Effect of External Electric Field Stress on Gliadin Protein Conformation
Abstract
:1. Introduction
2. Experimental
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | GLN | GLN | TYR | PRO | SER | GLY | GLU | GLY | SER |
| 10 | PHE | GLN | PRO | SER | GLN | GLU | ASN | PRO | GLN |
3. Results and Discussion
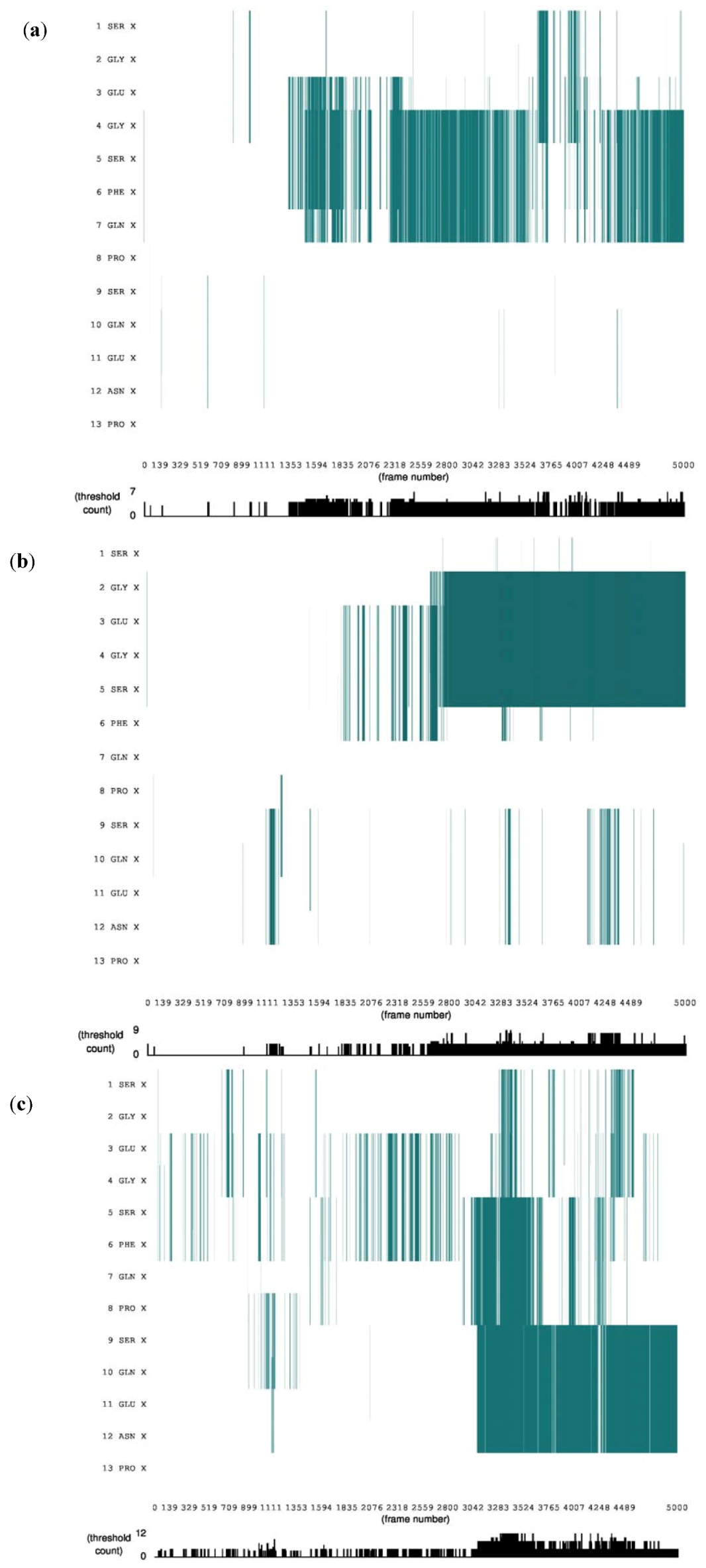
3.1. Secondary Structure Analysis
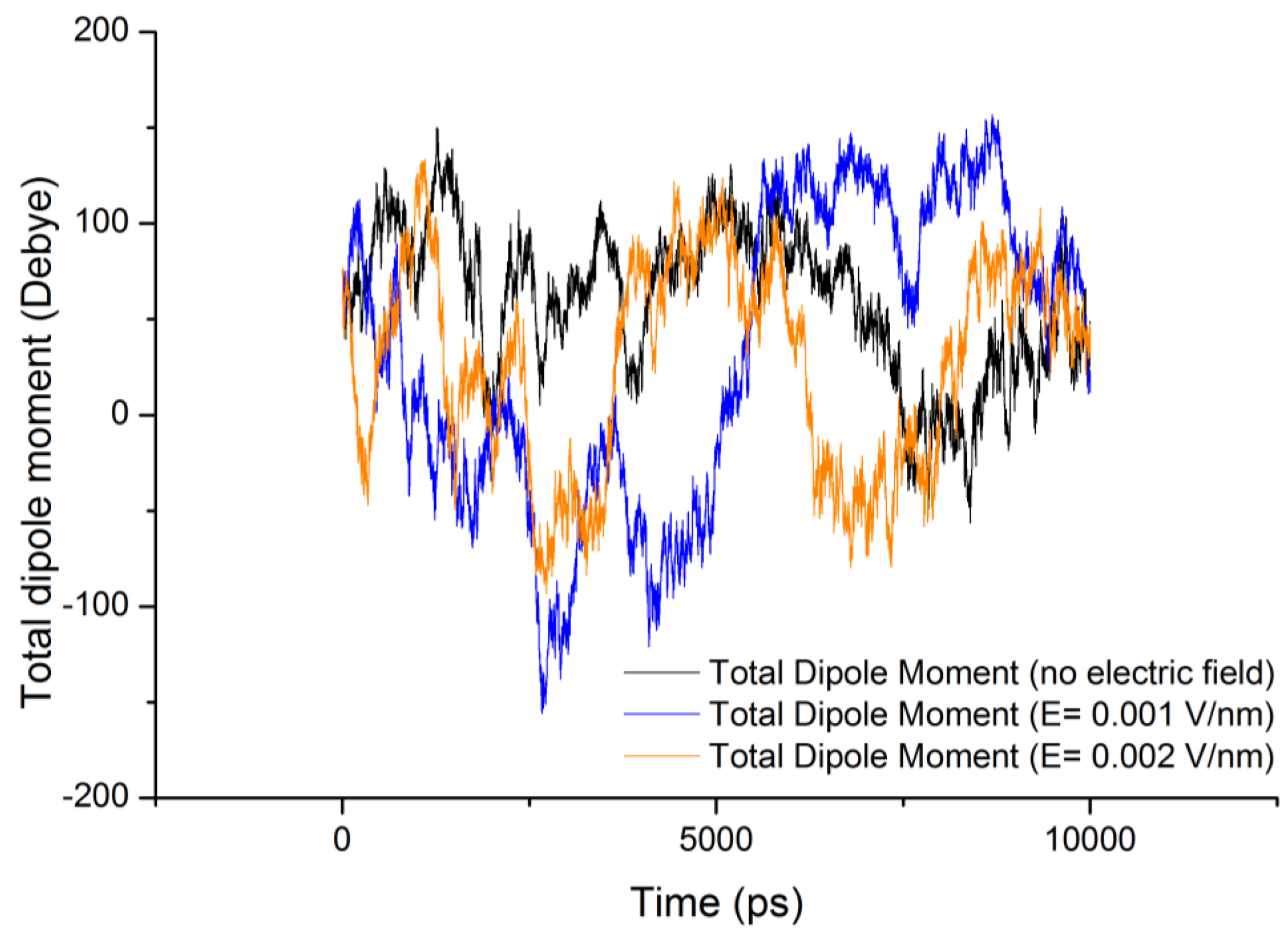
3.2. Dipole Moment Distribution

 is the dipole, qi is the charge of the atom i,
is the dipole, qi is the charge of the atom i,  is the directional vector of each atom and N is the number of atoms. In our study, the electric field was applied in the z-axis. Depending on the strength of the applied field, a change in the total dipole moment of the gliadin protein was observed (Figure 3).
is the directional vector of each atom and N is the number of atoms. In our study, the electric field was applied in the z-axis. Depending on the strength of the applied field, a change in the total dipole moment of the gliadin protein was observed (Figure 3). 
| Molecule | Electric field strength (V/nm) | RMSD average (nm) | Rg average (nm) | Total Dipole moment (Debye) |
|---|---|---|---|---|
| Gliadin protein | 0 | 0.536 ± 0.131 | 0.942 ± 0.095 | 59.8 ± 38.66 |
| Gliadin protein | 0.001 | 0.461 ± 0.102 | 1.030 ± 0.065 | 33.3 ± 78.48 |
| Gliadin protein | 0.002 | 0.617 ± 0.137 | 0.911 ± 0.094 | 26.9 ± 53.41 |
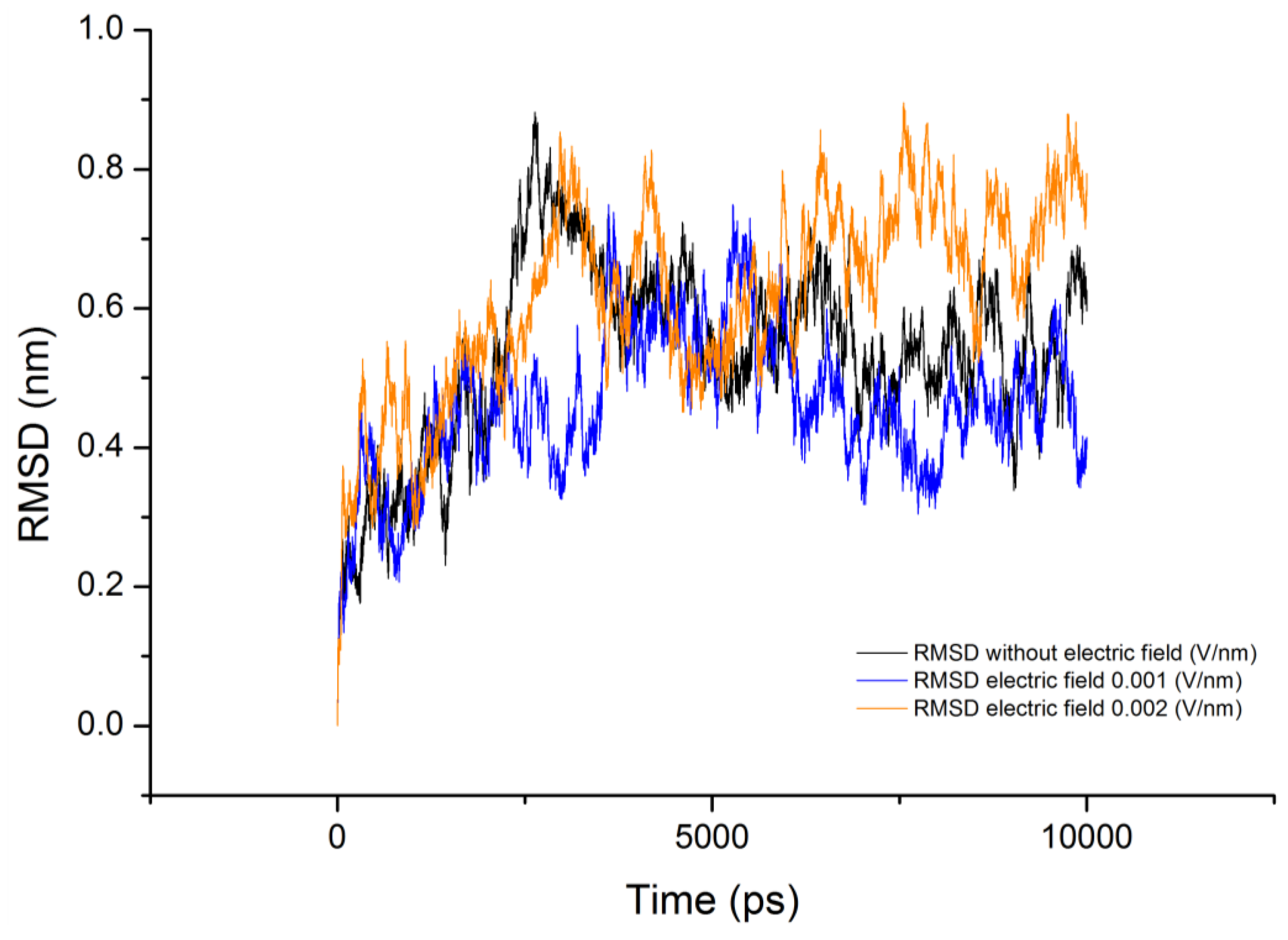
3.3. Root Mean Square Deviation (RMSD)

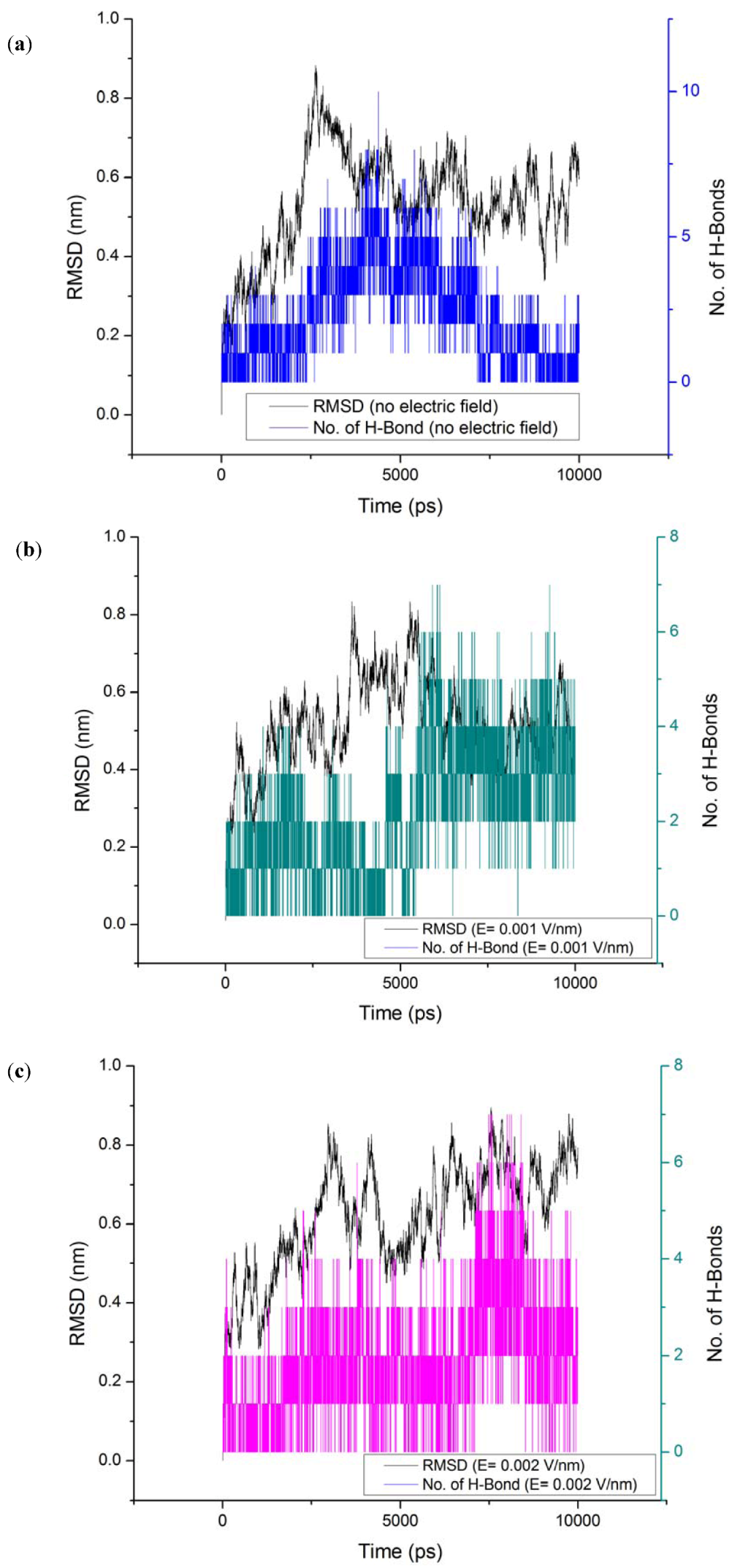
| Without electric field (52 Hbonds) | E = 0.001 V/nm (50 Hbonds) | E = 0.002 V/nm (54 Hbonds) | ||||||
|---|---|---|---|---|---|---|---|---|
| Donor | Acceptor | Occupancy | Donor | Acceptor | Occupancy | Donor | Acceptor | Occupancy |
| GLY | GLU | 5.14% | GLY | GLU | 3.12% | GLY | GLU | 11.86% |
| GLN | PRO | 2.12% | GLN | PRO | 8.08% | SER | GLU | 21.30% |
| ASN | GLN | 5.54% | GLU | SER | 25.13% | GLN | PRO | 3.10% |
| GLU | SER | 9.50% | SER | GLU | 40.05% | GLN | SER | 24.04% |
| GLN- | SER | 3.24% | GLU | GLU | 6.08% | GLN | ASN | 2.92% |
| GLN | ASN | 3.44% | SER | GLY | 2.02% | SER | ASN | 2.38% |
| SER | GLU | 22.00% | SER | GLY | 8.26% | GLU | SER | 2.02% |
| GLN | GLU | 4.46% | SER | ASN | 4.50% | |||
| SER | GLU | 25.39% | GLU | SER | 5.22% | |||
| GLU | GLU | 9.78% | ||||||
| SER | GLN | 2.06% | ||||||
3.4. Radius of Gyration (Rg)

3.5. Solvent Accessible Surface Area (SASA)



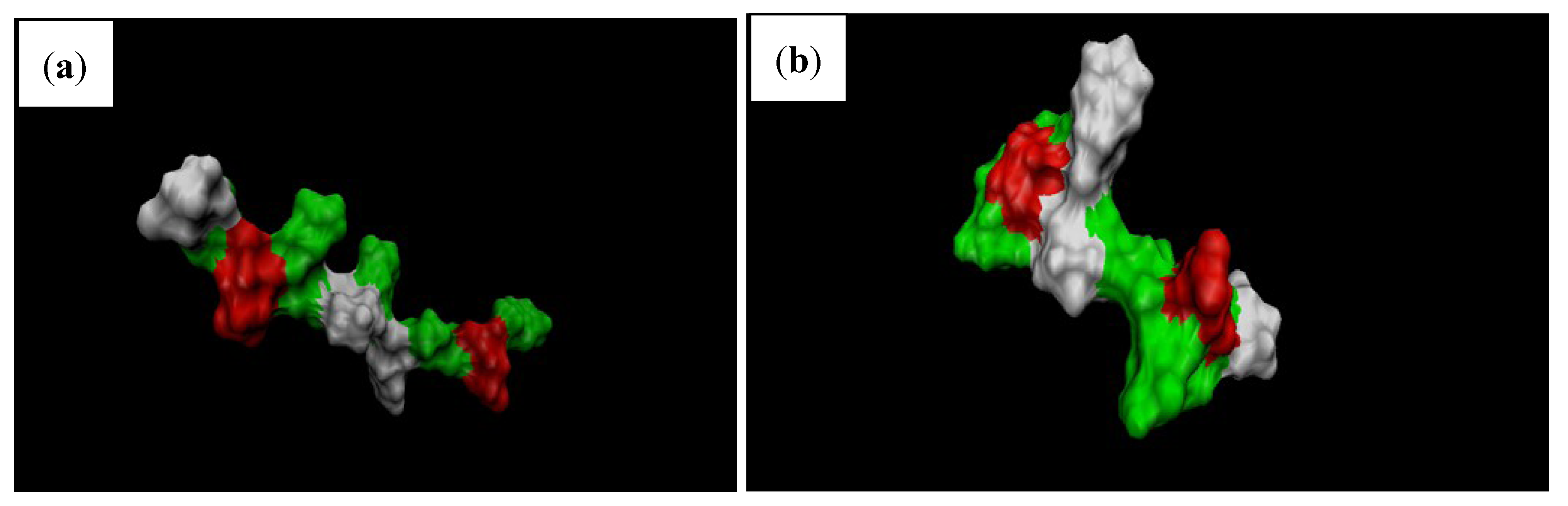
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef]
- Vader, L.W.; Stepniak, D.T.; Bunnik, E.M.; Kooy, Y.M.; de Haan, W.; van Veelen, P.A.; Koning, F. Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology 2003, 125, 1105–1113. [Google Scholar] [CrossRef]
- Stoven, S.; Murray, J.A.; Marietta, E. Celiac disease: Advances in treatment via gluten modification. Clin. Gastroenterol. Hepatol. 2012, 10, 859–862. [Google Scholar] [CrossRef]
- Mazzarella, G.; Salvati, V.M.; Iaquinto, G.; Stefanile, R.; Capobianco, F.; Luongo, D.; Bergamo, P.; Maurano, F.; Giardullo, N.; Malamisura, B.; et al. Reintroduction of gluten following flour transamidation in adult celiac patients: A randomized, controlled clinical study. Clin. Dev. Immun. 2012, 2012, e329150. [Google Scholar]
- Miśkiewicz, P.; Kępczyńska-Nyk, A.; Bednarczuk, T. Coeliac disease in endocrine diseases of autoimmune origin. Endokrynol. Polska 2012, 63, 240–249. [Google Scholar]
- Singh, A.; Orsat, V.; Raghavan, V. A comprehensive review on electrohydrodynamic drying and high voltage electric field in the context of food and bioprocessing. Drying Tech. 2012, 30, 1812–1820. [Google Scholar] [CrossRef]
- van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comp. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Wellner, N.; Mills, E.N.C.; Brownsey, G.; Wilson, R.H.; Brown, N.; Freeman, J.; Halford, N.G.; Shewry, P.R.; Belton, P.S. Changes in protein secondary structure during gluten deformation studied by dynamic fourier transform infrared spectroscopy. Biomacromolecules 2005, 6, 255–261. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Astrakas, L.; Gousias, C.; Tzaphlidou, M. Electric field effects on chignolin conformation. J. Appl. Phys. 2011, 109, e094702. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Purcell, J.M.; Kasarda, D.D.; Wu, C.S.C. Secondary structures of wheat α- and ω-gliadin proteins: Fourier transform infrared spectroscopy. J. Cereal Sci. 1988, 7, 21–32. [Google Scholar] [CrossRef]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004, 32, W500–W502. [Google Scholar] [CrossRef]
- Budi, A.; Legge, F.S.; Treutlein, H.; Yarovsky, I. Effect of external stresses on protein conformation: A computer modelling study. Eur. Biophys. J. 2004, 33, 121–129. [Google Scholar] [CrossRef]
- Kortemme, T.; Morozov, A.V.; Baker, D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 2003, 326, 1239–1259. [Google Scholar] [CrossRef]
- Fabiola, F.; Bertam, R.; Korostelev, A.; Chapman, M.S. An improved hydrogen bond potential: Impact on medium resolution protein structures. Prot. Sci. 2002, 11, 1415–1423. [Google Scholar] [CrossRef]
- Mata, I.; Molins, E.; Alkorta, I.; Espinosa, E. Effect of an external electric field on the dissociation energy and the electron density properties: The case of the hydrogen bonded dimer HF⋯HF. J. Chem. Phys. 2009, 130, e044104. [Google Scholar] [CrossRef]
- Maiuri, L.; Ciacci, C.; Ricciardelli, I.; Vacca, L.; Raia, V.; Auricchio, S.; Picard, J.; Osman, M.; Quaratino, S.; Londei, M. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003, 362, 30–37. [Google Scholar] [CrossRef]
- Henderson, K.N.; Tye-Din, J.A.; Reid, H.H.; Chen, Z.; Borg, N.A.; Beissbarth, T.; Tatham, A.; Mannering, S.I.; Purcell, A.W.; Dudek, N.L.; et al. A Structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 2007, 27, 23–34. [Google Scholar] [CrossRef]
- Budi, A.; Legge, F.S.; Treutlein, H.; Yarovsky, I. Comparative study of insulin chain-B in isolated and monomeric environments under external stress. J. Phys. Chem. B 2008, 112, 7916–7924. [Google Scholar]
- Budi, A.; Legge, F.S.; Treutlein, H.; Yarovsky, I. Electric field effects on insulin chain-B conformation. J. Phys. Chem. B 2005, 109, 22641–22648. [Google Scholar] [CrossRef]
- Budi, A.; Legge, F.S.; Treutlein, H.; Yarovsky, I. Effect of frequency on insulin response to electric field stress. J. Phys. Chem. B 2007, 111, 5748–5756. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Singh, A.; Munshi, S.; Raghavan, V. Effect of External Electric Field Stress on Gliadin Protein Conformation. Proteomes 2013, 1, 25-39. https://doi.org/10.3390/proteomes1020025
Singh A, Munshi S, Raghavan V. Effect of External Electric Field Stress on Gliadin Protein Conformation. Proteomes. 2013; 1(2):25-39. https://doi.org/10.3390/proteomes1020025
Chicago/Turabian StyleSingh, Ashutosh, Shirin Munshi, and Vijaya Raghavan. 2013. "Effect of External Electric Field Stress on Gliadin Protein Conformation" Proteomes 1, no. 2: 25-39. https://doi.org/10.3390/proteomes1020025
APA StyleSingh, A., Munshi, S., & Raghavan, V. (2013). Effect of External Electric Field Stress on Gliadin Protein Conformation. Proteomes, 1(2), 25-39. https://doi.org/10.3390/proteomes1020025





