Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. PBMC Preparation
2.3. Polyclonal Stimulation of PBMC
2.4. Subcellular Fractionation of PBMC
2.5. Sample Digestion for Differential Proteome Analysis
2.6. Mass Spectrometric Analysis and Label-Free Quantification
2.7. Western Blots
2.8. Data Analysis
2.9. Data Availability
3. Results
3.1. The Bovine Peripheral Blood Derived Lymphocyte Proteome Consists of 2447 Proteins
3.2. The Proteome Shows Significant Differential Expression between Cow Immune Phenotypes after Polyclonal Immune Stimulation with Pokeweed Mitogen
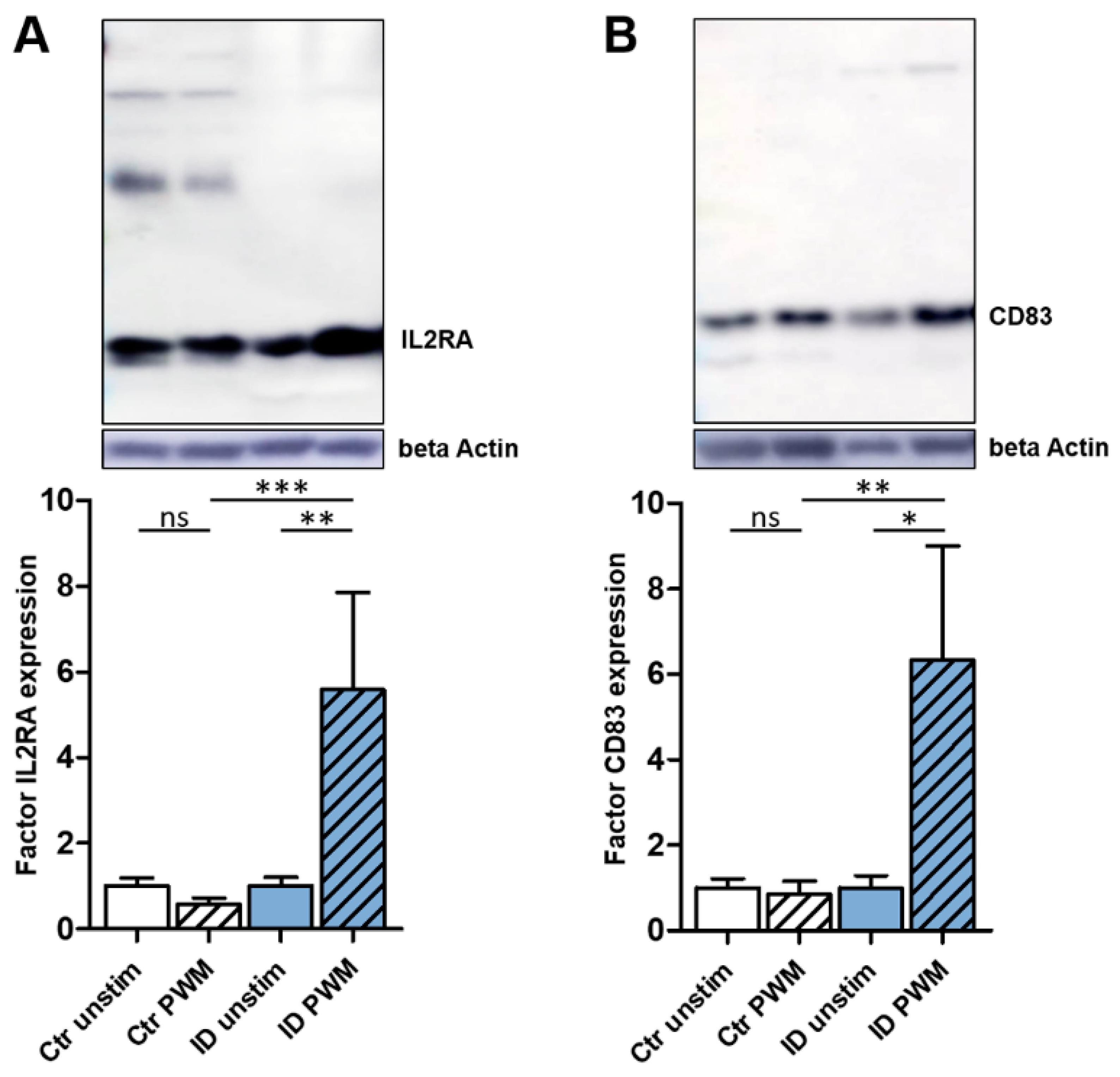
3.3. PWM Stimulation Leads to a Significantly Higher Expression of IL2RA and CD83 in ID Lymphocytes
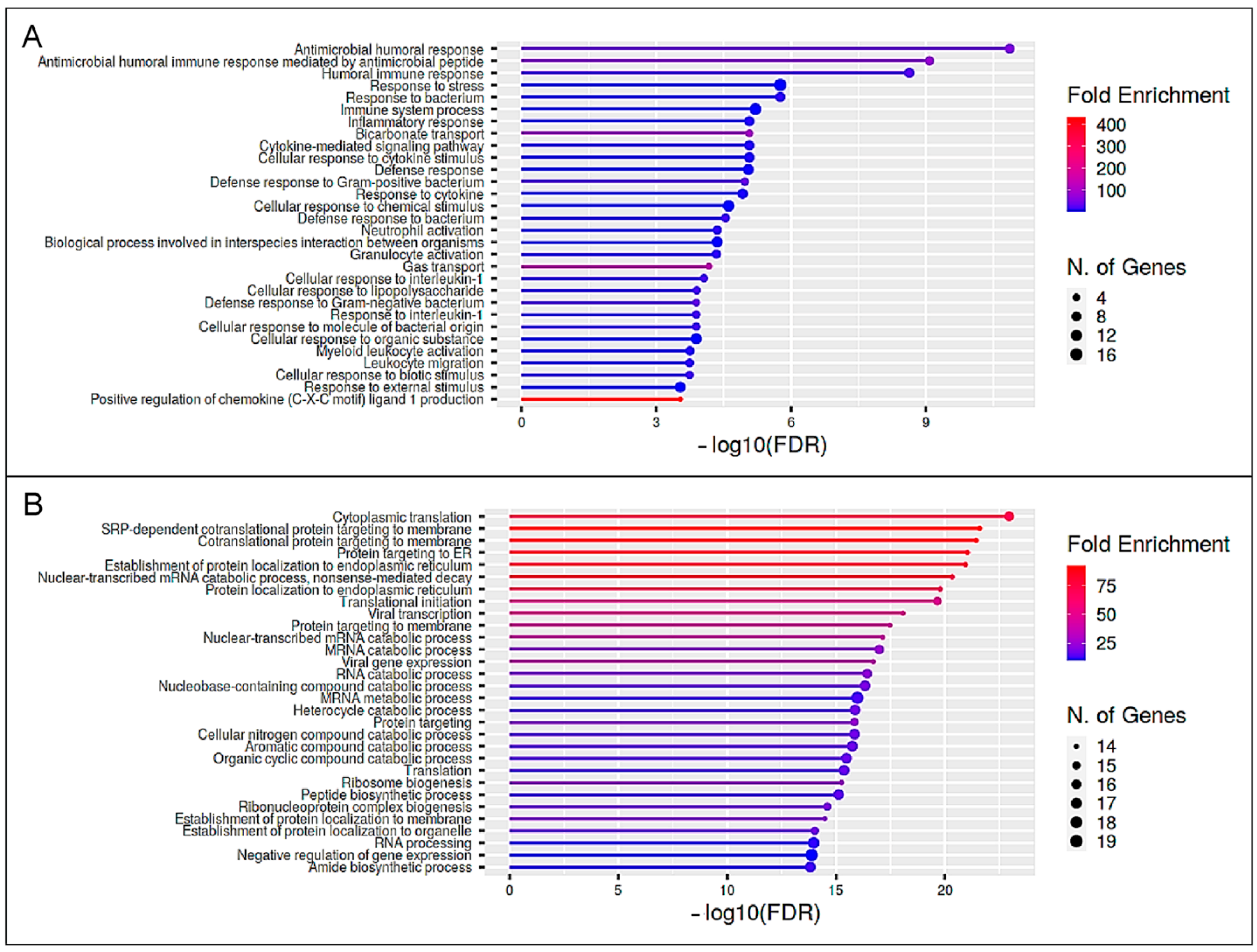
3.4. The Enriched Pathway Profile Reveals Functional Differences between the Lymphocytes of Bovine Immune Phenotypes after Immune Stimulation
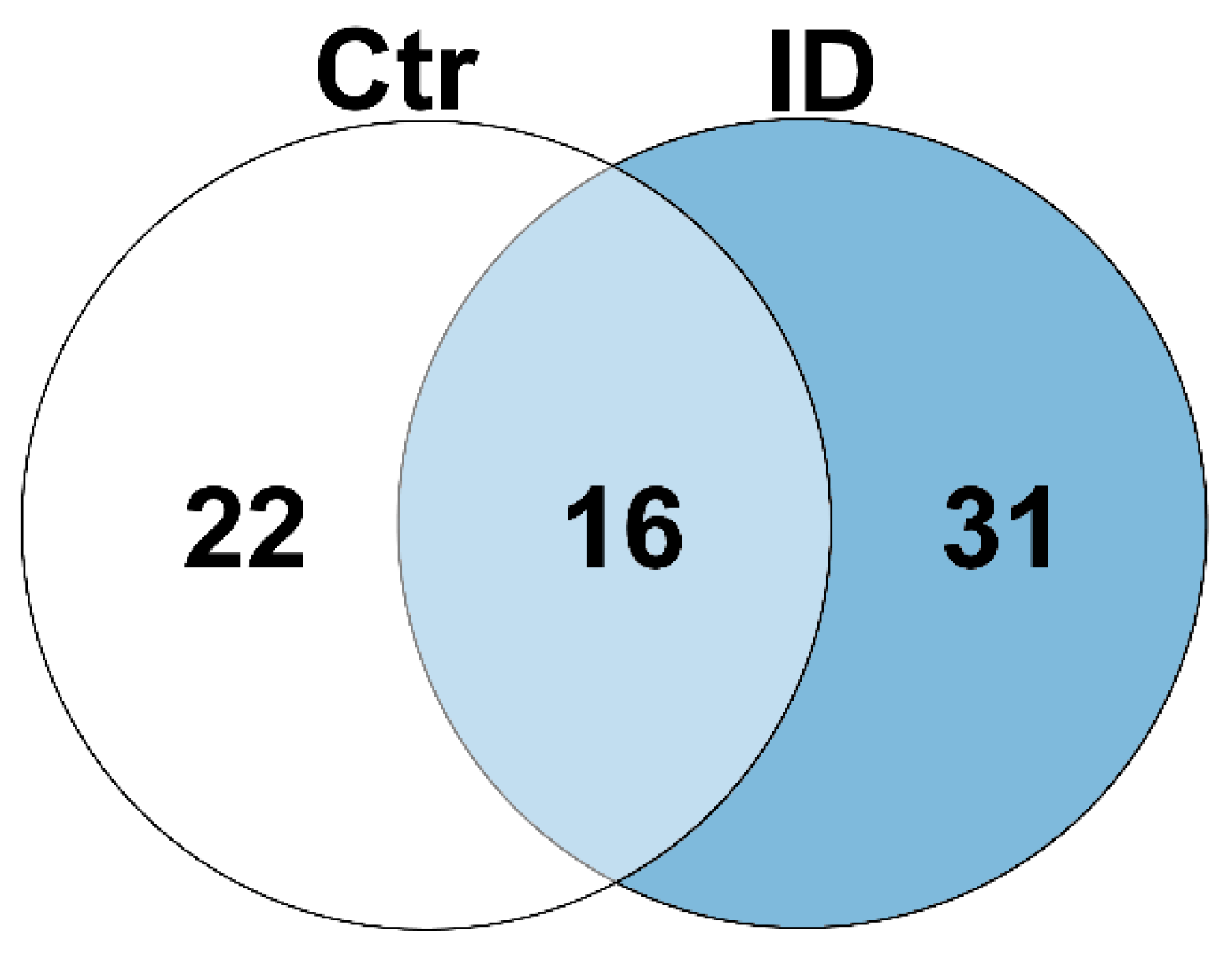
3.5. The Bovine Peripheral Blood Lymphocyte Secretome Is Composed of 1204 Proteins with a Changed Cytokine Profile between Cow Immune Phenotypes
3.6. The Enriched Pathway Profile Indicates a Functional Impact of Differentially Secreted Proteins of Bovine Immune Phenotypes after Immune Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bastian, M.; Holsteg, M.; Hanke-Robinson, H.; Duchow, K.; Cussler, K. Bovine Neonatal Pancytopenia: Is this alloimmune syndrome caused by vaccine-induced alloreactive antibodies? Vaccine 2011, 29, 5267–5275. [Google Scholar] [CrossRef]
- Sauter-Louis, C.; Carlin, A.; Friedrich, A.; Assad, A.; Reichmann, F.; Rademacher, G.; Heuer, C.; Klee, W. Case control study to investigate risk factors for bovine neonatal pancytopenia (BNP) in young calves in southern Germany. Prev. Vet. Med. 2012, 105, 49–58. [Google Scholar] [CrossRef]
- Reichmann, F.; Pfitzner, A.; Rademacher, G.; Schwedinger, E.; Cussler, K.; Sauter-Louis, C.M. Incidence of bovine neonatal pancytopenia in 243 farms in Germany. BMC Vet. Res. 2016, 12, 220. [Google Scholar] [CrossRef]
- Penny, C.D.; Bell, C.; Morrison, L.; Howie, F.; Willoughby, K. Pancytopenia and haemorrhage in young beef calves. Vet. Rec. 2009, 164, 762. [Google Scholar] [CrossRef]
- Lutterberg, K.; Kleinwort, K.J.H.; Hobmaier, B.F.; Hauck, S.M.; Nuske, S.; Scholz, A.M.; Deeg, C.A. A Functionally Different Immune Phenotype in Cattle Is Associated With Higher Mastitis Incidence. Front. Immunol. 2018, 9, 2884. [Google Scholar] [CrossRef]
- Mellstedt, H. In vitro activation of human T and B lymphocytes by Pokeweed mitogen. Clin. Exp. Immunol. 1975, 19, 75–82. [Google Scholar]
- Friedrich, A.; Buttner, M.; Rademacher, G.; Klee, W.; Weber, B.K.; Muller, M.; Carlin, A.; Assad, A.; Hafner-Marx, A.; Sauter-Louis, C.M. Ingestion of colostrum from specific cows induces Bovine Neonatal Pancytopenia (BNP) in some calves. BMC Vet. Res. 2011, 7, 10. [Google Scholar] [CrossRef]
- Jones, B.A.; Sauter-Louis, C.; Henning, J.; Stoll, A.; Nielen, M.; Van Schaik, G.; Smolenaars, A.; Schouten, M.; den Uijl, I.; Fourichon, C.; et al. Calf-level factors associated with bovine neonatal pancytopenia--a multi-country case-control study. PLoS ONE 2013, 8, e80619. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The Proteome of Native Adult Muller Glial Cells From Murine Retina. Mol. Cell Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zielinska, D.F.; Mann, M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 2011, 410, 307–309. [Google Scholar] [CrossRef]
- Kleinwort, K.J.H.; Hauck, S.M.; Degroote, R.L.; Scholz, A.M.; Holzel, C.; Maertlbauer, E.; Deeg, C. Peripheral blood bovine lymphocytes and MAP show distinctly different proteome changes and immune pathways in host-pathogen interaction. PeerJ 2019, 7, e8130. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Kalina, T.; Fiser, K.; Perez-Andres, M.; Kuzilkova, D.; Cuenca, M.; Bartol, S.J.W.; Blanco, E.; Engel, P.; van Zelm, M.C. CD Maps-Dynamic Profiling of CD1-CD100 Surface Expression on Human Leukocyte and Lymphocyte Subsets. Front. Immunol. 2019, 10, 2434. [Google Scholar] [CrossRef]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the Treatment of Cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef]
- Song, W.; Li, D.; Tao, L.; Luo, Q.; Chen, L. Solute carrier transporters: The metabolic gatekeepers of immune cells. Acta Pharm. Sin. B 2020, 10, 61–78. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, Z.; Bai, C.; Sun, H.; Wang, X. Proteomic profiling of lymphocytes in autoimmunity, inflammation and cancer. J. Transl. Med. 2014, 12, 6. [Google Scholar] [CrossRef]
- Stevenson, H.C.; Miller, P.J.; Waxdal, M.J.; Haynes, B.F.; Thomas, C.A.; Fauci, A.S. Interaction of Pokeweed mitogen with monocytes in the activation of human lymphocytes. Immunology 1983, 49, 633–640. [Google Scholar]
- Wallays, G.; Ceuppens, J.L. Human T lymphocyte activation by Pokeweed mitogen induces production of TNF-alpha and GM-CSF and helper signaling by IL-1 and IL-6 results in IL-2-dependent T cell growth. Eur. Cytokine Netw. 1993, 4, 269–277. [Google Scholar]
- Xian, H.; Yang, S.; Jin, S.; Zhang, Y.; Cui, J. LRRC59 modulates type I interferon signaling by restraining the SQSTM1/p62-mediated autophagic degradation of pattern recognition receptor DDX58/RIG-I. Autophagy 2020, 16, 408–418. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 2019, 20, 1445. [Google Scholar] [CrossRef]
- Bhattacharyya, M.; Madden, P.; Henning, N.; Gregory, S.; Aid, M.; Martinot, A.J.; Barouch, D.H.; Penaloza-MacMaster, P. Regulation of CD4 T cells and their effects on immunopathological inflammation following viral infection. Immunology 2017, 152, 328–343. [Google Scholar] [CrossRef]
- He, G.; Ma, Y.; Chou, S.Y.; Li, H.; Yang, C.; Chuang, J.Z.; Sung, C.H.; Ding, A. Role of CLIC4 in the host innate responses to bacterial lipopolysaccharide. Eur. J. Immunol. 2011, 41, 1221–1230. [Google Scholar] [CrossRef]
- Catalfamo, M.; Karpova, T.; McNally, J.; Costes, S.V.; Lockett, S.J.; Bos, E.; Peters, P.J.; Henkart, P.A. Human CD8+ T cells store RANTES in a unique secretory compartment and release it rapidly after TcR stimulation. Immunity 2004, 20, 219–230. [Google Scholar] [CrossRef]
- Schall, T.J.; Jongstra, J.; Dyer, B.J.; Jorgensen, J.; Clayberger, C.; Davis, M.M.; Krensky, A.M. A human T cell-specific molecule is a member of a new gene family. J. Immunol. 1988, 141, 1018–1025. [Google Scholar]
- French, S.W.; Mendoza, A.S.; Afifiyan, N.; Tillman, B.; Vitocruz, E.; French, B.A. The role of the IL-8 signaling pathway in the infiltration of granulocytes into the livers of patients with alcoholic hepatitis. Exp. Mol. Pathol. 2017, 103, 137–140. [Google Scholar] [CrossRef]
- Harrington, L.E.; Hatton, R.D.; Mangan, P.R.; Turner, H.; Murphy, T.L.; Murphy, K.M.; Weaver, C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005, 6, 1123–1132. [Google Scholar] [CrossRef]
- Kondo, T.; Takata, H.; Matsuki, F.; Takiguchi, M. Cutting edge: Phenotypic characterization and differentiation of human CD8+ T cells producing IL-17. J. Immunol. 2009, 182, 1794–1798. [Google Scholar] [CrossRef]
- Valeri, M.; Raffatellu, M. Cytokines IL-17 and IL-22 in the host response to infection. Pathog. Dis. 2016, 74, ftw111. [Google Scholar] [CrossRef]
- Rachitskaya, A.V.; Hansen, A.M.; Horai, R.; Li, Z.; Villasmil, R.; Luger, D.; Nussenblatt, R.B.; Caspi, R.R. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 2008, 180, 5167–5171. [Google Scholar] [CrossRef]
- Mangan, P.R.; Harrington, L.E.; O’Quinn, D.B.; Helms, W.S.; Bullard, D.C.; Elson, C.O.; Hatton, R.D.; Wahl, S.M.; Schoeb, T.R.; Weaver, C.T. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 2006, 441, 231–234. [Google Scholar] [CrossRef]
- Happel, K.I.; Dubin, P.J.; Zheng, M.; Ghilardi, N.; Lockhart, C.; Quinton, L.J.; Odden, A.R.; Shellito, J.E.; Bagby, G.J.; Nelson, S.; et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 2005, 202, 761–769. [Google Scholar] [CrossRef]
- Raffatellu, M.; Santos, R.L.; Verhoeven, D.E.; George, M.D.; Wilson, R.P.; Winter, S.E.; Godinez, I.; Sankaran, S.; Paixao, T.A.; Gordon, M.A.; et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 2008, 14, 421–428. [Google Scholar] [CrossRef]
- Paidipally, P.; Periasamy, S.; Barnes, P.F.; Dhiman, R.; Indramohan, M.; Griffith, D.E.; Cosman, D.; Vankayalapati, R. NKG2D-dependent IL-17 production by human T cells in response to an intracellular pathogen. J. Immunol. 2009, 183, 1940–1945. [Google Scholar] [CrossRef]
- Khader, S.A.; Bell, G.K.; Pearl, J.E.; Fountain, J.J.; Rangel-Moreno, J.; Cilley, G.E.; Shen, F.; Eaton, S.M.; Gaffen, S.L.; Swain, S.L.; et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 2007, 8, 369–377. [Google Scholar] [CrossRef]
- Bai, H.; Cheng, J.; Gao, X.; Joyee, A.G.; Fan, Y.; Wang, S.; Jiao, L.; Yao, Z.; Yang, X. IL-17/Th17 promotes type 1 T cell immunity against pulmonary intracellular bacterial infection through modulating dendritic cell function. J. Immunol. 2009, 183, 5886–5895. [Google Scholar] [CrossRef]
- Orgun, N.N.; Mathis, M.A.; Wilson, C.B.; Way, S.S. Deviation from a strong Th1-dominated to a modest Th17-dominated CD4 T cell response in the absence of IL-12p40 and type I IFNs sustains protective CD8 T cells. J. Immunol. 2008, 180, 4109–4115. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.J.; Zhang, C.; Chen, R.; Li, L.; He, J.; Xie, Y.; Chen, Y. Loss of neuronal CD200 contributed to microglial activation after acute cerebral ischemia in mice. Neurosci. Lett. 2018, 678, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Barclay, A.N.; Wright, G.J.; Brooke, G.; Brown, M.H. CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol. 2002, 23, 285–290. [Google Scholar] [CrossRef]
- Gorczynski, R.M. CD200 and its receptors as targets for immunoregulation. Curr. Opin. Investig. Drugs 2005, 6, 483–488. [Google Scholar] [PubMed]
- Hoek, R.M.; Ruuls, S.R.; Murphy, C.A.; Wright, G.J.; Goddard, R.; Zurawski, S.M.; Blom, B.; Homola, M.E.; Streit, W.J.; Brown, M.H.; et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science 2000, 290, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Foster-Cuevas, M.; Wright, G.J.; Puklavec, M.J.; Brown, M.H.; Barclay, A.N. Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor. J. Virol. 2004, 78, 7667–7676. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Azmy, E.; El-Gilany, A.H. Upregulation of CD200 is associated with regulatory T cell expansion and disease progression in multiple myeloma. Hematol. Oncol. 2017, 35, 51–57. [Google Scholar] [CrossRef]
- Elshal, M.F.; Aldahlawi, A.M.; Saadah, O.I.; McCoy, J.P. Reduced Dendritic Cells Expressing CD200R1 in Children with Inflammatory Bowel Disease: Correlation with Th17 and Regulatory T Cells. Int. J. Mol. Sci. 2015, 16, 28998–29010. [Google Scholar] [CrossRef]
- Zhu, B.; Yu, Y.; Liu, X.; Han, Q.; Kang, Y.; Shi, L. CD200 Modulates S. aureus-Induced Innate Immune Responses Through Suppressing p38 Signaling. Int. J. Mol. Sci. 2019, 20, 659. [Google Scholar] [CrossRef]
- Grosche, L.; Knippertz, I.; Konig, C.; Royzman, D.; Wild, A.B.; Zinser, E.; Sticht, H.; Muller, Y.A.; Steinkasserer, A.; Lechmann, M. The CD83 Molecule—An Important Immune Checkpoint. Front. Immunol. 2020, 11, 721. [Google Scholar] [CrossRef]
- Wild, A.B.; Krzyzak, L.; Peckert, K.; Stich, L.; Kuhnt, C.; Butterhof, A.; Seitz, C.; Mattner, J.; Gruner, N.; Gansbauer, M.; et al. CD83 orchestrates immunity toward self and non-self in dendritic cells. JCI Insight 2019, 4, e126246. [Google Scholar] [CrossRef]
- Doebbeler, M.; Koenig, C.; Krzyzak, L.; Seitz, C.; Wild, A.; Ulas, T.; Bassler, K.; Kopelyanskiy, D.; Butterhof, A.; Kuhnt, C.; et al. CD83 expression is essential for Treg cell differentiation and stability. JCI Insight 2018, 3, e99712. [Google Scholar] [CrossRef] [PubMed]
- Boyman, O.; Sprent, J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012, 12, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Lin, J.X.; Leonard, W.J. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 2013, 38, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.G.; Caudana, P.; Paillet, J.; Piaggio, E.; Kroemer, G. Effects of interleukin-2 in immunostimulation and immunosuppression. J. Exp. Med. 2020, 217, e20191247. [Google Scholar] [CrossRef] [PubMed]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Yamaguchi, T.; Nomura, T.; Ono, M. Regulatory T cells and immune tolerance. Cell 2008, 133, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.P.; Imbert, J.; Leonard, W.J. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006, 17, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Pollizzi, K.N.; Powell, J.D. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 2014, 14, 435–446. [Google Scholar] [CrossRef]
- Belot, M.P.; Castell, A.L.; Le Fur, S.; Bougneres, P. Dynamic demethylation of the IL2RA promoter during in vitro CD4+ T cell activation in association with IL2RA expression. Epigenetics 2018, 13, 459–472. [Google Scholar] [CrossRef]
- Perrin, P.J.; Davis, T.A.; Smoot, D.S.; Abe, R.; June, C.H.; Lee, K.P. Mitogenic stimulation of T cells reveals differing contributions for B7-1 (CD80) and B7-2 (CD86) costimulation. Immunology 1997, 90, 534–542. [Google Scholar] [CrossRef]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.T.; Gao, Y.; Lazarevic, V. Transcriptional regulation of CD4(+) TH cells that mediate tissue inflammation. J. Leukoc. Biol. 2018, 104, 1069–1085. [Google Scholar] [CrossRef] [PubMed]
- Iwata, S.; Mikami, Y.; Sun, H.W.; Brooks, S.R.; Jankovic, D.; Hirahara, K.; Onodera, A.; Shih, H.Y.; Kawabe, T.; Jiang, K.; et al. The Transcription Factor T-bet Limits Amplification of Type I IFN Transcriptome and Circuitry in T Helper 1 Cells. Immunity 2017, 46, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Koenecke, C.; Lee, C.W.; Thamm, K.; Fohse, L.; Schafferus, M.; Mittrucker, H.W.; Floess, S.; Huehn, J.; Ganser, A.; Forster, R.; et al. IFN-gamma production by allogeneic Foxp3+ regulatory T cells is essential for preventing experimental graft-versus-host disease. J. Immunol. 2012, 189, 2890–2896. [Google Scholar] [CrossRef] [PubMed]
- Dudemaine, P.L.; Fecteau, G.; Lessard, M.; Labrecque, O.; Roy, J.P.; Bissonnette, N. Increased blood-circulating interferon-gamma, interleukin-17, and osteopontin levels in bovine paratuberculosis. J. Dairy Sci. 2014, 97, 3382–3393. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, J.M.; Liao, W.J.; Lu, H. Scission of the p53-MDM2 Loop by Ribosomal Proteins. Genes Cancer 2012, 3, 298–310. [Google Scholar] [CrossRef]
- Li, S. Regulation of Ribosomal Proteins on Viral Infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Zhou, Y. Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res. 2018, 247, 15–20. [Google Scholar] [CrossRef]
- Cervantes-Salazar, M.; Angel-Ambrocio, A.H.; Soto-Acosta, R.; Bautista-Carbajal, P.; Hurtado-Monzon, A.M.; Alcaraz-Estrada, S.L.; Ludert, J.E.; Del Angel, R.M. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: This interaction is required for viral translation and replication in Huh-7 cells. Virology 2015, 484, 113–126. [Google Scholar] [CrossRef]
- Wang, B.; Duan, X.; Fu, M.; Liu, Y.; Wang, Y.; Li, X.; Cao, H.; Zheng, S.J. The association of ribosomal protein L18 (RPL18) with infectious bursal disease virus viral protein VP3 enhances viral replication. Virus Res. 2018, 245, 69–79. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Zhou, P.; Han, H. Cloning of mouse genomic ribosomal protein L6 gene and analysis of its promoter. Biochim. Biophys. Acta 2002, 1576, 219–224. [Google Scholar] [CrossRef]
- Mekdad, H.E.; Boutant, E.; Karnib, H.; Biedma, M.E.; Sharma, K.K.; Malytska, I.; Laumond, G.; Roy, M.; Real, E.; Paillart, J.C.; et al. Characterization of the interaction between the HIV-1 Gag structural polyprotein and the cellular ribosomal protein L7 and its implication in viral nucleic acid remodeling. Retrovirology 2016, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.H.; Ma, F.F.; Guan, G.K.; Wang, X.F.; Li, C.; Huang, J. White spot syndrome virus VP51 interact with ribosomal protein L7 of Litopenaeus vannamei. Fish. Shellfish Immunol. 2015, 44, 382–388. [Google Scholar] [CrossRef] [PubMed]
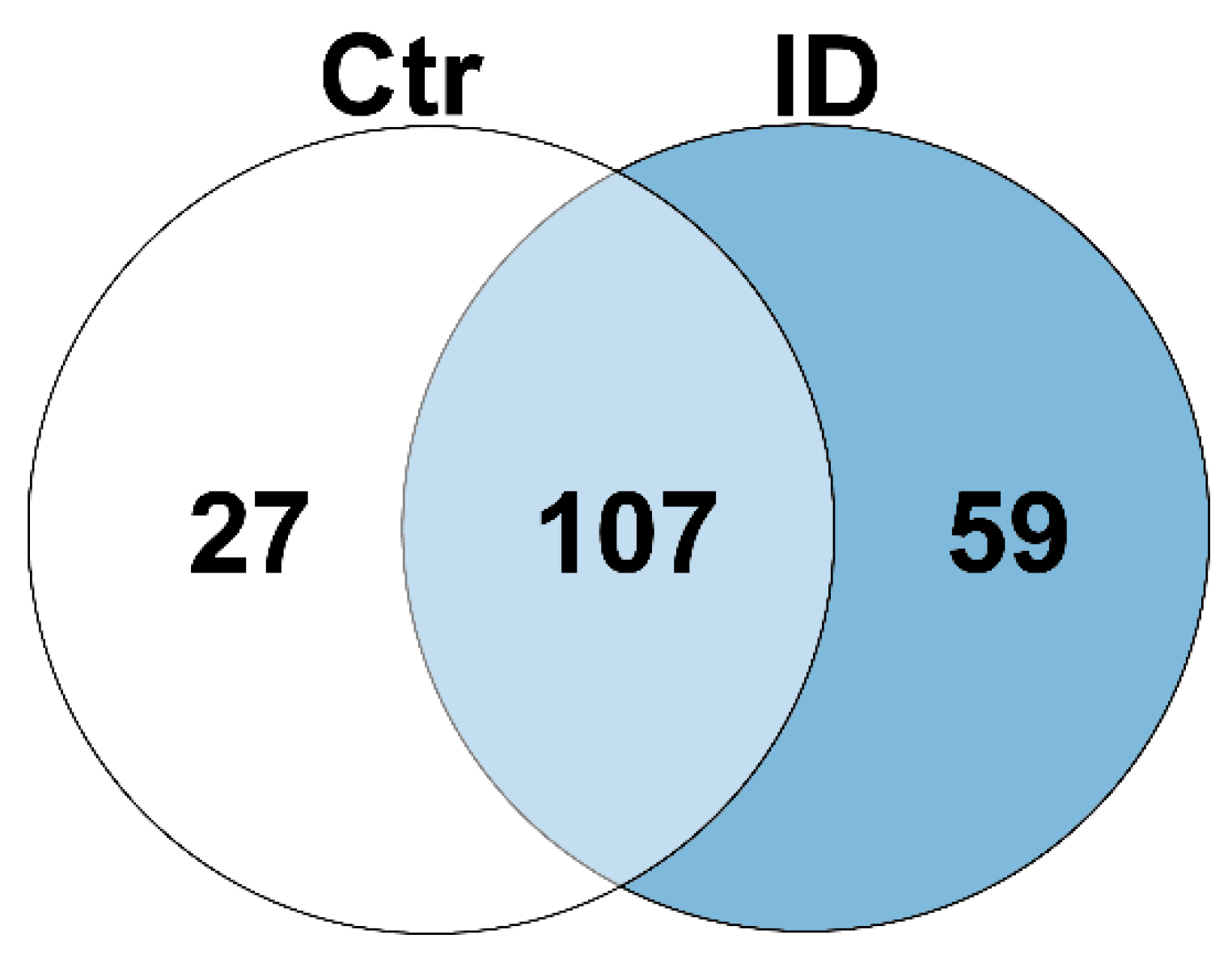

| Protein Abundance per Phenotype | Up in Controls | Up in Controls and Immune Deviant Cows | Up in Immune Deviant Cows | ||||
|---|---|---|---|---|---|---|---|
| No. of proteins | 27 | 107 | 59 | ||||
| Gene names | CACYBP | KPNA2 | ISG20 | GBP4 | CARS1 | CD200 | DPP4 |
| LAP3 | GBP6 | MCM6 | TNFSF8 | IARS1 | RFC5 | IPO5 | |
| ADK | CBS | IFI44L | SLC1A5 | SLC38A1 | IPO4 | ABCF1 | |
| LRRC59 | MTHFD2 | NOC3L | AASDHPPT | PCNA | SEMA4A | HAT1 | |
| CLIC4 | PYCR1 | AARS1 | EED | RPL23 | UTP3 | ALDH18A1 | |
| PPAT | PABPC4 | DNMT1 | SLC3A2 | TARS1 | NAT10 | EIF4A1 | |
| H1-2 | G3BP1 | PCK2 | MDN1 | ATP1B3 | DDX21 | AHSA1 | |
| U2AF1 | PSAT1 | RPL23 | SLC1A4 | TBCD | RFC4 | TF | |
| H1-1 | TNFRSF4 | UBA7 | RAP1A | RPL27A | LRPPRC | ABCE1 | |
| ELAVL1 | RPL37A | PSPH | P2RX7 | RPL27 | FAS | GPR183 | |
| HNRNPA1 | GBP1 | CCT6A | SLAMF1 | CAD | GTPBP1 | MGAT2 | |
| SYNCRIP | WARS1 | RRM1 | ICAM3 | FNBP1 | NOP2 | DDX3X | |
| NAMPT | GBP6 | MCM7 | STAT3 | ACAD10 | HPF1 | HSPD1 | |
| TIMM50 | UHRF1 | PAF1 | SHMT2 | NUDC | GNE | YARS1 | |
| OLA1 | HERC6 | B2M | PYCR2 | EIF4G1 | HLA-E | RPL8 | |
| CPOX | PPA1 | SMC4 | ACOT7 | SLC29A1 | RPL3 | GSPT1 | |
| RALY | RPS10 | HSP90AB1 | UBA5 | DOCK10 | TMEM87A | JAML | |
| IMPDH2 | MCM4 | BCAT1 | CD69 | STX11 | PMPCB | ||
| HARS1 | MTHFD1L | ICAM1 | ALCAM | TFRC | PHGDH | ||
| TRAP1 | G3BP2 | SMC2 | EIF3H | GMPS | ATAD3A | ||
| HYOU1 | ISG15 | GARS1 | MOV10 | HLA-E | NMT1 | ||
| PARK7 | ASNS | CDK1 | POLD1 | PPID | SWAP70 | ||
| RPL36 | NUB1 | IL2RA | FKBP4 | TSR1 | RPS27 | ||
| PNP | MCM5 | CD83 | RPL21 | BSG | IPO7 | ||
| MX1 | FDPS | DNAJA1 | LARS1 | EIF5 | HLA-E | ||
| PABPC1 | MCM2 | MCM3 | ABCB4 | CCDC47 | KTN1 | ||
| NAA50 | GBP1 | TIAL1 | EIF5B | RPL35 | CD44 | ||
| DHX30 | EIF4G2 | EEF1D | HSPH1 | RPS8 | |||
| GBP4 | IFI44 | RPS7 | MTHFD1 | RPS23 | |||
| EIF2S2 | TNFRSF18 | ICOS | RPL15 | ||||
| Protein Abundance per Phenotype | Up in Controls | Up in Controls and Immune Deviant Cows | Up in Immune Deviant Cows | ||
|---|---|---|---|---|---|
| No. of proteins | 22 | 16 | 31 | ||
| Gene names | MMP1 | IL17F | CCL5 | RPS7 | OAS1 |
| IL17A | LYZ | INHBA | RPL3 | RPL18 | |
| MMP3 | IL1RN | IL2 | SF3B1 | RPL6 | |
| CXCL8 | EIF4G1 | GBP1 | NUCB1 | RPL30 | |
| CXCL1 | LCN2 | IDO1 | PRMT1 | RPL7A | |
| CXCL2 | SSB | GZMA | RPL5 | MVP | |
| CAMP | TGM2 | CTSB | HNRNPR | ||
| HBA1 | NOS2 | HNRNPM | SYNCRIP | ||
| PGLYRP1 | ICAM1 | XDH | ARPC3 | ||
| ARMC10 | IFNG | AARS1 | SULT1A1 | ||
| CA2 | PPA1 | RPL18A | HNRNPA3 | ||
| HBB | WARS1 | RPS8 | RPL14 | ||
| SLC4A1 | LTF | RPS5 | CORO7 | ||
| CAMP | PTX3 | RPS9 | RPS10 | ||
| UPP1 | SDS | EIF4A1 | MACROH2A1 | ||
| GLG1 | NUCB2 | RPL27 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinwort, K.J.H.; Degroote, R.L.; Hirmer, S.; Korbonits, L.; Lorenz, L.; Scholz, A.M.; Hauck, S.M.; Deeg, C.A. Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen. Proteomes 2022, 10, 7. https://doi.org/10.3390/proteomes10010007
Kleinwort KJH, Degroote RL, Hirmer S, Korbonits L, Lorenz L, Scholz AM, Hauck SM, Deeg CA. Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen. Proteomes. 2022; 10(1):7. https://doi.org/10.3390/proteomes10010007
Chicago/Turabian StyleKleinwort, Kristina J. H., Roxane L. Degroote, Sieglinde Hirmer, Lucia Korbonits, Lea Lorenz, Armin M. Scholz, Stefanie M. Hauck, and Cornelia A. Deeg. 2022. "Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen" Proteomes 10, no. 1: 7. https://doi.org/10.3390/proteomes10010007
APA StyleKleinwort, K. J. H., Degroote, R. L., Hirmer, S., Korbonits, L., Lorenz, L., Scholz, A. M., Hauck, S. M., & Deeg, C. A. (2022). Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen. Proteomes, 10(1), 7. https://doi.org/10.3390/proteomes10010007







