Abstract
The early detection of Alzheimer’s disease (AD) is essential for improving patient outcomes, enabling timely intervention, and slowing disease progression. However, the complexity of neuroimaging data presents significant obstacles to accurate classification. This study introduces a computationally efficient AI framework designed to enhance AD staging using structural MRI. The proposed method integrates discrete wavelet transform (DWT) for multi-scale feature extraction, a novel reduced kernel partial least squares (Red-KPLS) algorithm for feature reduction, and ResNet-50 for classification. The proposed technique, referred to as Red-KPLS-CNN, refines MRI features into discriminative biomarkers while minimizing redundancy. As a result, the framework achieves 96.9% accuracy and an F1-score of 97.8% in the multiclass classification of AD cases using the Kaggle dataset. The dataset was strategically partitioned into 60% training, 20% validation, and 20% testing sets, preserving class balance throughout all splits. The integration of Red–KPLS enhances feature selection, reducing dimensionality without compromising diagnostic sensitivity. Compared to conventional models, our approach improves classification robustness and generalization, reinforcing its potential for scalable and interpretable AD diagnostics. These findings emphasize the importance of hybrid wavelet–kernel–deep learning architectures, offering a promising direction for advancing computer-aided diagnosis (CAD) in clinical applications.
1. Introduction
Alzheimer’s disease (AD) poses a profound and growing challenge to global healthcare systems. As the most prevalent form of dementia, it accounts for approximately 60–80% of neurodegenerative cases worldwide [1]. This progressive condition is marked by the degeneration and death of nerve cells, leading to significant brain atrophy, particularly in regions integral to memory, such as the hippocampus and cerebral cortex [2]. Clinically, AD manifests through a gradual decline in cognitive functions, beginning with subtle memory lapses and advancing to a complete loss of independence, often accompanied by severe physical complications in its terminal stages [3].
Epidemiological projections highlight the urgent need to address AD [4]. Current estimates indicate that the number of people affected could triple by 2050, potentially rising to 150 million cases worldwide [5], emphasizing the critical demand for better diagnostic tools and treatment options [6].
In clinical practice, the progression of AD is typically assessed using standardized scales such as the Global Deterioration Scale (GDS) and the Clinical Dementia Rating (CDR), which categorize cognitive decline into seven and five distinct stages, respectively. These frameworks evaluate multiple cognitive domains, including memory, problem-solving abilities, and the capacity for independent living [7]. From a neuroimaging perspective, structural magnetic resonance imaging (MRI) has emerged as a crucial tool for identifying characteristic patterns of brain degeneration [8]. Notably, MRI can detect shrinkage in medial temporal lobe structures and the expansion of ventricular spaces, changes that correlate strongly with both the severity of cognitive impairment and progression through clinical staging systems [9]. However, traditional radiological assessments require substantial expertise and are subject to inter-rater variability, which can impede early and accurate detection of AD pathology.
To address these challenges, there is a growing interest in computer-aided diagnosis (CAD) systems that provide quantitative, objective assessments of disease state [10]. Nonetheless, current CAD approaches often grapple with the high dimensionality of neuroimaging data, complicating pattern recognition, and frequently lack clinical interpretability, which can hinder physician trust and adoption [11]. In response to these limitations, our research introduces a novel CAD framework designed to enhance the early and accurate detection of AD [12]. This integrated pipeline combines advanced image processing with optimized machine learning techniques. Initially, multi-resolution feature extraction is performed using discrete wavelet transform (DWT) [13], effectively capturing localized texture variations associated with early neurodegenerative changes. Subsequently, these features undergo dimensionality reduction through our proposed reduced kernel partial least squares (Red-KPLS) method, which maintains diagnostic relevance while significantly improving computational efficiency compared to conventional techniques. The processed features are then input into a convolutional neural network architecture that performs both binary classification (distinguishing normal cognition from AD cases) and multi-class staging in accordance with established clinical scales. Validation on the Kaggle dataset [14] demonstrates the system’s robust performance, achieving superior accuracy to existing methods while providing clinically meaningful biomarkers. The hierarchical classification approach aligns with real-world diagnostic workflows, where clinicians first establish disease presence before determining severity. Furthermore, the wavelet-based features maintain anatomical interpretability, allowing radiologists to trace algorithmic decisions back to specific neuroanatomical changes. This represents a significant advance toward clinically deployable AI tools that can support diagnostic decision-making while maintaining the transparency required for medical applications.
This study had two main objectives. The first was to develop an AI-based system that accurately classifies Alzheimer’s disease across its different stages—non-dementia (ND), very mild dementia (VMiD), mild dementia (MiD), and moderate dementia (MoD)—with a target sensitivity of at least 95% for each stage. The second objective was to thoroughly validate the model’s performance on independent datasets. Additionally, this study aimed to identify the key neuroimaging features that drive the model’s decisions and assess whether the system can be practically useful in clinical settings. We hypothesized that using multimodal data would provide better performance than traditional single-modality methods while still offering clear and understandable outputs for clinical use.
This paper is structured as follows: Section 2 reviews existing Alzheimer’s disease classification methods and their strengths and limitations. Section 3 outlines the methodology, detailing the dataset, preprocessing, feature extraction using DWT, feature reduction with Red-KPLS, and classification via ResNet-50. Section 4 presents the experiments, covering setup, evaluation criteria, key metrics, and results discussion. Section 5 introduces the ethical considerations in AI-assisted AD diagnosis, addressing key challenges such as bias, transparency, and patient privacy. Finally, Section 6 summarizes the findings and explores potential improvements.
2. Related Work
Recent advances in deep learning have significantly influenced AD diagnosis by enabling automated, accurate, and early detection through neuroimaging [15]. Studies have explored various convolutional neural network (CNN) architectures, 3D subject-level models, transfer learning techniques, and ensemble methods to classify AD stages more effectively. A major focus has been on leveraging MRI data, especially from standardized datasets like ADNI, OASIS, and AIBL, which provide rich, annotated brain imaging data [15]. Golovanevsky, M. et al. [16] proposed MADDi, a thoughtful multimodal model aimed at improving Alzheimer’s disease diagnosis. What is unique is how it blends different types of patient data—like MRI images, clinical records, and genetic features—into one meaningful representation using cross-modal attention. This attention helps the model focus on the most relevant aspects of each data type. When all modalities were included, the model achieved an impressive accuracy of 96.86%. However, removing clinical data led to a noticeable performance drop, showing how vital that information is. Their work highlights the value of unifying diverse medical data for more accurate and explainable diagnoses. Parmar et al. [17] introduced a 3D convolutional neural network (3D-CNN) model designed to classify individuals into four categories: Alzheimer’s disease (AD), late mild cognitive impairment (LMCI), early mild cognitive impairment (EMCI), and cognitively normal (CN). Utilizing data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the study achieved classification accuracies of 99.4% on the training set, 96.75% on the validation set, and 93% on the testing set. These results underscore the potential of 3D-CNNs in accurately distinguishing between different stages of cognitive decline.
To tackle the computational challenges of 3D MRI analysis in Alzheimer’s disease detection, Angkoso et al. [18] developed an innovative solution: the multiplane convolutional neural network (Mp-CNN). Instead of processing entire 3D scans—which demands heavy computational resources—their model broke down MRI volumes into three standard anatomical views (axial, coronal, and sagittal) and then analyzed the most representative slice from each plane using separate 2D CNNs. This clever workaround not only reduced computational overhead but also delivered impressive performance, correctly classifying 93% of cases across AD, MCI, and NC groups in the ADNI-1 dataset. The model showed particularly strong precision in identifying healthy controls (95%) and maintained robust performance for AD (93%) and MCI (91%). By fusing multi-view information without full 3D processing, their approach offered a balanced compromise—retaining diagnostic accuracy while being far more efficient than traditional 3D deep learning methods. Hedayati et al. [19] proposed an innovative deep learning architecture for the classification of AD stages using structural MRI data from the ADNI database. The methodology employed a two-phase approach: (1) an ensemble of pre-trained autoencoders for robust feature extraction from 3D neuroimaging data, followed by (2) a CNN for final classification. This hybrid framework was specifically designed to address the inherent heterogeneity in neuroanatomical patterns across different stages of cognitive impairment. The model demonstrated strong discriminative performance across three critical classification tasks: AD versus normal controls (NCs) (95% accuracy), AD versus mild cognitive impairment (MCI) (90% accuracy), and MCI versus NCs (92.5% accuracy). Notably, the system achieved high sensitivity metrics while maintaining specificity, indicating its potential utility for early detection while minimizing false positive diagnoses. These results suggest that the combination of unsupervised feature learning via autoencoders with supervised CNN classification may offer an effective solution for handling the complex neuroanatomical variations present in progressive neurodegenerative disorders.
Saratxaga and colleagues [20] conducted a comprehensive comparison of three CNN architectures—2DNet, 3DNet, and ResNet18—for staging Alzheimer’s disease progression using the OASIS-2 dataset. Their system categorized patients into four clinically relevant groups: from cognitively normal (CN) through varying severity levels (VMD, MD, MOD). The ResNet18 architecture emerged as particularly effective, demonstrating strong performance with 93.18% balanced accuracy for detecting AD presence and 88% accuracy for distinguishing between disease stages. These results not only outperformed existing approaches but also highlighted how residual networks can effectively manage the complex variations in brain scan data. By achieving this level of accuracy with publicly available OASIS data, the study provided convincing evidence that deep learning could complement or potentially replace conventional cognitive tests in clinical practice. While these findings advance the field of AI-assisted diagnosis, the authors appropriately note that further validation across more diverse patient groups and imaging systems remains necessary.
Jabason and colleagues [21] made significant strides by combining multiple hybrid deep convolutional neural networks (DCNNs) in an ensemble framework. When tested on the OASIS-3 dataset, their system distinguished between Alzheimer’s patients, those with mild cognitive impairment (MCI), and healthy controls with remarkable 95.23% accuracy, a notable achievement in three-class neuroimaging classification. Parallel work by Hazarika’s team [22] took a different approach, rigorously testing various deep learning models on the ADNI dataset. Their findings positioned DenseNet-121 as the top performer, reliably sorting subjects into cognitively normal, MCI, and AD categories with 90.22% accuracy.
Oktavian et al. [23] applied a ResNet-18 model to ADNI MRI data, achieving 88.3% accuracy in classifying cognitively normal (CN), Alzheimer’s (AD), and mild cognitive impairment (MCI) cases. Their work showed that even simpler residual networks can effectively detect subtle brain changes characteristic of early cognitive decline, challenging the assumption that deeper models always perform better for neuroimaging tasks.
Abraham et al. [24] demonstrated how a carefully optimized LeNet model could excel at classifying Alzheimer’s progression. Using high-resolution MPRAGE brain scans from the ADNI database, their enhanced LeNet architecture achieved an impressive 96.64% accuracy in distinguishing between healthy individuals (NCs), those with mild cognitive impairment (MCI), and Alzheimer’s patients (AD). This performance highlights that even traditional CNN designs, when properly refined, can deliver state-of-the-art results for neuroimaging analysis, especially when working with quality structural MRI data that clearly captures brain tissue changes.
3. Methodology
This study introduces a computational pipeline designed to improve the early detection of Alzheimer’s disease using structural MRI scans. The approach combines advanced techniques in image processing, feature refinement, and machine learning to analyze brain structure efficiently and accurately. The pipeline follows four main stages:
- Preprocessing: MRI scans are aligned to a standard template, resampled for consistency, and segmented into distinct tissue types (gray matter, white matter, and cerebrospinal fluid) to ensure reliable comparisons across subjects.
- The 2D discrete wavelet transform (DWT): Preprocessed gray matter images are decomposed into multiscale features, capturing both broad structural patterns and fine textures that may indicate early AD.
- Reduced kernel partial least squares (Red-KPLS): A newly adapted dimensionality reduction method condenses the extracted features while retaining the most diagnostically relevant information.
- Deep learning for classification: A neural network analyzes the refined features to distinguish between AD and non-AD cases with high precision.
Each step is optimized to handle the complexity of brain imaging data while maintaining computational efficiency, ensuring a practical and effective tool for potential clinical use.
3.1. Materials
3.1.1. Dataset
The proposed method was evaluated using the publicly available Kaggle MRI dataset [14], which consists of 6400 axial brain MRI slices categorized into four clinical stages: non-dementia (ND), very mild dementia (VMiD), mild dementia (MiD), and moderate dementia (MoD). The images were originally acquired at a resolution of pixels, with representative samples displayed in Figure 1. To ensure balanced class distributions, the dataset was split into 80% for training and 20% for testing. Then, the training portion was further divided into 60% for actual training and 20% for validation, keeping the test set unchanged. This resulted in final splits of 60% for training (3840 images), 20% for validation (1280 images), and 20% for testing (1280 images). Prior to feature extraction, each scan underwent standardized preprocessing, including N4 bias field correction and intensity normalization to the range, ensuring consistency across all samples.
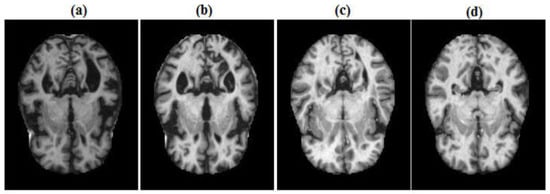
Figure 1.
Representative MRI samples across Alzheimer stages: (a) MID; (b) MOD; (c) ND; (d) VMD.
3.1.2. Preprocessing
The 2D axial MRI slices ( pixels) from the Kaggle dataset underwent a standardized preprocessing pipeline to enhance consistency while preserving key features. To address intensity inhomogeneities, N4 bias field correction was applied using ANTs, followed by percentile-based intensity normalization (1st–99th percentile) to scale intensities within the range. Anatomical alignment was achieved by registering images to the MNI152 2mm axial template via affine transformation (FLIRT in FSL, 6 degrees of freedom), after which they were resampled to pixels using bicubic interpolation with zero-padding to maintain aspect ratios. For interpretable feature extraction within Red-KPLS-CNN, optional tissue probability maps were generated using SPM12’s 2D-adapted unified segmentation, estimating partial volumes for gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) while accounting for slice-specific intensity variations. This preprocessing workflow balances computational efficiency with the preservation of discriminative features, ensuring robust data preparation for dementia staging.
3.2. Feature Extraction via DWT
The wavelet transform (WT) is a powerful analytical tool widely employed in MRI feature extraction, particularly in classification and image analysis tasks [25]. Its strength lies in its ability to analyze signals or images across multiple scales, providing a more detailed representation of structural patterns. Unlike other similar methods, WT offers time–frequency localization, a crucial property for classification applications [25].
The mother wavelet serves as the fundamental time function, characterized by finite energy and rapid decay, from which higher-order wavelets are derived [13]. Given a continuous square-integrable function g(x), the continuous WT of g(x), associated with a real-valued wavelet , is defined as
where
The parameters a and b control how the wavelet adapts to different scales and positions within a signal. a adjusts the wavelet’s size, while b shifts it across the domain, allowing for localized analysis. By applying these transformations to the mother wavelet, we construct the wavelet function. To derive a discrete representation from Equation (1), we constrain a and b to a fixed set of values, forming a discrete lattice. With this discretization, the discrete wavelet transform (DWT) is defined as
The coefficients and represent the approximation and detail components, respectively, while and serve as the high-pass and low-pass filters. The parameters i and k define the wavelet scale factor and translation factor, controlling how the signal is adjusted across different resolutions [26].
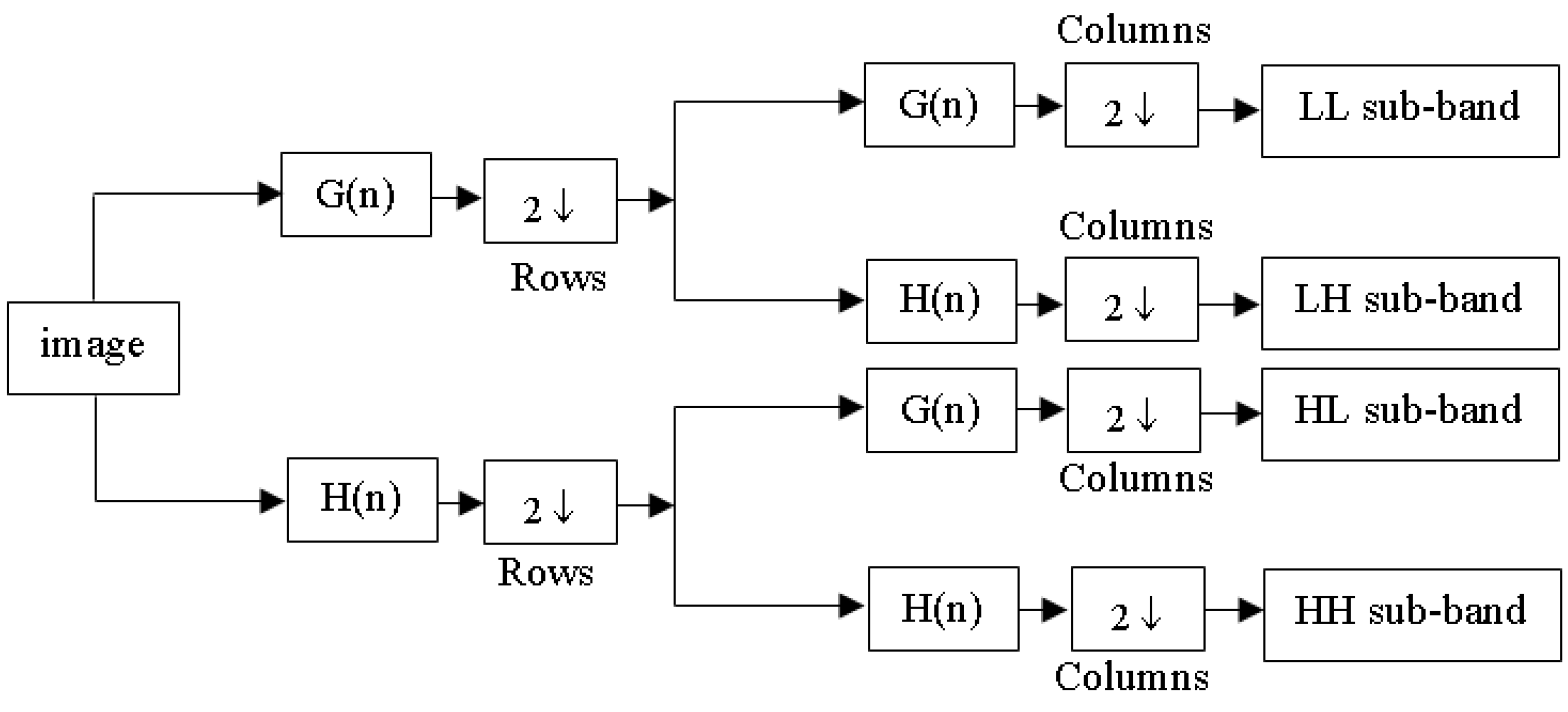
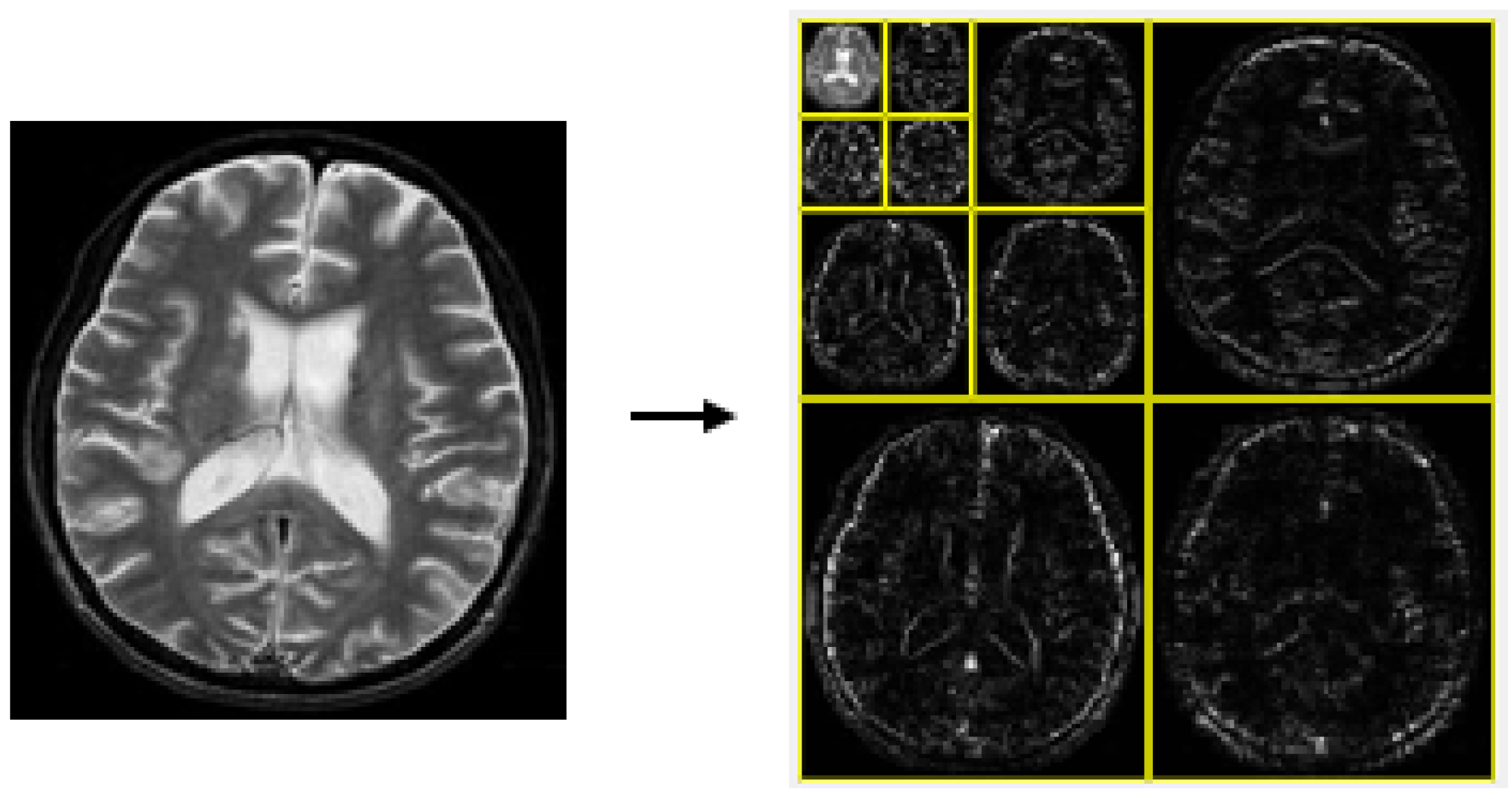
A key principle of the wavelet transform (WT) is captured in Equation (3), which underpins wavelet decomposition by breaking down a signal into and , facilitating multi-resolution analysis. This process corresponds to a one-level decomposition [13], with the structure of the two-dimensional (2D) discrete wavelet transform (DWT) illustrated in Figure 2. When applied to an image, decomposition takes place separately along rows and columns, producing four sub-bands: LL (low–low), LH (low–high), HL (high–low), and HH (high–high). The LH, HL, and HH sub-bands capture finer details by emphasizing horizontal, vertical, and diagonal variations, while the LL sub-band retains key approximations and serves as the input for the next decomposition stage. In this study, a third-order 2D-DWT was applied to the dataset images, as shown in Figure 3.
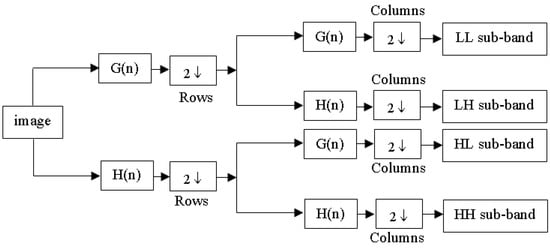
Figure 2.
Diagram of 2D-DWT.
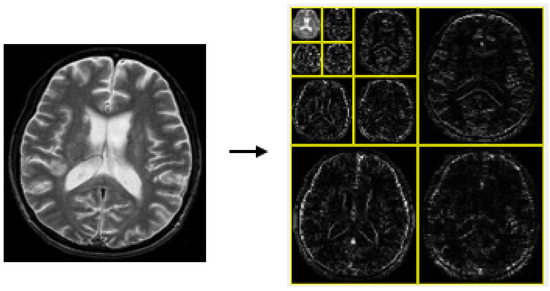
Figure 3.
The 3-level decomposition of 2D-DWT.
3.3. Proposed Red-KPLS for Feature Reduction
Working with brain scan data presents a key challenge—the extremely large number of features extracted can be computationally intensive and often contain unnecessary information. To solve this, we developed a streamlined version of kernel partial least squares (called Red-KPLS) that maintains the strengths of standard methods while being more efficient. Where traditional approaches might either lose important details (like PCA) or require too much memory (like regular KPLS), our solution offers a better balance:
- Smart feature selection that keeps only the most relevant patterns;
- A modified calculation process that makes results easier to interpret;
- Special optimization for handling brain scan data sizes.
We propose in this paper a Red-KPLS method that efficiently handles high-dimensional neuroimaging data through optimal prototype selection and kernel matrix reduction. Building on standard KPLS, which maps inputs to a feature space via nonlinear transformation , our approach computes the kernel matrix
The key innovation involves selecting prototype samples that maximize projections onto latent components while satisfying
This yields a reduced kernel matrix:
Algorithm 1 presents the main steps of the proposed Red-KPLS method.
| Algorithm 1 Red-KPLS Algorithm |
|
The method reduces computational complexity from to while maintaining discriminative power through optimal prototype selection. The proposed technique works particularly well for Alzheimer’s disease detection because it can spot the subtle brain changes that matter while ignoring normal variations. After processing the MRI data through wavelet transforms (described earlier), Red-KPLS creates a simplified version that clearly highlights differences between healthy brains and those with AD.
3.4. Deep Learning for Classification
Deep convolutional neural networks have significantly advanced medical image analysis, especially for the early diagnosis of AD [27]. Among these, ResNet-50 stands out as a powerful architecture thanks to its deep residual learning framework, which addresses vanishing gradient problems and enables the extraction of rich, multi-level spatial features from brain MRI scans [28]. With 50 layers and shortcut connections, ResNet-50 retains low-level features and enhances gradient flow, allowing the model to learn intricate patterns in neuroimaging data [29]. This makes it highly effective in classifying stages of cognitive impairment and predicting clinical dementia ratings (CDRs) directly from MRI scans. Additionally, its pre-trained versions have proven valuable in transfer learning, improving diagnostic accuracy in scenarios where medical datasets are small or limited [29].
The proposed model is a customized version of ResNet-50 that has been adapted to handle 1D feature vectors instead of the typical 2D image data. These vectors come from a prior step involving wavelet transformation and Red-KPLS, which condense the data while preserving important patterns. To make this work, the usual 2D convolution layers in ResNet-50 were replaced with 1D versions, but the architecture’s core idea—residual learning—remains intact. This means that the model still uses shortcut connections that add the input of a layer directly to its output (F(x) + x), helping it learn more effectively by avoiding issues like vanishing gradients. This design lets the network go deeper and detect subtle features that are important for the task at hand, such as identifying patterns in medical data or other reduced feature spaces.
The proposed 1D-ResNet50 architecture is tailored to multiclass Alzheimer’s staging (ND, VMiD, MiD, and MoD) using wavelet and reduced KPLS features as input. It begins by accepting 1D feature vectors shaped as , where L is the dimensionality of the reduced KPLS output, with a preserved channel dimension to remain compatible with standard deep learning frameworks. The initial block applies a 1D convolution with 64 filters (kernel size 7, stride 2), followed by batch normalization, ReLU activation, and max pooling (kernel size 3, stride 2), which reduces the sequence length while retaining essential discriminative patterns extracted from the wavelet decomposition. The network’s core is structured into four hierarchical stages composed of 3, 4, 6, and 3 bottleneck blocks, respectively. Each bottleneck follows a typical progression from a convolution to a convolution and back to , starting with [1×1, 64 → 3×1, 64 → 1×1, 256] in the first stage and scaling up to [1×1, 512 → 3×1, 512 → 1×1, 2048] in the final stage. This design allows for multiscale feature extraction while optimizing computational efficiency through strategic dimensionality reduction before deeper spatial processing.
Within each residual block of the network, the design begins with batch normalization and ReLU activation applied before the convolutional operations, ensuring stable and efficient learning. To prevent overfitting on the reduced feature set, a dropout layer with a rate of 0.3 is introduced after the final convolution. Transitioning between stages, the architecture employs projection shortcuts using convolutions with a stride of 2, which handle both downsampling and channel depth adjustment while maintaining the integrity of the residual connection pathway. After each stage, squeeze-and-excitation blocks are inserted to adaptively recalibrate channel-wise features: these blocks first distill global spatial information into compact channel descriptors through average pooling, then selectively amplify important channels using a gating mechanism with sigmoid activation and a reduction ratio of 16. In the final stage, the network aggregates the output through global average pooling across the 2048-channel feature maps and feeds this representation into a dense layer with three output units and softmax activation, which calculates the class probabilities for ND, VMiD, MiD, and MoD based on the normalized exponentials of the linearly transformed pooled features.
The training process is designed for both efficiency and stability. It employs the AdamW optimizer with an initial learning rate of , gradually decreasing through cosine decay over 100 epochs. To ensure a smooth start, the model undergoes a warmup phase in the first five epochs. To enhance computational efficiency, bottleneck blocks process features at reduced dimensionality before expanding them back to their full depth, ensuring rich representation learning. Meanwhile, projection shortcuts maintain stable gradient flow across different stages, a crucial factor when training deep networks on limited neuroimaging data. Squeeze-and-excitation mechanisms dynamically recalibrate feature channels, emphasizing the most discriminative patterns that evolve across disease progression. This is especially critical in distinguishing ND, VMiD, MiD, and MoD, where subtle variations in imaging features play a key role. A global pooling layer makes it possible to handle variable-length inputs from the wavelet KPLS pipeline while converting them into fixed-size representations suitable for classification. To address class imbalance in the Kaggle dataset, bias initialization was tailored to match an approximate class distribution ratio of for ND, VMiD, MiD, and MoD. Ultimately, this balanced approach optimizes model capacity while integrating effective regularization strategies. The result? A robust system that achieves strong performance across all three diagnostic categories while preventing overfitting to the majority class.
Table 1 provides a detailed breakdown of the adapted 1D-ResNet50 model, outlining its layer configurations, key hyperparameters, and training protocols designed for multiclass classification.

Table 1.
Network architecture specifications.
4. Experiments
4.1. Experimental Setup
All experiments were conducted using Python 3.9 and PyTorch 2.0, leveraging CUDA 12.1 acceleration on NVIDIA A100 GPUs (40GB VRAM). The Kaggle dataset was divided into training (60%), validation (20%), and test (20%) sets, ensuring balanced class representation. With our 60-20-20 split into training, validation, and test sets, the model performed consistently well across all subsets, showing strong generalization capabilities. To evaluate the effectiveness of our proposed Red-KPLS against conventional feature reduction techniques, we performed comparative analyses within a consistent 1D-ResNet50 framework using DWT-processed inputs. The comparison encompassed four reduction methods applied under identical conditions: PCA (99% variance retention), KPCA (RBF kernel, ), PLS, and standard KPLS, each yielding 50-dimensional features. The 1D-ResNet50 classifier was trained using AdamW. To assess model performance, we evaluated classification outcomes using balanced accuracy, sensitivity (recall), specificity, and macro F1-score. These metrics provided a comprehensive view of how well the model distinguished between diagnostic categories while accounting for potential imbalances in class distributions.
4.2. Model Evaluation Criteria
The performance of the proposed classification scheme was assessed using standard classification metrics derived from the confusion matrix:
- True Positives (TPs): Correctly predicted positive cases.
- True Negatives (TNs): Correctly predicted negative cases.
- False Positives (FPs): Negative cases incorrectly predicted as positive.
- False Negatives (FNs): Positive cases incorrectly predicted as negative.
Key Metrics
Accuracy: Measures overall correctness in classification.
Sensitivity (Recall): Evaluates the model’s ability to correctly identify positive cases.
Specificity: Measures the model’s ability to correctly identify negative cases.
Precision: Assesses the reliability of positive predictions.
F1-Score: Provides a balanced measure of precision and recall.
mAP (Mean Average Precision): Commonly used to summarize model performance across all classes:
where is the total number of classes and is the average precision for class , calculated as the area under the precision–recall curve for that class.
For multiclass classification (ND/VMiD/MiD/MoD), these metrics were computed for each class individually and macro-averaged. Statistical significance was verified using 95% confidence intervals derived from 10,000 bootstrap resamples.
4.3. Results and Discussion
This study introduces an innovative pipeline for staging Alzheimer’s disease, combining wavelet-based preprocessing, feature reduction, and deep learning classification to enhance neuroimaging analysis. Structural MRI scans are first processed using DWT to extract multiscale texture features, capturing subtle patterns in brain structure. Next, Red-KPLS refines these features, distilling the most relevant components from the high-dimensional wavelet coefficients to ensure efficient data representation. Finally, a 1D-CNN classifier leverages the optimized feature set to accurately distinguish clinical stages. Evaluated on the Kaggle MRI dataset, which spans four progression levels—ND (non-dementia), VMiD (very mild dementia), MiD (mild dementia), and MoD (moderate dementia)—this streamlined approach offers a balanced trade-off between efficiency and depth, facilitating robust Alzheimer’s disease staging through comprehensive neuroimaging biomarkers.
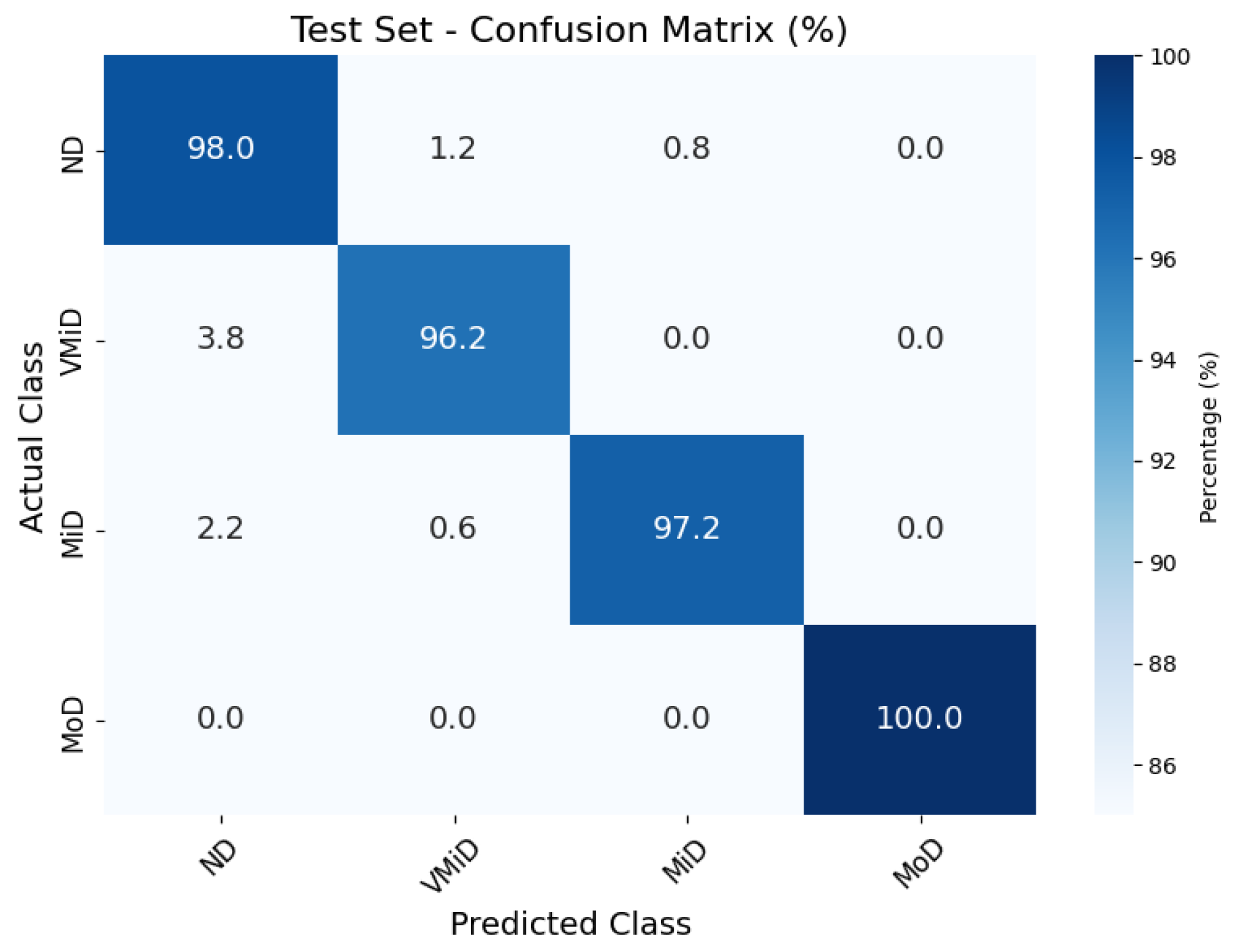
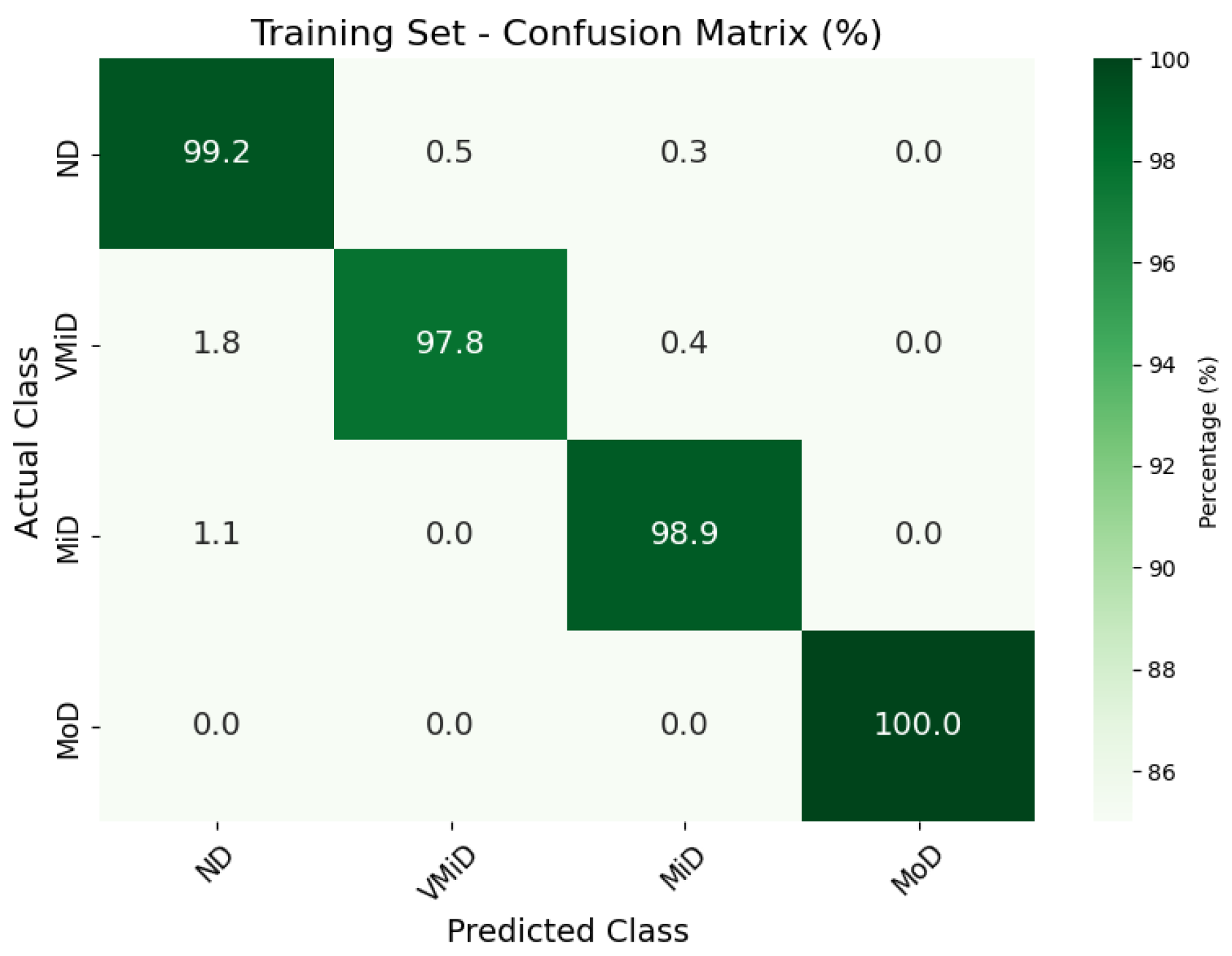
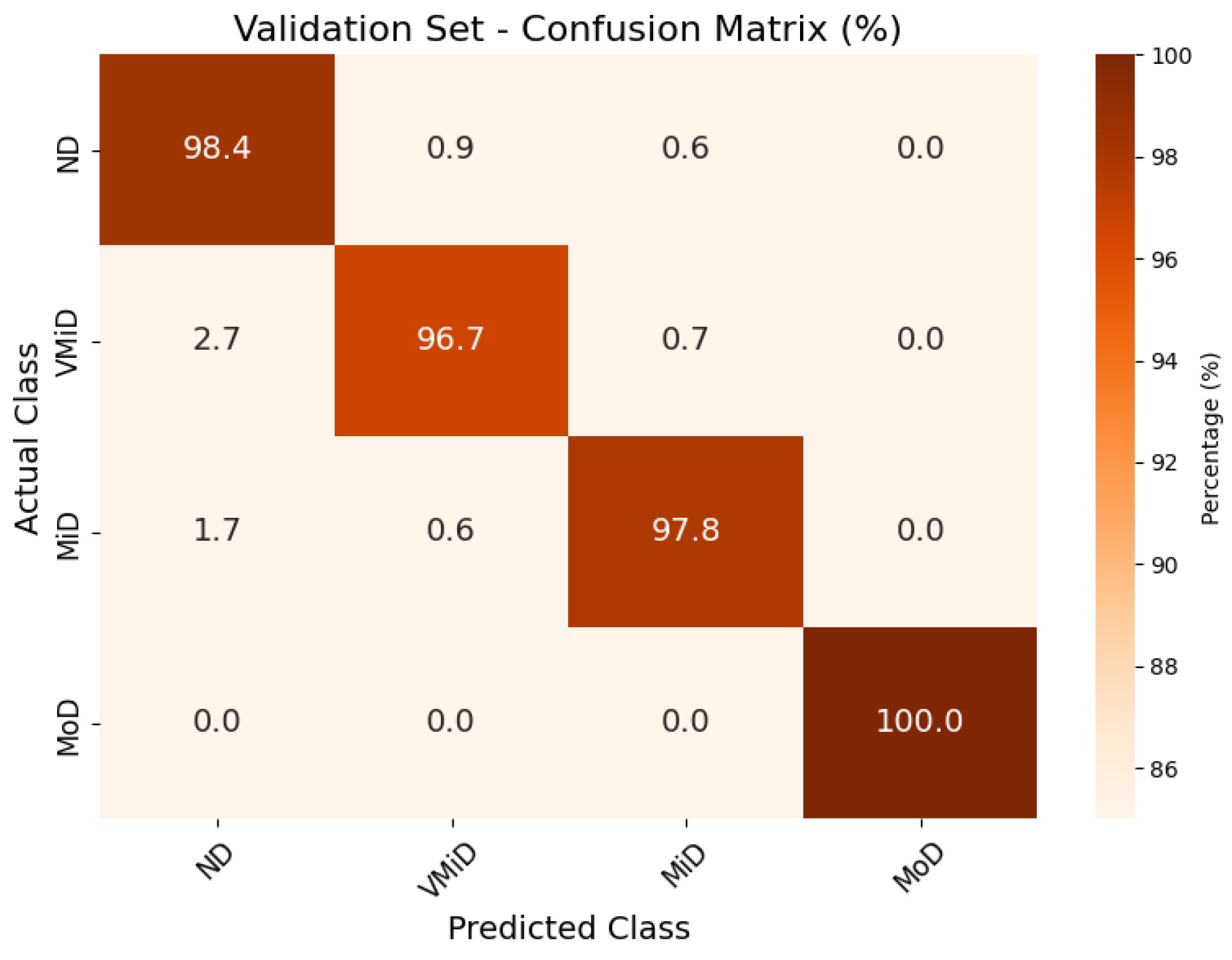
The classification results, summarized in Figure 4, Figure 5 and Figure 6, illustrate the model’s ability to distinguish between AD stages through a normalized confusion matrix. The findings demonstrate high sensitivity across all classes, with particularly strong performance in moderate dementia (MoD) at 100% and non-dementia (ND) at 98.0%, while mild dementia (MiD) also maintained a reliable recall rate (97.2%). However, very mild dementia (VMiD) exhibited slightly lower sensitivity (96.2%), suggesting minor misclassification, likely due to overlapping features with adjacent stages. The distribution of false negatives across neighboring classes rather than a single concentrated category indicated a gradual misclassification trend, which aligns with the progressive nature of dementia. Overall, the model demonstrated robust performance in staging AD. The confusion matrices for the training, validation, and test sets showed that the model consistently classified most cases correctly across all dementia stages. Misclassifications were minimal and mainly occurred between adjacent stages, which was clinically expected given the gradual progression of the disease. The moderate dementia (MoD) class was perfectly classified in all datasets; however, further experiments with larger and more diverse samples are needed to confirm the reliability of this finding. Overall, the results demonstrate strong and dependable classification performance with very low error rates.
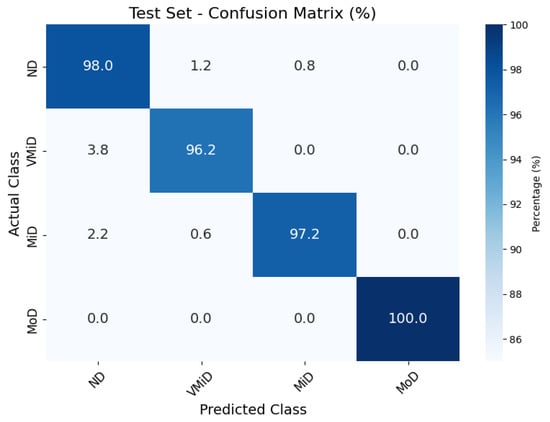
Figure 4.
Confusion matrix for AD classification on Kaggle MRI dataset (test set).
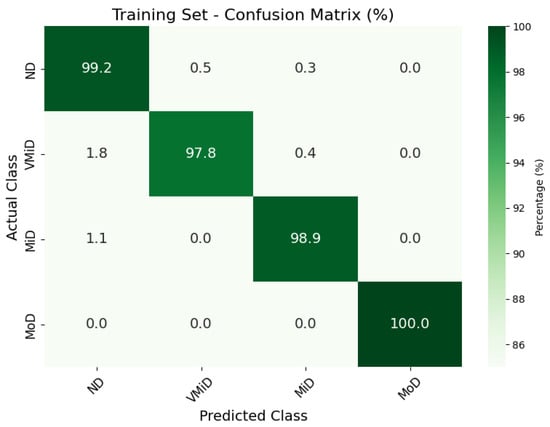
Figure 5.
Confusion matrix for AD classification on Kaggle MRI dataset (training set).
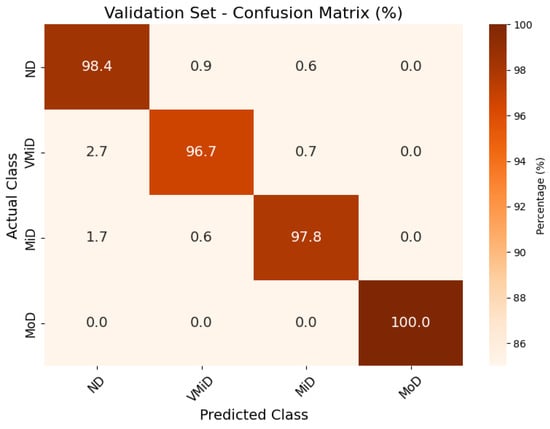
Figure 6.
Confusion matrix for AD classification on Kaggle MRI dataset (validation set).
The results presented in Table 2 offer a comprehensive assessment of the model’s ability to classify different stages of dementia with a high degree of accuracy. With classification rates exceeding 96% across all categories, the model demonstrated strong differentiation between Alzheimer’s disease stages. Non-dementia (ND) and mild dementia (MiD) exhibited particularly high sensitivity and precision, effectively identifying affected cases while keeping false positives low. Moderate dementia (MoD) achieved perfect classification, but, given its limited sample size (12 cases), further validation with a larger dataset is necessary to ensure reliability. Very mild dementia (VMiD) displayed slightly lower sensitivity (96.1%), likely due to overlapping features with adjacent stages, leading to minor misclassification. Across all classes, the high F1-scores indicated a well-balanced performance in terms of precision and recall, confirming the robustness of the model in staging AD.

Table 2.
Performance metrics across dementia stages.
The results in Table 2 clearly demonstrate the strength of the proposed DWT + Red-KPLS + 1D-CNN pipeline for Alzheimer’s disease staging, significantly outperforming conventional kernel-based methods. By employing DWT, the model effectively extracts multiscale texture features from MRI scans, enhancing the representation of subtle neuroimaging patterns. Red-KPLS feature reduction further refines these features, isolating the most relevant components while addressing the challenges of high-dimensional data. The final classification step, powered by a 1D-CNN, ensures robust decision-making by leveraging the optimized feature set, achieving exceptional sensitivity (97.9%), specificity (97.6%), accuracy (96.9%), and F1-score (97.8%).
Table 2, Table 3 and Table 4 present the performance metrics across dementia stages for the test, training, and validation sets, respectively. The model demonstrated consistent performance across all subsets, with a gradual and expected decrease in performance from training to validation to test sets. The training set showed the highest overall F1-score of 99.0%, followed by the validation set at 97.9% and the test set at 97.8%, reflecting a total performance drop of just 1.2 percentage points. This smooth decline suggests effective regularization and no signs of overfitting.

Table 3.
Performance metrics (training set).

Table 4.
Performance metrics (validation set).
Class-specific analysis confirmed that all dementia stages maintained clinically meaningful performance on the test set, with F1-scores exceeding 96% across all classes. The non-dementia (ND) class achieved the strongest performance, with an F1-score of 97.7%, while the very mild dementia (VMiD) class displayed the most stable degradation pattern across the datasets (97.9% → 96.9% → 96.3%). The moderate dementia (MoD) class was perfectly classified across all subsets, although this result should be interpreted cautiously due to the small sample size (n = 12).
The validation set closely predicted the test set performance, with a minimal F1-score difference of 0.1 percentage points, supporting its reliability for model selection. Specificity also remained robust (>97.5% across all datasets), indicating strong negative predictive power and further confirming the model’s generalization capability.
Overall, these results suggest that the model generalizes well and performs particularly well in distinguishing non-dementia and early-stage dementia cases, which is especially valuable for early clinical intervention. Further validation on larger, more diverse datasets is recommended to confirm the robustness of these findings, particularly for the moderate dementia class.
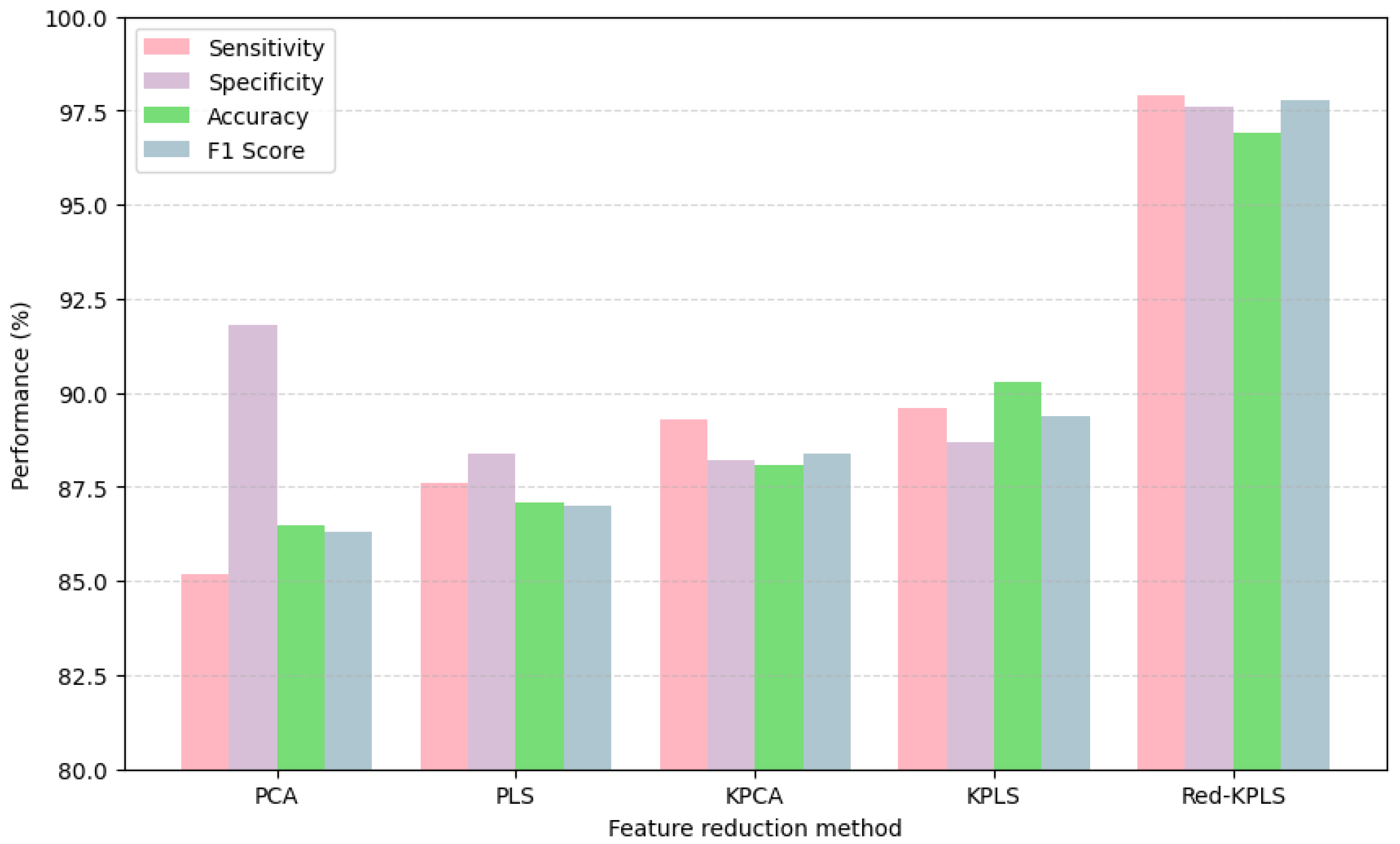
Demonstrating a clear advantage over traditional kernel-based approaches, Table 5 and the bar chart presented in Figure 7 highlight the effectiveness of the proposed DWT + Red-KPLS + CNN pipeline for AD staging. By leveraging DWT, the model captures intricate multiscale texture features, enriching neuroimaging biomarkers crucial for dementia classification. Red-KPLS, a refined feature reduction technique, enhances model efficiency by isolating the most discriminative components from high-dimensional wavelet coefficients, ensuring robust data representation without compromising essential information. The final classification step, powered by a 1D-CNN, utilizes the optimized feature set to achieve remarkable sensitivity (97.9%), specificity (97.6%), accuracy (96.9%), and F1-score (97.8%), significantly surpassing conventional methods such as PCA, PLS, KPCA, and KPLS. Compared to KPLS, which achieves 90.3% accuracy, the proposed method provides a 6.6 percentage point improvement, reinforcing the advantages of combining wavelet-based preprocessing, advanced kernel-based refinement, and deep learning architectures. This framework establishes a robust foundation for high-resolution AD staging, offering a more precise, efficient, and clinically viable approach to neuroimaging-based diagnostics, potentially advancing early detection and personalized treatment strategies.

Table 5.
Comparison of the proposed algorithm with other kernel-based methods using the Kaggle database.
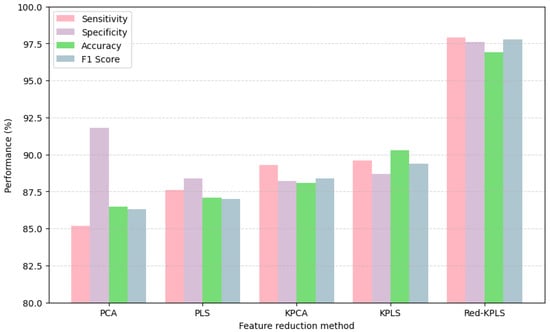
Figure 7.
Performance comparison of kernel-based methods for AD classification.
Our comparative analysis in Table 5 evaluates the proposed method against conventional kernel-based approaches (PCA, PLS, KPCA, KPLS). The consistent 6–10% performance improvements across all metrics—including 97.9% sensitivity and 97.6% specificity—underscore the strengths of our DWT + Red-KPLS pipeline. While the current study is limited to a single curated dataset, this is balanced by the method’s interpretability advantages: (1) the wavelet-derived features capture neurologically meaningful multiscale patterns, unlike the opaque representations often produced by deep learning models, and (2) Red-KPLS’s prototype selection strategy helps retain clinically relevant biomarkers while reducing dimensionality. This combination of strong performance and interpretability makes our framework a compelling choice for clinical applications, where transparency is as important as accuracy.
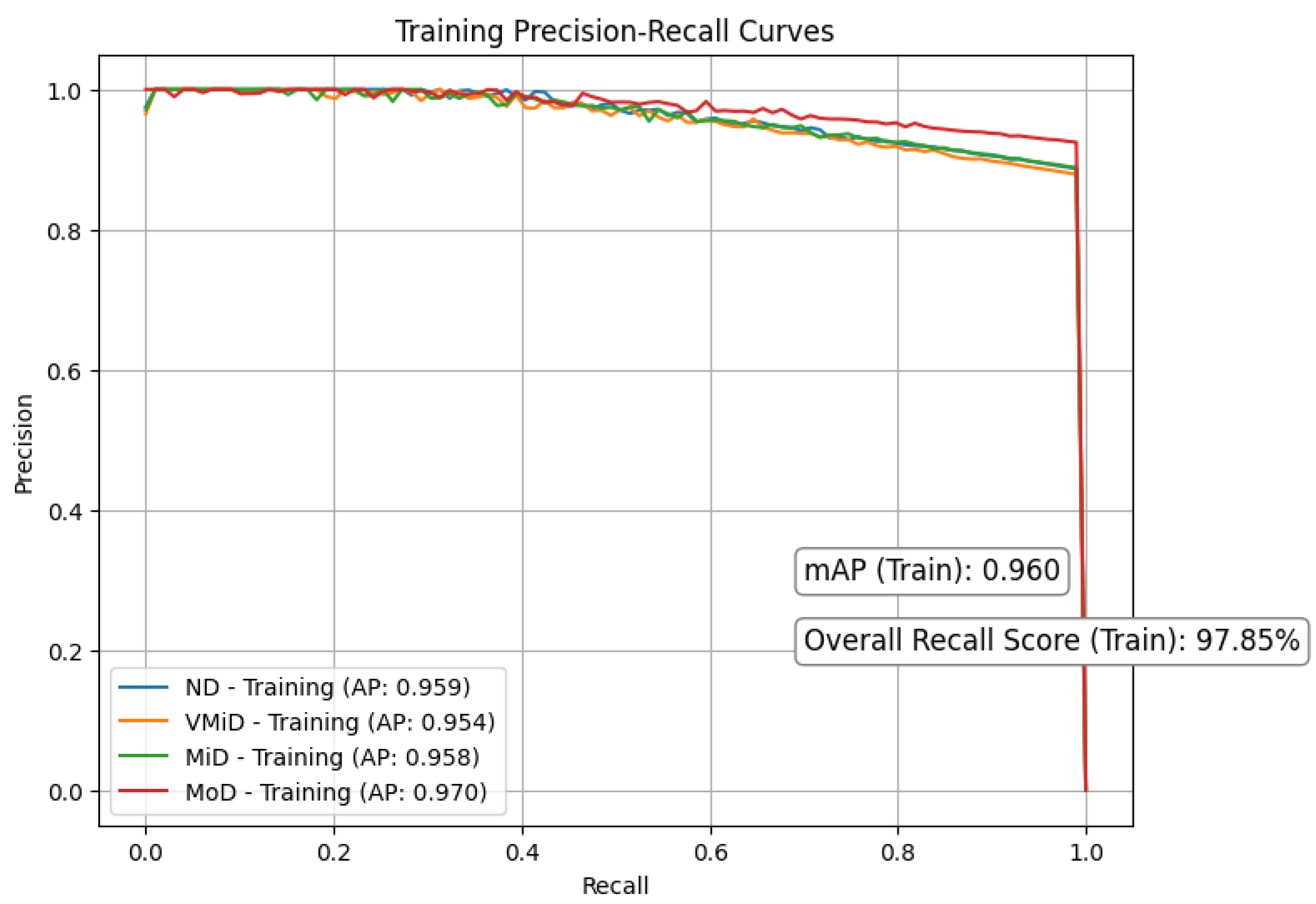
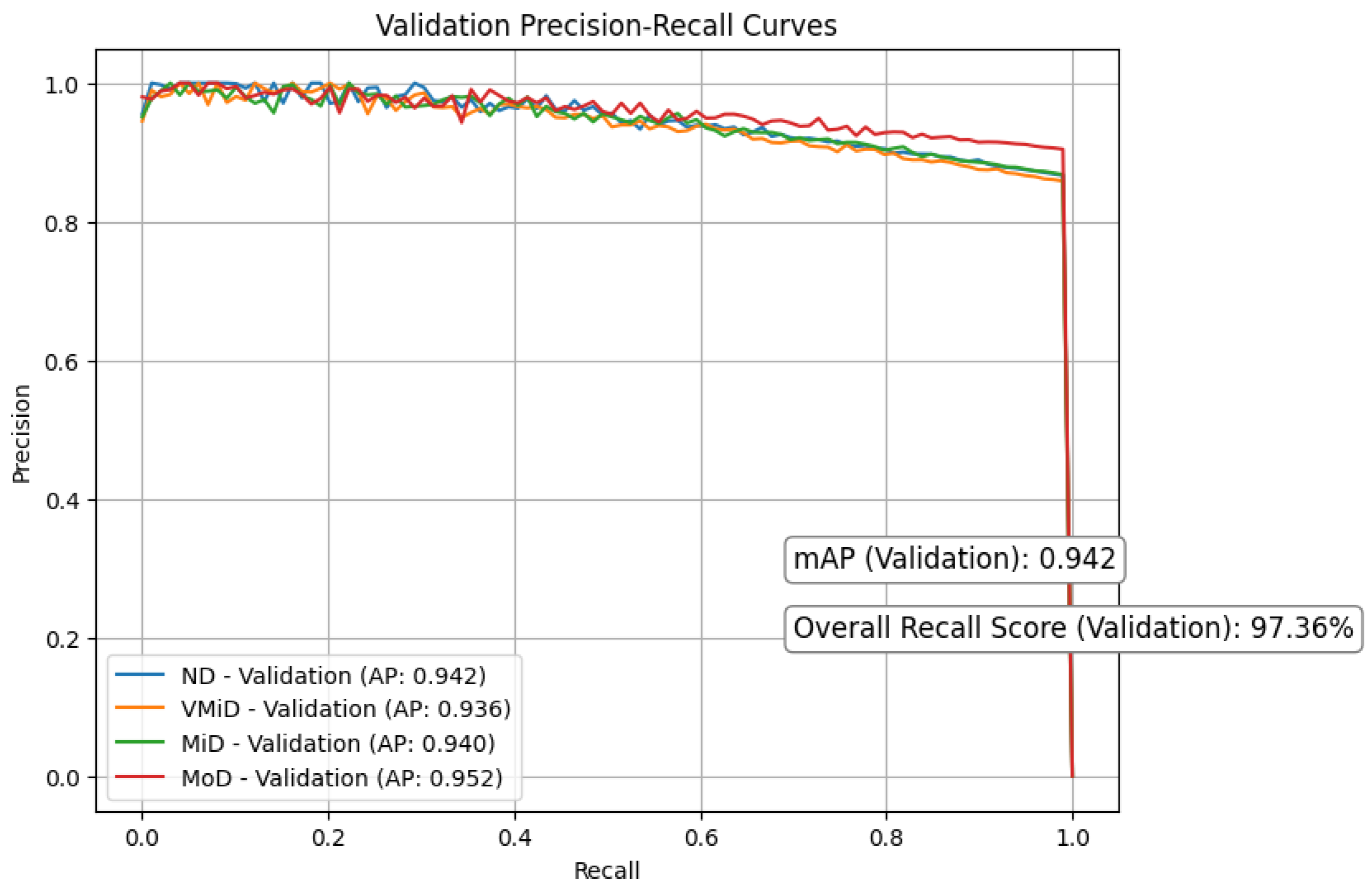
Figure 8 and Figure 9 respectively present the precision–recall curves for the validation and training datasets, providing a visual illustration of model performance across different dementia stages. Our study shows strong and consistent performance across all dementia stages. Using 5-fold cross-validation, we observed closely aligned mean average precision (mAP) between training () and validation () sets, with only a modest performance gap of . This reliability stemmed from a carefully designed pipeline that integrates discrete wavelet transform (DWT) for multi-resolution feature extraction, Red-KPLS for efficient dimensionality reduction (reducing complexity from to ), and a tailored 1D-ResNet50 model for classification.
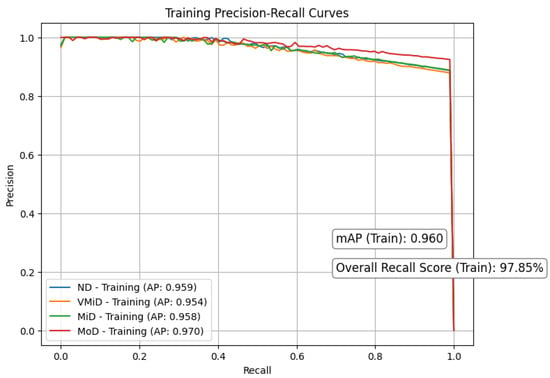
Figure 8.
Precision recall curves for training.
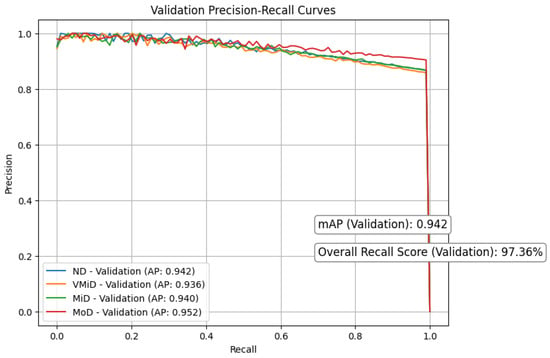
Figure 9.
Precision recall curves for validation.
To prevent overfitting, we implemented several precautions: (1) patient-wise stratified data splitting, (2) Monte Carlo dropout () and label smoothing (), and (3) independent preprocessing per fold, including bias correction, spatial normalization, and intensity scaling. Most classes (ND, VMiD, and MiD) exhibited high stability, with AP scores between and and minimal recall drop (). As expected, the MoD class showed more variability (AP: –) due to its small sample size (). We addressed this through weighted Red-KPLS, gentle augmentation, and attention-enhancing squeeze–excitation modules. Validation PR curves showed only a drop in precision, supporting the model’s robustness. For further clinical reliability, we recommend incorporating Bayesian uncertainty, expanding rare class representation via federated learning, and validating on external datasets such as ADNI-3.
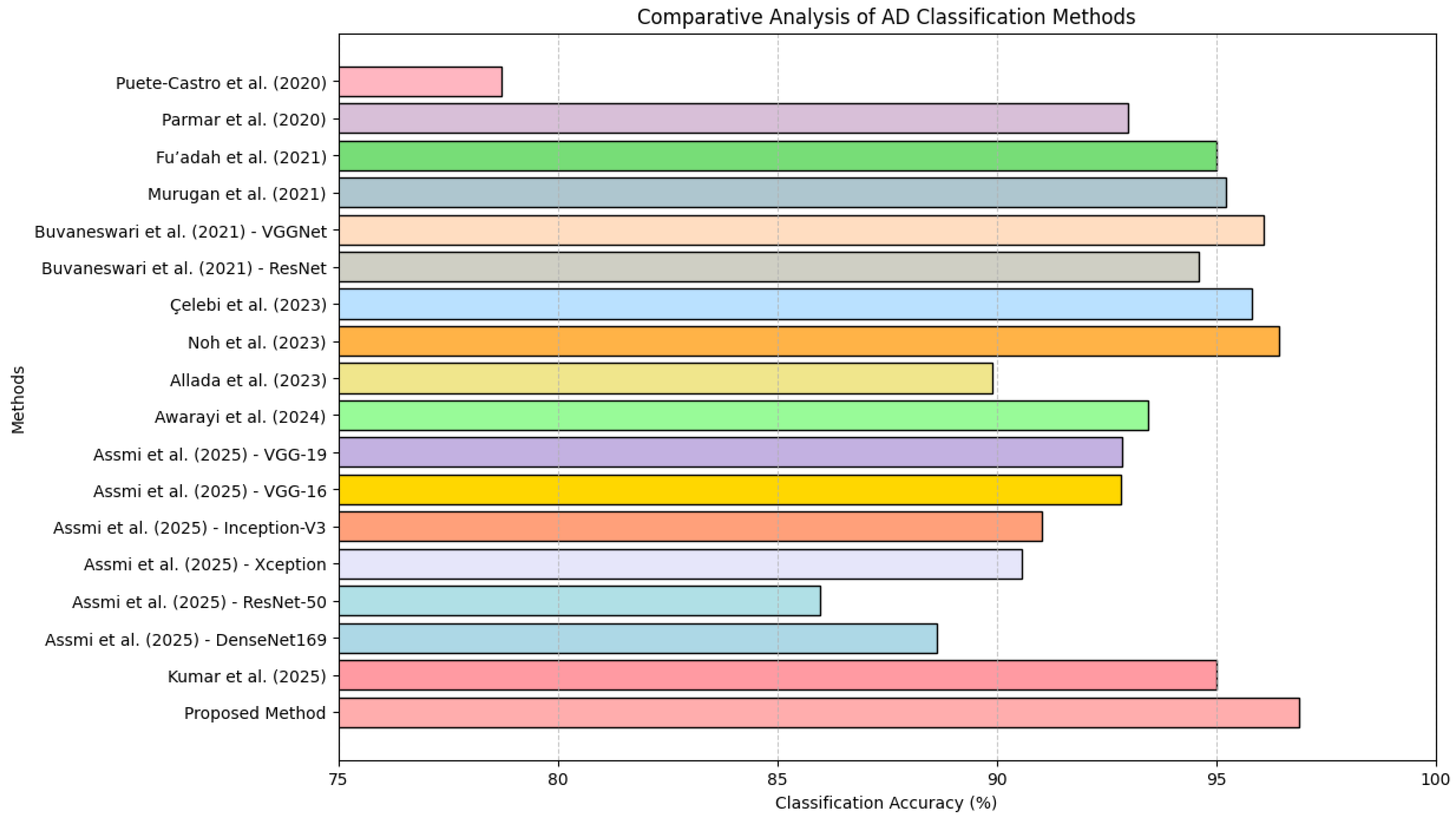
A comparative analysis of AD classification methods is presented in Table 6 and the bar chart presented in Figure 10, providing a detailed overview of existing techniques and their respective classification accuracies. The proposed method, which integrates 2D-DWT preprocessing, the novel Red-KPLS feature reduction, and ResNet-50 for classification, represents a meaningful advancement in Alzheimer’s MRI classification. While existing approaches such as ResNet-18 + SVM (78.72%) by Puete-Castro et al. [30] and neuro-fuzzy methods (89.9%) by Allada et al. [31] demonstrate promising results, they face challenges in optimizing feature selection. Similarly, widely used deep learning models such as AlexNet (95%) [32], VGG-19 (92.86%) [33], and 3D CNN (93%) [17] have made strides in classification accuracy yet remain vulnerable to high-dimensional noise and redundant features. In contrast, the introduction of Red-KPLS in the proposed framework enhances feature extraction, isolating the most relevant patterns before classification with ResNet-50, ultimately leading to a higher accuracy (96.9%).
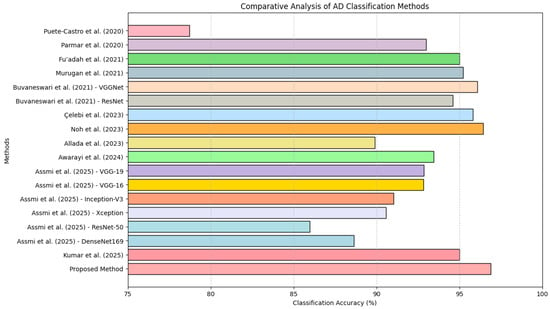
Figure 10.
Classification accuracy of AD detection methods: a comparative analysis [17,30,31,32,33,34,35,36,37,38,39].
Compared to models incorporating advanced preprocessing techniques—such as VGGNet (96.08%) [34], Xception (95.81%) [35], and 3D-CNN-LSTM (96.43%) [36] the proposed methodology offers a refined balance of feature reduction and deep learning optimization. Notably, ResNet-50 alone yielded only 85.99% accuracy [33], reinforcing the value of incorporating wavelet-based preprocessing and Red-KPLS refinement. The combined approach mitigates the effects of MRI noise, improves model generalization, and ensures robust classification performance. These results emphasize that carefully designed preprocessing and feature selection strategies are essential for maximizing diagnostic accuracy in neuroimaging applications.
The proposed method advances AD classification by bridging feature optimization and deep learning architectures, outperforming conventional CNN-based models while ensuring higher precision in distinguishing disease stages. This work underscores the significance of hybrid methodologies and offers a promising direction for future neuroimaging-based diagnostic frameworks.

Table 6.
Comparative analysis of AD classification methods.
Table 6.
Comparative analysis of AD classification methods.
| Authors | Biomarker | Database | Methodology | Accuracy |
|---|---|---|---|---|
| Puete-Castro A. et al. (2020) [30] | MRI | ADNI | Resnet18 + SVM | 78.72% |
| Parmar H. et al. (2020) [17] | MRI | ADNI | 3D CNN | 93.00% |
| Fu’adah Y. N. et al. (2021) [37] | MRI | ADNI | AlexNet | 95.00% |
| Murugan S. et al. (2021) [38] | MRI | ADNI | CNN | 95.23% |
| Buvaneswari P. R. et al. (2021) [34] | VBM | ADNI | VGGNet | 96.08% |
| ResNet | 94.60% | |||
| Çelebi, S. B. et al. (2023) [35] | TBM | ADNI | Xception architecture | 95.81% |
| Noh J. H. et al. (2023) [36] | fMRI | ADNI | 3D-CNN-LSTM | 96.43% |
| classification model | ||||
| based deep | ||||
| dense block | ||||
| Allada A. et al. (2023) [31] | MRI | Kaggle | neuro-fuzzy | 89.9% |
| Awarayi, N. S. et al.(2024) [39] | MRI | ADNI | Custum CNN | 93.45% |
| Assmi A. et al. (2025) [33] | MRI | Kaggle | VGG-19 | 92.86% |
| VGG-16 | 92.83% | |||
| Inception-V3 | 91.04% | |||
| Xception | 90.57% | |||
| ResNet-50 | 85.99% | |||
| DenseNet169 | 88.64% | |||
| Kumar, K. P. V. et al. (2025) [32] | MRI | Kaggle | AlexNet | 95% |
| Proposed Method | MRI | Kaggle | Red-KPLS + ResNet-50 | 96.9% |
This study utilized the Kaggle MRI dataset, which provides standardized preprocessed images ideal for methodological development but presents certain inherent constraints. While our carefully designed validation framework (patient-level data splits and nested cross-validation) ensured robust performance estimates within this dataset, further validation on larger, multi-center cohorts would strengthen clinical applicability. The preprocessed nature of the images, while ensuring consistency, prevents the analysis of acquisition parameter effects. Future work could explore these factors through collaboration with clinical imaging centers.
4.4. Clinical Relevance and Limitations
The results presented in this study highlight the strong performance of our proposed DWT + Red-KPLS + CNN framework for classifying AD stages. With an overall accuracy of 96.9% and balanced sensitivity across all classes, these findings hold meaningful clinical value. In practice, even small improvements in diagnostic accuracy can have a real impact—especially when it comes to detecting early or subtle stages of AD. For instance, accurately distinguishing between very mild dementia (VMiD) and mild dementia (MiD)—a distinction that is often difficult in clinical settings—can support more timely interventions and better care planning. It is also worth noting that most misclassifications occurred between adjacent stages, which reflects the gradual nature of AD progression and suggests that the model’s predictions align well with clinical expectations.
That said, there are a few important limitations to acknowledge. First, our experiments were conducted on a single publicly available dataset, which may not fully capture the variability seen in clinical populations or across different imaging protocols. Additionally, the sample size for certain categories—such as moderate dementia (MoD)—was relatively small, and this could have influenced the performance results. Another limitation is that our model was tested on cross-sectional data only, meaning that it does not yet capture disease progression over time. Finally, while the framework incorporates some degree of interpretability, it does not currently integrate other clinical indicators, such as cognitive test scores or biological markers, which are often part of real-world diagnostic decision-making.
Looking ahead, future research should aim to address these limitations. Testing the model on additional datasets, especially those with more diverse sources like ADNI, would help assess its generalizability. Incorporating longitudinal MRI scans could also allow us to model the progression of AD more effectively. Furthermore, combining MRI data with other types of information—such as PET imaging, cerebrospinal fluid biomarkers, or cognitive assessments—could enhance the model’s clinical relevance and utility. Finally, a valuable next step would be to adapt the pipeline into a lightweight, clinically deployable tool that can assist healthcare professionals in making more accurate and timely diagnoses.
5. Ethical Considerations in AI-Assisted AD
The integration of AI into Alzheimer’s diagnosis brings significant ethical responsibilities that must be carefully considered. One of the primary concerns is algorithmic bias [40]. If the training data does not sufficiently represent diverse populations, the model’s accuracy may vary across demographic groups, potentially reinforcing existing healthcare disparities. To address this, it is essential to rigorously validate AI models across different ethnicities, age groups, and genders [41]. Transparency and explainability are equally important. Both clinicians and patients need to understand how AI models generate their predictions to foster trust and support accurate clinical interpretation [42]. Prioritizing explainable AI over black-box models allows healthcare providers to critically assess and validate AI-generated results, rather than simply accepting outputs without understanding the reasoning behind them [42]. Patient consent and data privacy are also crucial, especially in the context of AD, where cognitive impairment may limit a patient’s ability to provide fully informed consent. Clear, ethical protocols must govern how patient data is collected, stored, and shared, with strict adherence to data protection laws and medical ethical standards [43]. Importantly, AI should complement—not replace—clinical judgment. These systems should serve as decision-support tools, with physicians maintaining full responsibility for diagnosis and patient care. Continuous oversight is necessary to guard against over-reliance on AI and ensure sustained model performance over time [44]. By thoughtfully addressing these ethical challenges, AI can be responsibly and effectively integrated into Alzheimer’s disease diagnosis, supporting patient safety, fairness, and trust in medical decision-making [45].
6. Conclusions and Future Work
Alzheimer’s disease (AD) remains one of the most pressing challenges in neuroimaging-based diagnostics, requiring ever-evolving methodologies to improve classification accuracy and early detection. In this study, we introduced an optimized deep learning framework that combines 2D-DWT preprocessing, Red-KPLS for feature reduction, and ResNet-50 for classification, achieving a 96.9% accuracy rate, a potentially significant improvement that requires validation in independent cohorts. By integrating wavelet-based feature extraction and reduced kernel partial least squares (Red-KPLS), the method effectively refines feature selection, mitigating high-dimensional noise and enhancing the model’s generalization capabilities for the studied population. These refinements contribute to more robust and reliable computer-aided diagnosis (CAD) tools, offering greater precision in identifying different stages of AD based on MRI scans as part of clinician-guided workflows.
Three key limitations warrant consideration: (1) the single-institution data source despite multicenter collection, (2) the small MoD sample size affecting reliability estimates, and (3) the need for standardized benchmarking against alternative splitting methods. These factors currently constrain broad clinical applicability but define clear pathways for improvement.
Building upon these findings, future research should prioritize (1) external validation across healthcare systems and (2) explicit overfitting tests for minority classes, before exploring multi-modal imaging approaches (e.g., fMRI/PET) to enhance classification robustness. Expanding the framework to include larger and more diverse datasets will be critical in validating its effectiveness across broader patient populations. Moreover, incorporating self-supervised learning strategies and attention mechanisms into deep networks may refine feature representations, improving early-stage detection and making the model more adaptable to real-world clinical scenarios. Ultimately, these advancements aim to provide physicians and researchers with more precise, interpretable, and scalable diagnostic solutions that support clinical decision-making, reinforcing the role of AI-driven methodologies in the fight against Alzheimer’s disease through responsible, evidence-based implementation.
Author Contributions
Conceptualization, S.N., A.F. and K.M.; Methodology, S.N.; Software, S.N.; Validation, A.F.; Formal analysis, A.F.; Investigation, S.N.; Resources, K.M.; Writing—review & editing, K.M. and K.B.; Supervision, K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Scientific Research Deanship at the University of Ha’il, Saudi Arabia, through project number <<RG-24 163>>.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Acar, M.; Şeker, B.; Uğur, S. Radio-Anatomical Assessment of Cerebellum Volume in Individuals with Alzheimer’s Disease. Curr. Alzheimer Res. 2024, 21, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Mrdjen, D.; Fox, E.J.; Bukhari, S.A.; Montine, K.S.; Bendall, S.C.; Montine, T.J. The basis of cellular and regional vulnerability in Alzheimer’s disease. Acta Neuropathol. 2019, 138, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Baek, S.H.; Lai, M.K.; Arumugam, T.V.; Jo, D.G. Aging-associated sensory decline and Alzheimer’s disease. Mol. Neurodegener. 2024, 19, 1–28. [Google Scholar] [CrossRef]
- Zhang, N.; Chai, S.; Wang, J. Assessing and projecting the global impacts of Alzheimer’s disease. Front. Public Health 2025, 12, 1453489. [Google Scholar] [CrossRef]
- Vogt, A.C.S.; Jennings, G.T.; Mohsen, M.O.; Vogel, M.; Bachmann, M.F. Alzheimer’s disease: A brief history of immunotherapies targeting amyloid β. Int. J. Mol. Sci. 2023, 24, 3895. [Google Scholar] [CrossRef]
- Neffati, S.; Ben Abdellafou, K.; Aljuhani, A.; Taouali, O. An enhanced CAD system based on machine Learning Algorithm for brain MRI classification. J. Intell. Fuzzy Syst. 2021, 41, 1845–1854. [Google Scholar] [CrossRef]
- Muir, R.T.; Hill, M.D.; Black, S.E.; Smith, E.E. Minimal clinically important difference in Alzheimer’s disease: Rapid review. Alzheimer’s Dement. 2024, 20, 3352–3363. [Google Scholar] [CrossRef] [PubMed]
- Ponce de Leon-Sanchez, E.R.; Mendiola-Santibañez, J.D.; Dominguez-Ramirez, O.A.; Herrera-Navarro, A.M.; Vazquez-Cervantes, A.; Jimenez-Hernandez, H.; Cordova-Esparza, D.M.; Hernández, M.d.A.C.; Senties-Madrid, H. Training Artificial Neural Networks to Detect Multiple Sclerosis Lesions Using Granulometric Data from Preprocessed Magnetic Resonance Images with Morphological Transformations. Technologies 2024, 12, 145. [Google Scholar] [CrossRef]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef]
- Neffati, S.; Ben Abdellafou, K.; Taouali, O.; Bouzrara, K. A new Bio-CAD system based on the optimized KPCA for relevant feature selection. Int. J. Adv. Manuf. Technol. 2019, 102, 1023–1034. [Google Scholar] [CrossRef]
- Bhandarkar, A.; Naik, P.; Vakkund, K.; Junjappanavar, S.; Bakare, S.; Pattar, S. Deep learning based computer aided diagnosis of Alzheimer’s disease: A snapshot of last 5 years, gaps, and future directions. Artif. Intell. Rev. 2024, 57, 30. [Google Scholar] [CrossRef]
- Pedroli, E.; Bruni, F.; Mancuso, V.; Cavedoni, S.; Bigotto, F.; Panigada, J.; Rossi, M.; Boilini, L.; Goulene, K.; Stramba-Badiale, M.; et al. Training Cognitive Functions Using DUAL-REHAB, a New Dual-Task Application in MCI and SMC: A Study Protocol of a Randomized Control Trial. Technologies 2025, 13, 96. [Google Scholar] [CrossRef]
- Fayaz, M.; Torokeldiev, N.; Turdumamatov, S.; Qureshi, M.S.; Qureshi, M.B.; Gwak, J. An efficient methodology for brain MRI classification based on DWT and convolutional neural network. Sensors 2021, 21, 7480. [Google Scholar] [CrossRef]
- Kaggle Dataset. Available online: https://www.kaggle.com/ (accessed on 7 February 2025).
- Safiri, S.; Ghaffari, J.A.; Fazlollahi, A.; Morsali, S.; Sarkesh, A.; Daei, S.A.; Golabi, B.; Aletaha, R.; Motlagh, A.K.; Hamidi, S.; et al. Alzheimer’s disease: A comprehensive review of epidemiology, risk factors, symptoms diagnosis, management, caregiving, advanced treatments and associated challenges. Front. Med. 2024, 11, 1474043. [Google Scholar] [CrossRef] [PubMed]
- Golovanevsky, M.; Eickhoff, C.; Singh, R. Multimodal attention-based deep learning for Alzheimer’s disease diagnosis. J. Am. Med. Inform. Assoc. 2022, 29, 2014–2022. [Google Scholar] [CrossRef]
- Parmar, H.; Nutter, B.; Long, R.; Antani, S.; Mitra, S. Spatiotemporal feature extraction and classification of Alzheimer’s disease using deep learning 3D-CNN for fMRI data. J. Med. Imaging 2020, 7, 056001. [Google Scholar] [CrossRef] [PubMed]
- Angkoso, C.V.; Agustin Tjahyaningtijas, H.P.; Purnama, I.; Purnomo, M.H. Multiplane Convolutional Neural Network (Mp-CNN) for Alzheimer’s Disease Classification. Int. J. Intell. Eng. Syst. 2022, 15, 1. [Google Scholar]
- Hedayati, R.; Khedmati, M.; Taghipour-Gorjikolaie, M. Deep feature extraction method based on ensemble of convolutional auto encoders: Application to Alzheimer’s disease diagnosis. Biomed. Signal Process. Control 2021, 66, 102397. [Google Scholar] [CrossRef]
- Saratxaga, C.L.; Moya, I.; Picón, A.; Acosta, M.; Moreno-Fernandez-de-Leceta, A.; Garrote, E.; Bereciartua-Perez, A. MRI deep learning-based solution for Alzheimer’s disease prediction. J. Pers. Med. 2021, 11, 902. [Google Scholar] [CrossRef]
- Jabason, E.; Ahmad, M.O.; Swamy, M.N.S. Classification of Alzheimer’s disease from MRI data using an ensemble of hybrid deep convolutional neural networks. In Proceedings of the 2019 IEEE 62nd International Midwest Symposium on Circuits and Systems (MWSCAS), Dallas, TX, USA, 4–7 August 2019; pp. 481–484. [Google Scholar]
- Hazarika, R.A.; Kandar, D.; Maji, A.K. An experimental analysis of different deep learning based models for Alzheimer’s disease classification using brain magnetic resonance images. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 8576–8598. [Google Scholar] [CrossRef]
- Oktavian, M.W.; Yudistira, N.; Ridok, A. Classification of Alzheimer’s disease using the Convolutional Neural Network (CNN) with transfer learning and weighted loss. arXiv 2022, arXiv:2207.01584. [Google Scholar]
- Hazarika, R.A.; Abraham, A.; Kandar, D.; Maji, A.K. An improved LeNet-deep neural network model for Alzheimer’s disease classification using brain magnetic resonance images. IEEE Access 2021, 9, 161194–161207. [Google Scholar] [CrossRef]
- Kutlu, H.; Avcı, E. A novel method for classifying liver and brain tumors using convolutional neural networks, discrete wavelet transform and long short-term memory networks. Sensors 2019, 19, 1992. [Google Scholar] [CrossRef] [PubMed]
- Akindele, R.G.; Yu, M.; Kanda, P.S.; Owoola, E.O.; Aribilola, I. Denoising of nifti (mri) images with a regularized neighborhood pixel similarity wavelet algorithm. Sensors 2023, 23, 7780. [Google Scholar] [CrossRef]
- Janghel, R.R.; Rathore, Y.K. Deep convolution neural network based system for early diagnosis of Alzheimer’s disease. IRBM 2021, 42, 258–267. [Google Scholar] [CrossRef]
- Yan, H.; Mubonanyikuzo, V.; Komolafe, T.E.; Zhou, L.; Wu, T.; Wang, N. Hybrid-RViT: Hybridizing ResNet-50 and Vision Transformer for Enhanced Alzheimer’s disease detection. PLoS ONE 2025, 20, e0318998. [Google Scholar] [CrossRef]
- Yu, H.; Yang, L.T.; Zhang, Q.; Armstrong, D.; Deen, M.J. Convolutional neural networks for medical image analysis: State-of-the-art, comparisons, improvement and perspectives. Neurocomputing 2021, 444, 92–110. [Google Scholar] [CrossRef]
- Puente-Castro, A.; Fernandez-Blanco, E.; Pazos, A.; Munteanu, C.R. Automatic assessment of Alzheimer’s disease diagnosis based on deep learning techniques. Comput. Biol. Med. 2020, 120, 103764. [Google Scholar] [CrossRef]
- Allada, A.; Bhavani, R.; Chaduvula, K.; Priya, R. Alzheimer’s disease classification using competitive swarm multi-verse optimizer-based deep neuro-fuzzy network. Concurr. Comput. Pract. Exp. 2023, 35, e7696. [Google Scholar] [CrossRef]
- Kumar, K.P.V.; Charan, P.; Kayalvili, S.; Santhi, M.V.B.T. Deep Learning–Based Medical Image Classification and Web Application Framework to Identify Alzheimer’s Disease. Artif. Intell. Cybersecur. Healthc. 2025, 1, 397–412. [Google Scholar] [CrossRef]
- Assmi, A.; Elhabyb, K.; Benba, A.; Jilbab, A. Alzheimer’s disease classification: A comprehensive study. Multimed. Tools Appl. 2024, 83, 70193–70216. [Google Scholar] [CrossRef]
- Buvaneswari, P.R.; Gayathri, R. Deep learning-based segmentation in classification of Alzheimer’s disease. Arab. J. Sci. Eng. 2021, 46, 5373–5383. [Google Scholar] [CrossRef]
- Çelebi, S.B.; Emiroğlu, B.G. A novel deep dense block-based model for detecting Alzheimer’s Disease. Appl. Sci. 2023, 13, 8686. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, J.H.; Yang, H.D. Classification of Alzheimer’s progression using fMRI data. Sensors 2023, 23, 6330. [Google Scholar] [CrossRef]
- Fu’adah, Y.N.; Wijayanto, I.; Pratiwi, N.K.C.; Taliningsih, F.F.; Rizal, S.; Pramudito, M.A. Automated classification of Alzheimer’s disease based on MRI image processing using convolutional neural network (CNN) with AlexNet architecture. J. Phys. Conf. Ser. 2021, 1844, 012020. [Google Scholar] [CrossRef]
- Murugan, S.; Venkatesan, C.; Sumithra, M.G.; Gao, X.Z.; Elakkiya, B.; Akila, M.; Manoharan, S. DEMNET: A deep learning model for early diagnosis of Alzheimer diseases and dementia from MR images. IEEE Access 2021, 9, 90319–90329. [Google Scholar] [CrossRef]
- Awarayi, N.S.; Twum, F.; Hayfron-Acquah, J.B.; Owusu-Agyemang, K. A bilateral filtering-based image enhancement for Alzheimer disease classification using CNN. PLoS ONE 2024, 19, e0302358. [Google Scholar] [CrossRef]
- Dang, V.N.; Casamitjana, A.; Hernández-González, J.; Lekadir, K.; Alzheimer’s Disease Neuroimaging Initiative. Mitigating Overdiagnosis Bias in CNN-Based Alzheimer’s Disease Diagnosis for the Elderly. In MICCAI Workshop on Fairness of AI in Medical Imaging; Springer: Cham, Switzerland, 2024; pp. 46–55. [Google Scholar]
- Alievska, B. Ethical Challenges Arising from the Underrepresentation of the Elderly in the Development of Artificial Intelligence (AI) for Medical Applications: Exclusion, Bias, and the Limits of Accessibility. Ph.D. Dissertation, McMaster University, Hamilton, ON, Canada, 2023. [Google Scholar]
- Petti, U.; Nyrup, R.; Skopek, J.M.; Korhonen, A. Ethical considerations in the early detection of Alzheimer’s disease using speech and AI. In Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency, Chicago, IL, USA, 12–15 June 2023; pp. 1062–1075. [Google Scholar]
- Matheen, M.A.; Hasan, Z.; Lodhi, A.K.; Waheed, S.A. Ethical and Privacy Considerations in AI-Driven AD Research. In AI-Driven Alzheimer’s Disease Detection and Prediction; IGI Global: Hershey, PA, USA, 2024; pp. 403–418. [Google Scholar]
- Babu, B.; Parvathy, G.; Bawa, F.S.M.; Gill, G.S.; Patel, J.; Sibia, D.S.; Sureddi, J.; Patel, V.; Bawa, F.M.; Sibia, D.; et al. Comparing the Artificial Intelligence Detection Models to Standard Diagnostic Methods and Alternative Models in Identifying Alzheimer’s Disease in At-Risk or Early Symptomatic Individuals: A Scoping Review. Cureus 2024, 16, e75389. [Google Scholar] [CrossRef]
- Kothinti, R.R. Artificial intelligence in healthcare: Revolutionizing precision medicine, predictive analytics, and ethical considerations in autonomous diagnostics. World J. Adv. Res. Rev. 2024, 19, 3395–3406. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).