, T.W.J. Antibodies to Citrullinated Proteins and Differences in Clinical Progression of Rheumatoid Arthritis. Arthritis Res. Ther. 2005, 7, R949–R958. [Google Scholar] [CrossRef]
- Vossenaar, E.R.; Zendman, A.J.W.; van Venrooij, W.J.; Pruijn, G.J.M. PAD, a Growing Family of Citrullinating Enzymes: Genes, Features and Involvement in Disease. BioEssays 2003, 25, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Makrygiannakis, D.; af Klint, E.; Lundberg, I.E.; Lofberg, R.; Ulfgren, A.-K.; Klareskog, L.; Catrina, A.I. Citrullination Is an Inflammation-Dependent Process. Ann. Rheum. Dis. 2006, 65, 1219–1222. [Google Scholar] [CrossRef] [PubMed]
- Wegner, N.; Lundberg, K.; Kinloch, A.; Fisher, B.; Malmström, V.; Feldmann, M.; Venables, P.J. Autoimmunity to Specific Citrullinated Proteins Gives the First Clues to the Etiology of Rheumatoid Arthritis. Immunol. Rev. 2010, 233, 34–54. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.; Colasanti, T.; Barbati, C.; Pecani, A.; Sabatinelli, D.; Pendolino, M.; Truglia, S.; Massaro, L.; Mancini, R.; Miranda, F.; et al. The Role of Posttranslational Protein Modifications in Rheumatological Diseases: Focus on Rheumatoid Arthritis. J. Immunol. Res. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, B.; Wu, W.; Jin, Y. Comparison of Enzymatic Saccharification and Lignin Structure of Masson Pine and Poplar Pretreated by P-Toluenesulfonic Acid. Int. J. Biol. Macromol. 2020, 151, 861–869. [Google Scholar] [CrossRef]
- Bang, H.; Egerer, K.; Gauliard, A.; Lüthke, K.; Rudolph, P.E.; Fredenhagen, G.; Berg, W.; Feist, E.; Burmester, G.-R. Mutation and Citrullination Modifies Vimentin to a Novel Autoantigen for Rheumatoid Arthritis. Arthritis Rheum. 2007, 56, 2503–2511. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and Testing of the GROMOS Force-Field Versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843. [Google Scholar] [CrossRef]
- Petrov, D.; Margreitter, C.; Grandits, M.; Oostenbrink, C.; Zagrovic, B. A Systematic Framework for Molecular Dynamics Simulations of Protein Post-Translational Modifications. PLoS Comput. Biol. 2013, 9, e1003154. [Google Scholar] [CrossRef]
- Nandel, F.S.; Garla, R. Conformational Behavior of Stereo Regular Substituted Polyglycolides Is Side Chain Dependent. J. Biophys. Chem. 2011, 2, 285–299. [Google Scholar] [CrossRef][Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1998, 18, 1463–1472. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.; Mark, A.E.; Peggion, E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Drew, E.D.; Janes, R.W. 2StrucCompare: A Webserver for Visualizing Small but Noteworthy Differences between Protein Tertiary Structures through Interrogation of the Secondary Structure Content. Nucleic Acids Res. 2019, 47, W477–W481. [Google Scholar] [CrossRef]
- Heinig, M.; Frishman, D. STRIDE: A Web Server for Secondary Structure Assignment from Known Atomic Coordinates of Proteins. Nucleic Acids Res. 2004, 32 (Suppl. S2), W500–W502. [Google Scholar] [CrossRef]
- Bennike, T.; Lauridsen, K.B.; Olesen, M.K.; Andersen, V.; Birkelund, S.; Stensballe, A. Optimizing the Identification of Citrullinated Peptides by Mass Spectrometry: Utilizing the Inability of Trypsin to Cleave after Citrullinated Amino Acids. J. Proteom. Bioinform. 2013, 6. [Google Scholar] [CrossRef]
- De Ceuleneer, M.; De Wit, V.; Van Steendam, K.; Van Nieuwerburgh, F.; Tilleman, K.; Deforce, D. Modification of Citrulline Residues with 2,3-Butanedione Facilitates Their Detection by Liquid Chromatography/Mass Spectrometry. Rapid Commun. Mass Spectrom. RCM 2011, 25, 1536–1542. [Google Scholar] [CrossRef]
- Andrade, F.; Darrah, E.; Gucek, M.; Cole, R.N.; Rosen, A.; Zhu, X. Autocitrullination of Human Peptidyl Arginine Deiminase Type 4 Regulates Protein Citrullination during Cell Activation. Arthritis Rheum. 2010, 62, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Okeke, N.L.; Sharpe, O.; Batliwalla, F.M.; Lee, A.T.; Ho, P.P.; Tomooka, B.H.; Gregersen, P.K.; Robinson, W.H. Circulating Immune Complexes Contain Citrullinated Fibrinogen in Rheumatoid Arthritis. Arthritis Res. Ther. 2008, 10, R94. [Google Scholar] [CrossRef] [PubMed]
- van Beers, J.J.B.C.; Schwarte, C.M.; Stammen-Vogelzangs, J.; Oosterink, E.; Božič, B.; Pruijn, G.J.M. The Rheumatoid Arthritis Synovial Fluid Citrullinome Reveals Novel Citrullinated Epitopes in Apolipoprotein E, Myeloid Nuclear Differentiation Antigen, and β-Actin. Arthritis Rheum. 2013, 65, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between Stability of a Protein and Its Dipeptide Composition: A Novel Approach for Predicting in Vivo Stability of a Protein from Its Primary Sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef] [PubMed]
- López-Alemany, R.; Longstaff, C.; Hawley, S.; Mirshahi, M.; Fábregas, P.; Jardí, M.; Merton, E.; Miles, L.A.; Félez, J. Inhibition of Cell Surface Mediated Plasminogen Activation by a Monoclonal Antibody against Alpha-Enolase. Am. J. Hematol. 2003, 72, 234–242. [Google Scholar] [CrossRef]
- Sugahara, T.; Nakajima, H.; Shirahata, S.; Murakami, H. Purification and Characterization of Immunoglobulin Production Stimulating Factor-II Beta Derived from Namalwa Cells. Cytotechnology 1992, 10, 137–146. [Google Scholar] [CrossRef]
- Neeli, I.; Khan, S.N.; Radic, M. Histone Deimination as a Response to Inflammatory Stimuli in Neutrophils. J. Immunol. 2008, 180, 1895–1902. [Google Scholar] [CrossRef]
- Flick, M.J.; Du, X.; Degen, J.L. Fibrin(Ogen)-Alpha M Beta 2 Interactions Regulate Leukocyte Function and Innate Immunity in Vivo. Exp. Biol. Med. Maywood NJ 2004, 229, 1105–1110. [Google Scholar] [CrossRef]
- Smit, M.J.; Verdijk, P.; van der Raaij-Helmer, E.M.H.; Navis, M.; Hensbergen, P.J.; Leurs, R.; Tensen, C.P. CXCR3-Mediated Chemotaxis of Human T Cells Is Regulated by a Gi- and Phospholipase C-Dependent Pathway and Not via Activation of MEK/P44/P42 MAPK nor Akt/PI-3 Kinase. Blood 2003, 102, 1959–1965. [Google Scholar] [CrossRef]
- Gao, J.-M.; Xiang, R.-L.; Jiang, L.; Li, W.-H.; Feng, Q.-P.; Guo, Z.-J.; Sun, Q.; Zeng, Z.-P.; Fang, F. Sulfated Tyrosines 27 and 29 in the N-Terminus of Human CXCR3 Participate in Binding Native IP-10. Acta Pharmacol. Sin. 2009, 30, 193–201. [Google Scholar] [CrossRef]
- Nishikawa, O.; Yokoyama, S.; Okabe, H.; Yamamoto, A. Enhancement of Non-Polar Lipid Transfer Reaction through Stabilization of Substrate Lipid Particles with Apolipoproteins. J. Biochem. 1988, 103, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Rudnev, V.R.; Kulikova, L.I.; Kaysheva, A.L.; Efimov, A.V.; Tikhonov, D.A. Use of the Molecular Dynamics Method to Investigate the Stability of α-α-Corner Structural Motifs in Proteins. Symmetry 2021, 13, 1193. [Google Scholar] [CrossRef]
- Efimov, A.V. Structure of Coiled β-β-Hairpins and β-β-Corners. FEBS Lett. 1991, 284, 288–292. [Google Scholar] [CrossRef]
- Sternberg, M.J.; Thornton, J.M. On the Conformation of Proteins: The Handedness of the Beta-Strand-Alpha-Helix-Beta-Strand Unit. J. Mol. Biol. 1976, 105, 367–382. [Google Scholar] [CrossRef]
- Efimov, A.V. A Structural Tree for Proteins Containing 3beta-Corners. FEBS Lett. 1997, 407, 37–41. [Google Scholar] [CrossRef]
- Efimov, A.V. Standard Structures in Proteins. Prog. Biophys. Mol. Biol. 1993, 60, 201–239. [Google Scholar] [CrossRef]
- Tsai, F.C.; Sherman, J.C. Circular Dichroism Analysis of a Synthetic Peptide Corresponding to the Alpha, Alpha-Corner Motif of Hemoglobin. Biochem. Biophys. Res. Commun. 1993, 196, 435–439. [Google Scholar] [CrossRef]
- Rudnev, V.R. Recognition and Stability Analysis of Structural Motifs of α-α-Corner Type in Globular Proteins. Mat. Biol. Bioinformatika 2013, 8, 398–406. [Google Scholar] [CrossRef]
- Yang, M.-L.; Sodré, F.M.C.; Mamula, M.J.; Overbergh, L. Citrullination and PAD Enzyme Biology in Type 1 Diabetes–Regulators of Inflammation, Autoimmunity, and Pathology. Front. Immunol. 2021, 12, 678953. [Google Scholar] [CrossRef]
- van Venrooij, W.J.; Pruijn, G.J.M. Citrullination: A Small Change for a Protein with Great Consequences for Rheumatoid Arthritis. Arthritis Res. 2000, 2, 249–251. [Google Scholar] [CrossRef]
- Witalison, E.E.; Thompson, P.R.; Hofseth, L.J. Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Curr. Drug Targets 2015, 16, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Vasilopoulos, Y.; Gkretsi, V.; Armaka, M.; Aidinis, V.; Kollias, G. Actin Cytoskeleton Dynamics Linked to Synovial Fibroblast Activation as a Novel Pathogenic Principle in TNF-driven Arthritis. Ann. Rheum. Dis. 2007, 66 (Suppl. S3), iii23–iii28. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Kim, M.Y.; Kim, K.H.; Lee, I.H.; Shin, S.H.; Lee, J.S.; Kim, J.H. Synovial Fluid of Patients with Rheumatoid Arthritis Induces α-Smooth Muscle Actin in Human Adipose Tissue-Derived Mesenchymal Stem Cells through a TGF-Β1-Dependent Mechanism. Exp. Mol. Med. 2010, 42, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Won, P.; Kim, Y.; Jung, H.; Rim, Y.A.; Sohn, D.H.; Robinson, W.H.; Moon, S.-J.; Ju, J.H. Pathogenic Role of Circulating Citrullinated Antigens and Anti-Cyclic Monoclonal Citrullinated Peptide Antibodies in Rheumatoid Arthritis. Front. Immunol. 2021, 12, 2389. [Google Scholar] [CrossRef] [PubMed]
- Atmani, F.; Khan, S.R. Characterization of Uronic-Acid-Rich Inhibitor of Calcium Oxalate Crystallization Isolated from Rat Urine. Urol. Res. 1995, 23, 95–101. [Google Scholar] [CrossRef]
- Gobezie, R.; Kho, A.; Krastins, B.; Sarracino, D.A.; Thornhill, T.S.; Chase, M.; Millett, P.J.; Lee, D.M. High Abundance Synovial Fluid Proteome: Distinct Profiles in Health and Osteoarthritis. Arthritis Res. Ther. 2007, 9, R36. [Google Scholar] [CrossRef]
- Sancandi, M.; Uysal-Onganer, P.; Kraev, I.; Mercer, A.; Lange, S. Protein Deimination Signatures in Plasma and Plasma-EVs and Protein Deimination in the Brain Vasculature in a Rat Model of Pre-Motor Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 2743. [Google Scholar] [CrossRef]
- D’Alessio, S.; Thorgeirsdóttir, S.; Kraev, I.; Skírnisson, K.; Lange, S. Post-Translational Protein Deimination Signatures in Plasma and Plasma EVs of Reindeer (Rangifer Tarandus). Biology 2021, 10, 222. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Matsumoto, I.; Osada, A.; Kurata, I.; Ebe, H.; Tanaka, Y.; Inoue, A.; Umeda, N.; Kondo, Y.; Tsuboi, H.; et al. Identification of Novel Biomarker as Citrullinated Inter-Alpha-Trypsin Inhibitor Heavy Chain 4, Specifically Increased in Sera with Experimental and Rheumatoid Arthritis. Arthritis Res. Ther. 2018, 20, 66. [Google Scholar] [CrossRef]
- Blanco, F.; Ramírez-Alvarado, M.; Serrano, L. Formation and Stability of Beta-Hairpin Structures in Polypeptides. Curr. Opin. Struct. Biol. 1998, 8, 107–111. [Google Scholar] [CrossRef]
- Maynard, A.J.; Sharman, G.J.; Searle, M.S. Origin of β-Hairpin Stability in Solution: Structural and Thermodynamic Analysis of the Folding of a Model Peptide Supports Hydrophobic Stabilization in Water. J. Am. Chem. Soc. 1998, 120, 1996–2007. [Google Scholar] [CrossRef]
- Sivanesam, K.; Kier, B.L.; Whedon, S.D.; Chatterjee, C.; Andersen, N.H. Hairpin Structure Stability Plays a Role in the Activity of Two Antimicrobial Peptides. FEBS Lett. 2016, 590, 4480–4488. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Riley, B.T.; de Veer, S.J.; Hoke, D.E.; Van Haeften, J.; Leahy, D.; Swedberg, J.E.; Brattsand, M.; Hartfield, P.J.; Buckle, A.M.; et al. Potent, Multi-Target Serine Protease Inhibition Achieved by a Simplified β-Sheet Motif. PLoS ONE 2019, 14, e0210842. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, H.; Kobayashi, H.; Yagyu, T.; Wakahara, K.; Kondo, T.; Kurita, N.; Sekino, H.; Inagaki, K.; Suzuki, M.; Kanayama, N.; et al. Bikunin Inhibits Lipopolysaccharide-Induced Tumor Necrosis Factor Alpha Induction in Macrophages. Clin. Vaccine Immunol. 2004, 11, 1140–1147. [Google Scholar] [CrossRef]
- Matsuo, K.; Xiang, Y.; Nakamura, H.; Masuko, K.; Yudoh, K.; Noyori, K.; Nishioka, K.; Saito, T.; Kato, T. Identification of Novel Citrullinated Autoantigens of Synovium in Rheumatoid Arthritis Using a Proteomic Approach. Arthritis Res. Ther. 2006, 8, R175. [Google Scholar] [CrossRef]
- Harre, U.; Georgess, D.; Bang, H.; Bozec, A.; Axmann, R.; Ossipova, E.; Jakobsson, P.-J.; Baum, W.; Nimmerjahn, F.; Szarka, E.; et al. Induction of Osteoclastogenesis and Bone Loss by Human Autoantibodies against Citrullinated Vimentin. J. Clin. Investig. 2012, 122, 1791–1802. [Google Scholar] [CrossRef]
- Brentville, V.A.; Vankemmelbeke, M.; Metheringham, R.L.; Durrant, L.G. Post-Translational Modifications Such as Citrullination Are Excellent Targets for Cancer Therapy. Semin. Immunol. 2020, 47, 101393. [Google Scholar] [CrossRef]
- Rudnev, V.R.; Kulikova, L.I.; Nikolsky, K.S.; Malsagova, K.A.; Kopylov, A.T.; Kaysheva, A.L. Current Approaches in Supersecondary Structures Investigation. Int. J. Mol. Sci. 2021, 22, 11879. [Google Scholar] [CrossRef]
- von der Ecken, J.; Heissler, S.M.; Pathan-Chhatbar, S.; Manstein, D.J.; Raunser, S. Cryo-EM Structure of a Human Cytoplasmic Actomyosin Complex at near-Atomic Resolution. Nature 2016, 534, 724–728. [Google Scholar] [CrossRef]
- Baek, K.; Liu, X.; Ferron, F.; Shu, S.; Korn, E.D.; Dominguez, R. Modulation of Actin Structure and Function by Phosphorylation of Tyr-53 and Profilin Binding. Proc. Natl. Acad. Sci. USA 2008, 105, 11748–11753. [Google Scholar] [CrossRef]
- Risi, C.; Belknap, B.; Forgacs-Lonart, E.; Harris, S.P.; Schröder, G.F.; White, H.D.; Galkin, V.E. N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament. Structure 2018, 26, 1604–1611.e4. [Google Scholar] [CrossRef] [PubMed]
- Yogeswaran, A.; Troidl, C.; McNamara, J.W.; Wilhelm, J.; Truschel, T.; Widmann, L.; Aslam, M.; Hamm, C.W.; Sadayappan, S.; Lipps, C. The C0-C1f Region of Cardiac Myosin Binding Protein-C Induces Pro-Inflammatory Responses in Fibroblasts via TLR4 Signaling. Cells 2021, 10, 1326. [Google Scholar] [CrossRef] [PubMed]
- Wunder, A.; Müller-Ladner, U.; Stelzer, E.H.K.; Funk, J.; Neumann, E.; Stehle, G.; Pap, T.; Sinn, H.; Gay, S.; Fiehn, C. Albumin-Based Drug Delivery as Novel Therapeutic Approach for Rheumatoid Arthritis. J. Immunol. 2003, 170, 4793–4801. [Google Scholar] [CrossRef] [PubMed]
- Sebbag, M.; Moinard, N.; Auger, I.; Clavel, C.; Arnaud, J.; Nogueira, L.; Roudier, J.; Serre, G. Epitopes of Human Fibrin Recognized by the Rheumatoid Arthritis-Specific Autoantibodies to Citrullinated Proteins. Eur. J. Immunol. 2006, 36, 2250–2263. [Google Scholar] [CrossRef]
- Snir, O.; Widhe, M.; Hermansson, M.; von Spee, C.; Lindberg, J.; Hensen, S.; Lundberg, K.; Engström, A.; Venables, P.J.W.; Toes, R.E.M.; et al. Antibodies to Several Citrullinated Antigens Are Enriched in the Joints of Rheumatoid Arthritis Patients. Arthritis Rheum. 2010, 62, 44–52. [Google Scholar] [CrossRef]
- Joshua, V.; Schobers, L.; Titcombe, P.J.; Israelsson, L.; Rönnelid, J.; Hansson, M.; Catrina, A.I.; Pruijn, G.J.M.; Malmström, V. Antibody Responses to de Novo Identified Citrullinated Fibrinogen Peptides in Rheumatoid Arthritis and Visualization of the Corresponding B Cells. Arthritis Res. Ther. 2016, 18, 284. [Google Scholar] [CrossRef]
- Hill, J.A.; Bell, D.A.; Brintnell, W.; Yue, D.; Wehrli, B.; Jevnikar, A.M.; Lee, D.M.; Hueber, W.; Robinson, W.H.; Cairns, E. Arthritis Induced by Posttranslationally Modified (Citrullinated) Fibrinogen in DR4-IE Transgenic Mice. J. Exp. Med. 2008, 205, 967–979. [Google Scholar] [CrossRef]
- Ho, P.P.; Lee, L.Y.; Zhao, X.; Tomooka, B.H.; Paniagua, R.T.; Sharpe, O.; BenBarak, M.J.; Chandra, P.E.; Hueber, W.; Steinman, L.; et al. Autoimmunity against Fibrinogen Mediates Inflammatory Arthritis in Mice. J. Immunol. 2010, 184, 379–390. [Google Scholar] [CrossRef]
- Herman, R.A.; Veng-Pedersen, P.; Hoffman, J.; Koehnke, R.; Furst, D.E. Pharmacokinetics of Low-Dose Methotrexate in Rheumatoid Arthritis Patients. J. Pharm. Sci. 1989, 78, 165–171. [Google Scholar] [CrossRef]
- Bannwarth, B.; Péhourcq, F.; Schaeverbeke, T.; Dehais, J. Clinical Pharmacokinetics of Low-Dose Pulse Methotrexate in Rheumatoid Arthritis. Clin. Pharmacokinet. 1996, 30, 194–210. [Google Scholar] [CrossRef]
- Hefton, A.; Liang, S.Y.; Ni, K.; Carter, V.; Ukadike, K.; Lood, C.; Mustelin, T. Autoantibodies against Citrullinated Serum Albumin in Patients with Rheumatoid Arthritis. J. Transl. Autoimmun. 2019, 2, 100023. [Google Scholar] [CrossRef] [PubMed]
- Tutturen, A.E.V.; Fleckenstein, B.; de Souza, G.A. Assessing the Citrullinome in Rheumatoid Arthritis Synovial Fluid with and without Enrichment of Citrullinated Peptides. J. Proteome Res. 2014, 13, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Rethi, B.; Krishnamurthy, A.; Joshua, V.; Circiumaru, A.; Hensvold, A.H.; Ossipova, E.; Grönwall, C.; Liu, Y.; Engstrom, M.; et al. Anticitrullinated Protein Antibodies Facilitate Migration of Synovial Tissue-Derived Fibroblasts. Ann. Rheum. Dis. 2019, 78, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Titcombe, P.J.; Wigerblad, G.; Sippl, N.; Zhang, N.; Shmagel, A.K.; Sahlström, P.; Zhang, Y.; Barsness, L.O.; Ghodke-Puranik, Y.; Baharpoor, A.; et al. Pathogenic Citrulline-Multispecific B Cell Receptor Clades in Rheumatoid Arthritis. Arthritis Rheumatol. 2018, 70, 1933–1945. [Google Scholar] [CrossRef]
- Steen, J.; Forsström, B.; Sahlström, P.; Odowd, V.; Israelsson, L.; Krishnamurthy, A.; Badreh, S.; Mathsson Alm, L.; Compson, J.; Ramsköld, D.; et al. Recognition of Amino Acid Motifs, Rather Than Specific Proteins, by Human Plasma Cell–Derived Monoclonal Antibodies to Posttranslationally Modified Proteins in Rheumatoid Arthritis. Arthritis Rheumatol. 2019, 71, 196–209. [Google Scholar] [CrossRef]
- Bos, W.H.; Wolbink, G.J.; Boers, M.; Tijhuis, G.J.; de Vries, N.; van der Horst-Bruinsma, I.E.; Tak, P.P.; van de Stadt, R.J.; van der Laken, C.J.; Dijkmans, B.A.C.; et al. Arthritis Development in Patients with Arthralgia Is Strongly Associated with Anti-Citrullinated Protein Antibody Status: A Prospective Cohort Study. Ann. Rheum. Dis. 2010, 69, 490–494. [Google Scholar] [CrossRef]
- Kleyer, A.; Finzel, S.; Rech, J.; Manger, B.; Krieter, M.; Faustini, F.; Araujo, E.; Hueber, A.J.; Harre, U.; Engelke, K.; et al. Bone Loss before the Clinical Onset of Rheumatoid Arthritis in Subjects with Anticitrullinated Protein Antibodies. Ann. Rheum. Dis. 2014, 73, 854–860. [Google Scholar] [CrossRef]
- Wigerblad, G.; Bas, D.B.; Fernades-Cerqueira, C.; Krishnamurthy, A.; Nandakumar, K.S.; Rogoz, K.; Kato, J.; Sandor, K.; Su, J.; Jimenez–Andrade, J.M.; et al. Autoantibodies to Citrullinated Proteins May Induce Joint Pain Independent of Inflammation. Ann. Rheum. Dis. 2016, 75, 730–738. [Google Scholar] [CrossRef]
- Trier, N.; Houen, G. Epitope Specificity of Anti-Citrullinated Protein Antibodies. Antibodies 2017, 6, 5. [Google Scholar] [CrossRef]
- Alghamdi, M.; Alasmari, D.; Assiri, A.; Mattar, E.; Aljaddawi, A.A.; Alattas, S.G.; Redwan, E.M. An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [Google Scholar] [CrossRef]
- Tesija Kuna, A.; Zirovic, M. Antibodies to Citrullinated Proteins/Peptides in Rheumatoid Arthritis: What Have We Learned so Far? Biochem. Med. 2008, 18, 275–290. [Google Scholar] [CrossRef]

| Parameter | Cut-Offvalue, nm | SASA of Arg (Lys)/Cit, nm2 | SASA of PTM Site, nm2 | Major Conformationlifetime (%) | Secondarystructureassignment (STRIDE) | Distance between COM of Ist and Last Element, nm | Pairwise RMSD (PDB vs. PDB-MD), nm | Pairwise RMSD MD of (PDB vs. PDB-PTM-MD), nm |
|---|---|---|---|---|---|---|---|---|
| Protein AMBP (PDB ID 1BIK) | ||||||||
| PDB | – | 3.623 | 10.505 | – | CCCCCCCCCCCEEEEEEETTTTEEEEEEEC | 0.197 | 0.152 | 0.352 |
| PDB-MD | 0.27 | 3.647± 0.140 | 10.358± 0.425 | 68 | CCCCCCCTTTTEEEEEETTTTTCEEEEEEC | 0.326± 0.044 | ||
| PDB-PTM-MD | 3.531± 0.130 | 10.538± 0.467 | 41 | CTTTCCCCCEEEEEEEETTTTTCEEEEEEC | 0.289± 0.043 | |||
| Albumin(PDB ID 1N5U) | ||||||||
| PDB | – | 3.741 | 10.669 | – | CCCHHHHHHHHHHHCCHHHHHHGGGGGGGGCCHHHHHHHHHHCC | 1.029 | 0.403 | 0.221 |
| PDB-MD | 0.25 | 3.639± 0.139 | 10.348± 0.420 | 85 | CCCHHHHHHTTTTTTHHHHHHCHHHHHHHCCCHHHHHHHHHHHC | 0.969± 0.058 | ||
| PDB-PTM-MD | 3.544± 0.131 | 10.292± 0.403 | 70 | CCCHHHHHHHHHHTTHHHHHHCHHHHHHHHCCHHHHHHHHHHHC | 1.007± 0.062 | |||
| Fibrinogen beta chain (PDB ID 3E1I) | ||||||||
| PDB | – | 3.611 | 12.713 | – | CEEEEETTTTTTCCEEEEEETTTTTTCEECCCCCC | 1.207 | 0.416 | 0.187 |
| PDB-MD | 0.3 | 3.645± 0.133 | 13.426± 0.386 | 74 | CCEEEECCTTTTCCEEEEEETTTTTTTEEEEEEEC | 0.932± 0.062 | ||
| PDB-PTM-MD | 0.3 | 3.538± 0.128 | 13.349± 0.360 | 79 | CCEEEECCTTTTCCEEEEECCCTTTTTTEECCCCC | 0.882± 0.065 | ||
| Actin, cytoplasmic 1 (PDB ID 6ANU) | ||||||||
| PDB | – | 3.472 | 10.080 | – | CCHHHHHHHHHHHHHHHCCCCGGGCCTTTCCTTTTCHHHHHHHHHHHHHHC | 1.344 | 0.257 | 0.223 |
| PDB-MD | 0.27 | 3.645± 0.142 | 11.014± 0.374 | 68 | CCHHHHHHHHHHHHTTTTTCTTTTTCTTTTTTTTTCHHHHHHHHHHHHCCC | 1.061± 0.111 | ||
| PDB-PTM-MD | 0.27 | 3.563± 0.130 | 11.065± 0.220 | 60 | CCHHHHHHHHHHHHHHHCCCTTTTTTCTTTTTCCCCHHHHHHHHHHHHHHC | 1.202± 0.118 | ||
| Tubulin polymerization-promoting protein (AF-H9GXR8-F1) | ||||||||
| PDB | – | 3.272 | 10.347 | – | CHHHHHHHHHHHTTTTTTTTTHHHHHHHHHHHCTTTTCCCCHHHHHHHHHHHHHHHC | 1.300 | 0.294 | 0.104 |
| PDB-MD | 0.23 | 3.255± 0.122 | 10.937± 0.461 | 70 | CCHHHHHHHHHTTTCTTTTTTHHHHHHHHHTTTTTTTTTTCHHHHHHHHHHHHHHCC | 1.214± 0.074 | ||
| PDB-PTM-MD | 0.23 | 3.546± 0.132 | 11.028± 0.499 | 80 | CCHHHHHHHHHTTTCTTTTTTHHHHHHHHHHHTBTTBTTTCHHHHHHHHHHHHHHCC | 1.221± 0.068 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taldaev, A.; Rudnev, V.; Kulikova, L.; Nikolsky, K.; Efimov, A.; Malsagova, K.; Kaysheva, A. Molecular Dynamics Study of Citrullinated Proteins Associated with the Development of Rheumatoid Arthritis. Proteomes 2022, 10, 8. https://doi.org/10.3390/proteomes10010008
Taldaev A, Rudnev V, Kulikova L, Nikolsky K, Efimov A, Malsagova K, Kaysheva A. Molecular Dynamics Study of Citrullinated Proteins Associated with the Development of Rheumatoid Arthritis. Proteomes. 2022; 10(1):8. https://doi.org/10.3390/proteomes10010008
Chicago/Turabian StyleTaldaev, Amir, Vladimir Rudnev, Liudmila Kulikova, Kirill Nikolsky, Alexander Efimov, Kristina Malsagova, and Anna Kaysheva. 2022. "Molecular Dynamics Study of Citrullinated Proteins Associated with the Development of Rheumatoid Arthritis" Proteomes 10, no. 1: 8. https://doi.org/10.3390/proteomes10010008
APA StyleTaldaev, A., Rudnev, V., Kulikova, L., Nikolsky, K., Efimov, A., Malsagova, K., & Kaysheva, A. (2022). Molecular Dynamics Study of Citrullinated Proteins Associated with the Development of Rheumatoid Arthritis. Proteomes, 10(1), 8. https://doi.org/10.3390/proteomes10010008







