STAMINA: Bioinformatics Platform for Monitoring and Mitigating Pandemic Outbreaks
Abstract
:1. Introduction
2. Related Work
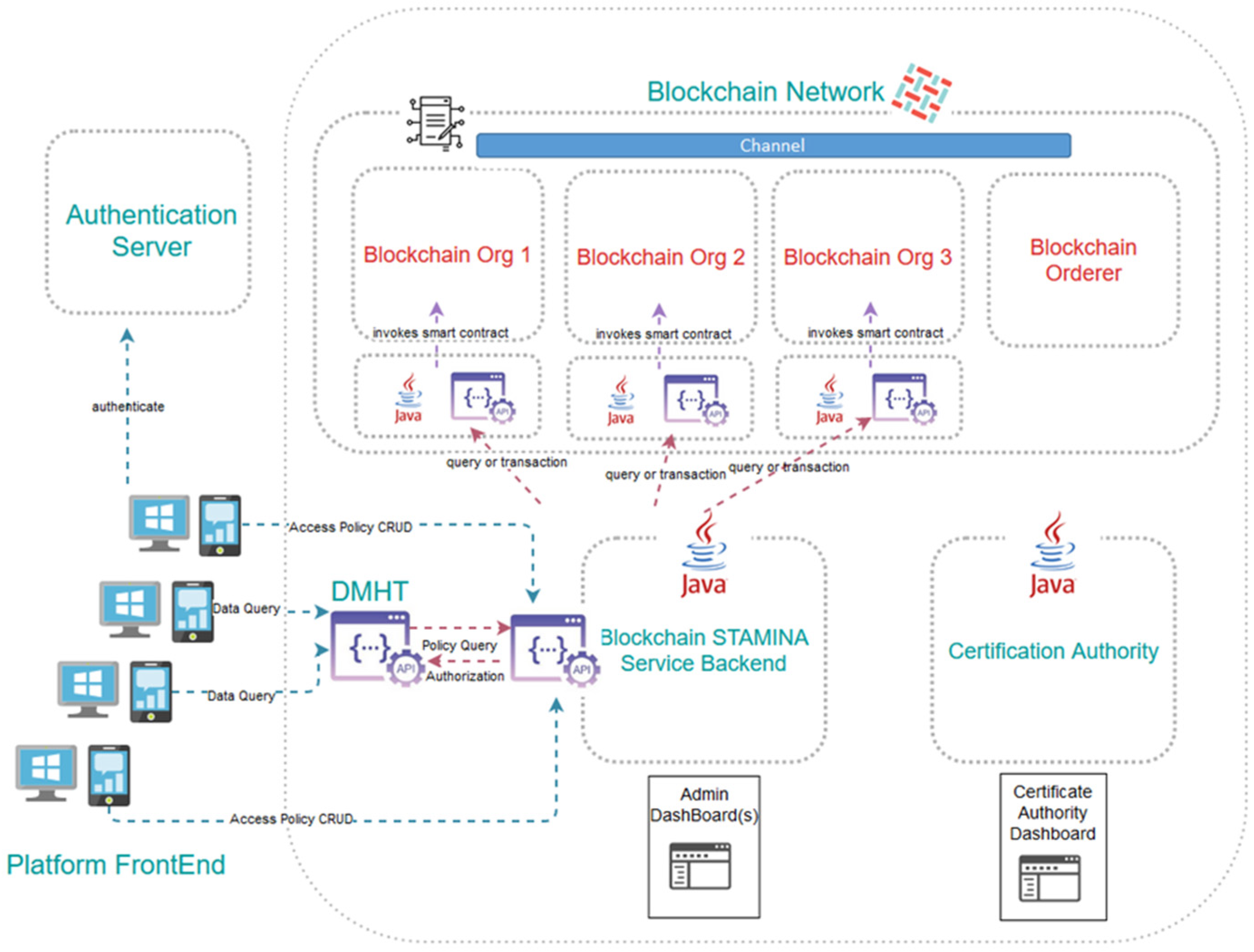
3. Overall System Architecture
4. Platform Components
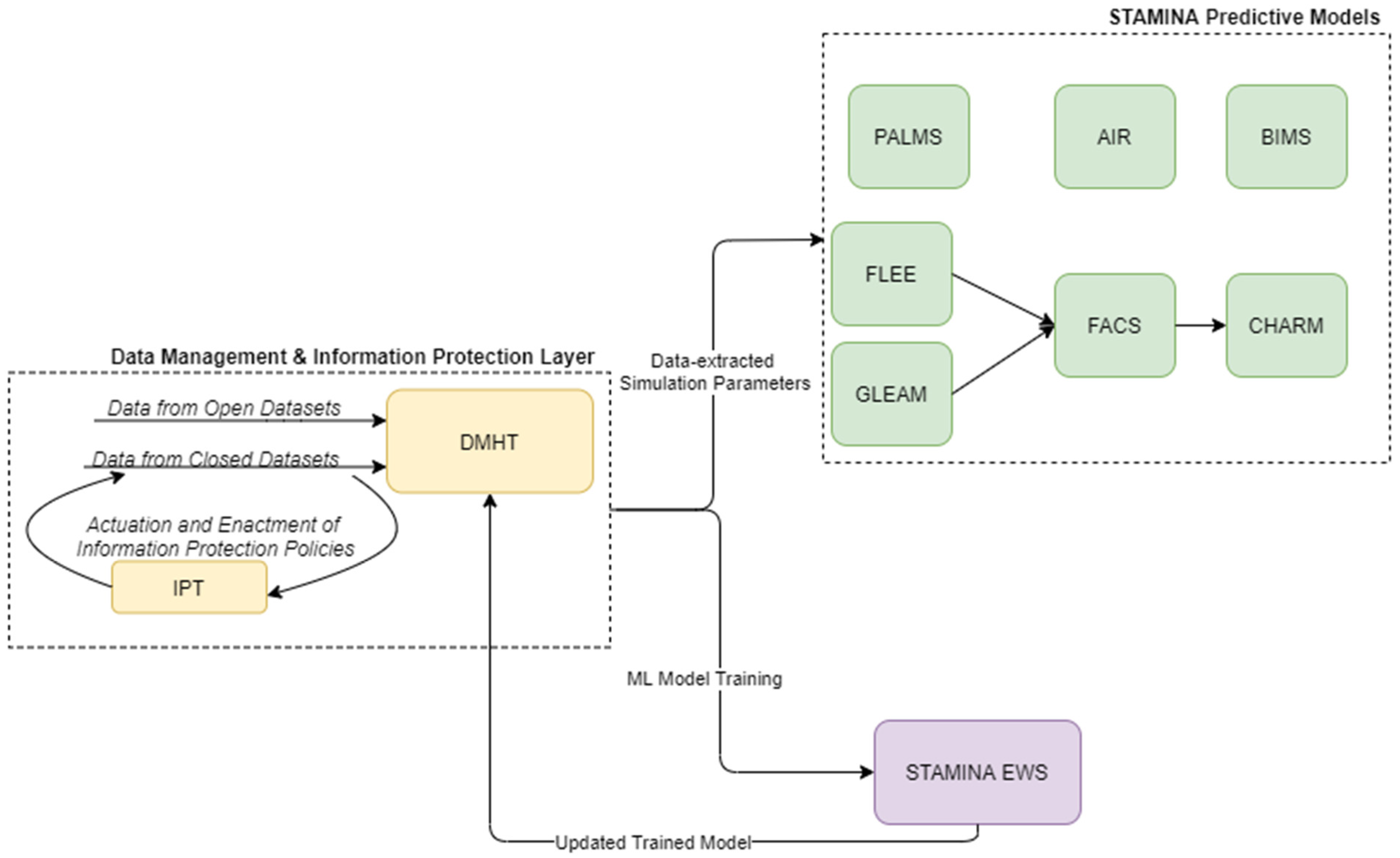
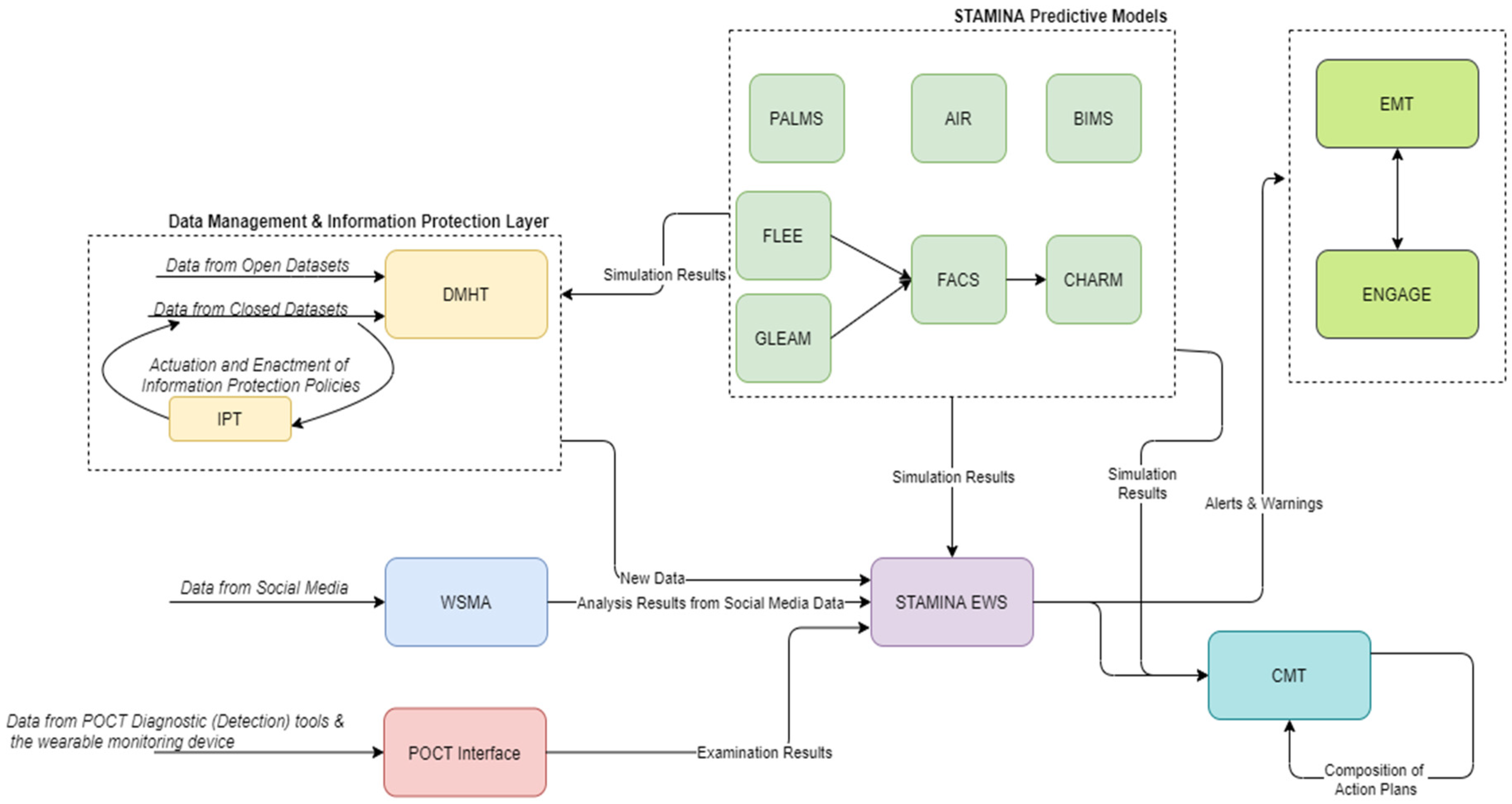
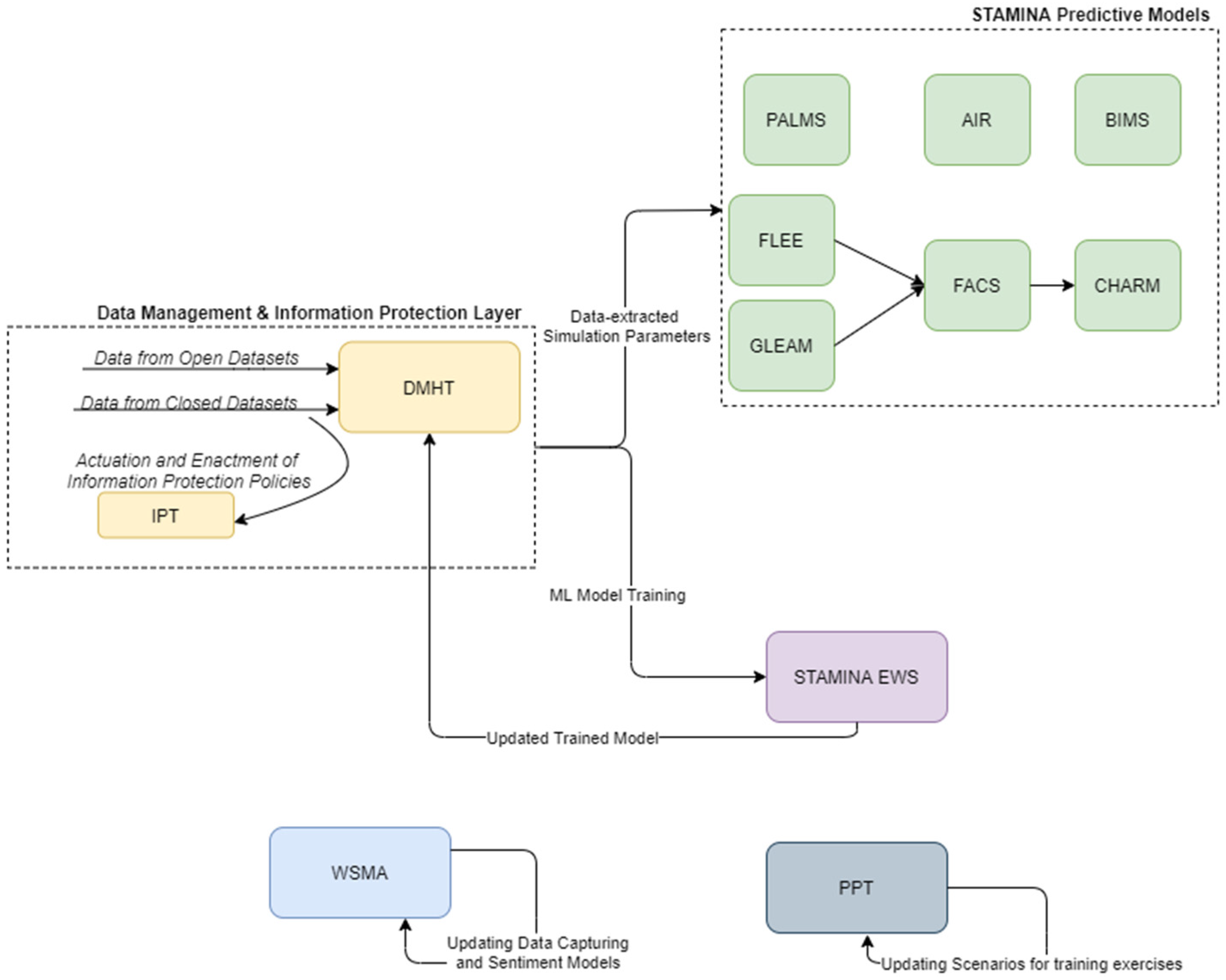
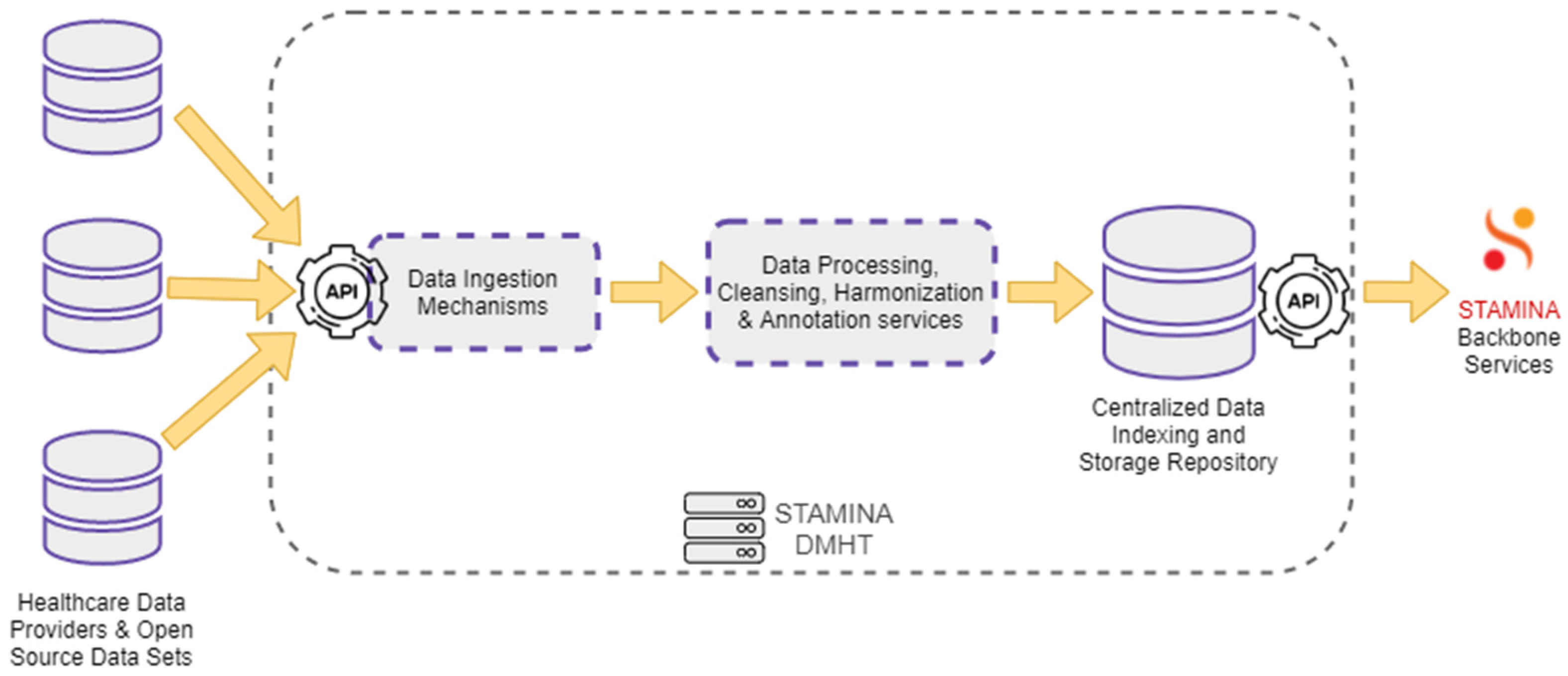
4.1. Data Management
4.2. Information Protection
4.3. Platform Intelligence, Prediction and Aleting Services
4.3.1. FACS
4.3.2. FLEE
4.3.3. CHARM
4.3.4. AIR
- To allow for the examination of the predicted excess mortality due to antimicrobial resistance. The AIR model will extend its design to present x conditions of Multi-Task Learning (MTL) model architectures to predict output estimates (i.e., number of deaths due to prolonged symptoms and mortality rates) for a specified time period.
- To demonstrate the usefulness of incorporating additional patients’ contextual epidemiological information.
- To perform an association analysis among the patients’ data so that when joined with the primary task (initial input information) it could perform better and thus provide an improved computationally predictive excess mortality model.
4.3.5. PALMS
4.3.6. BIMS
4.3.7. GLEAM
4.3.8. Web and Social Media Analytics
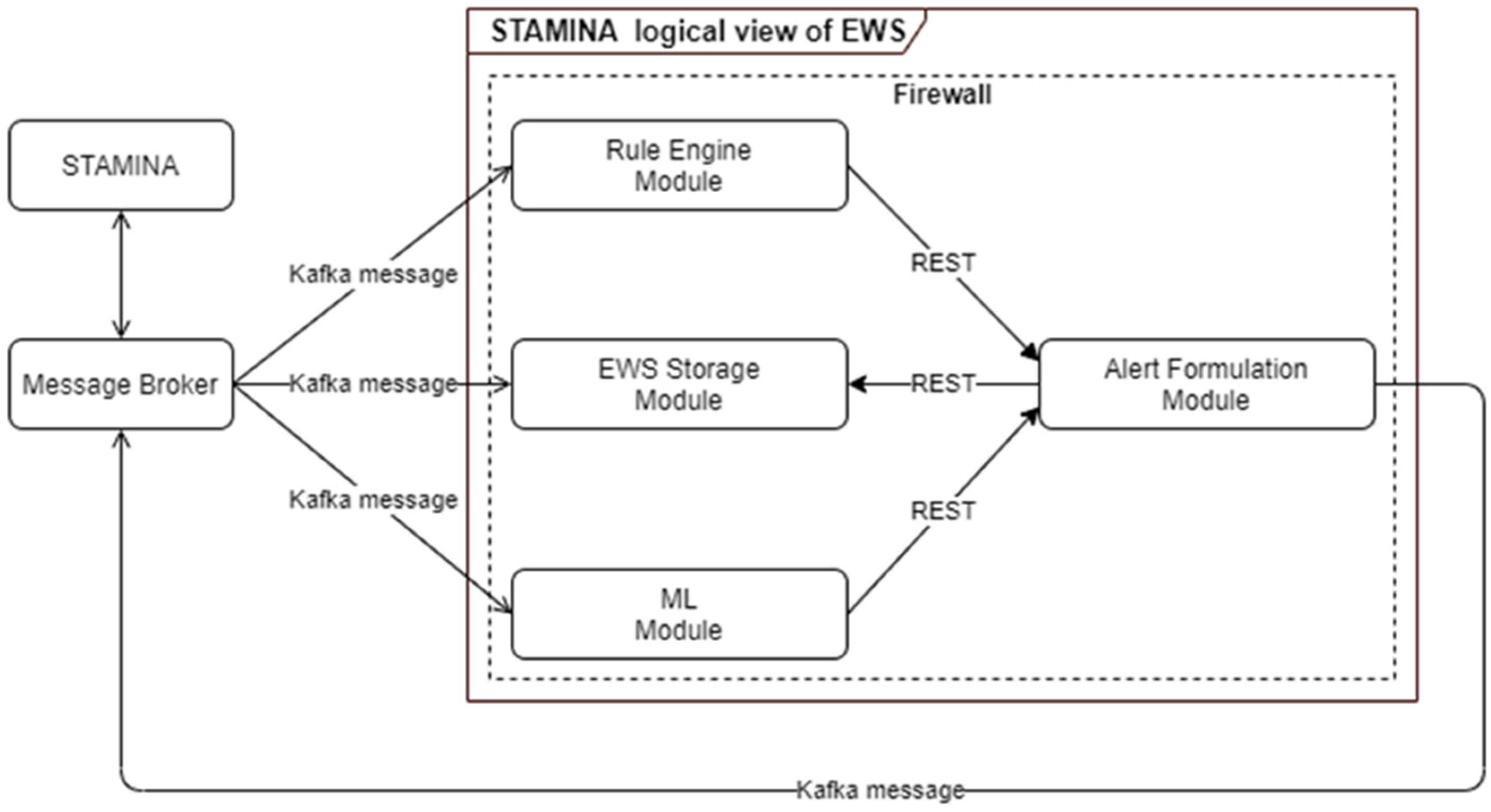
4.3.9. Machine-Learning Early Warning System
- To create alerts and warnings so that end users are informed in case of potential outbreaks, taking into consideration certain rules and patterns based on ML models;
- To identify needs regarding material or human resources;
- To monitor and assist in the validation of continued effectiveness of policies and measures.
4.4. Execution of Training Exercises
- User authentication;
- View of all saved scenarios;
- Creation of new scenario and/or modification of an existing one;
- Add-in script;
- Events that represent real-life situations;
- Messages that can be exchanged in real-time between participants;
- Social media posts;
- Data sources that can trigger a simulation or alert;
- Multilingual menu capability;
- Declaration of a scenario as private;
- Sharing of a scenario with specific user (s) and/or group (s);
- Map visualization;
- Scenario visualization as a timeline;
- Scenario execution.
4.5. Additional Information from Point-of-Care Points
4.6. External Interfaces
- The EMT will be the main user interface and will provide the common operational picture in a country-wide manner. The focus lies on map visualizations of georeferenced data. However, other types of visualizations such as graphs and tables are available. Moreover, messages can be included as a type of visualization, for which there is multilingual support, allowing for a more robust cooperation. The EMT is built to support decision-making by integrating and displaying the most recent and relevant data from diverse sources, such as sensor data, in a birds-eye view of a situation. Next to that, the overview can be complemented with basic data analysis functionalities for making short-term predictions and monitoring resources for incoming data. This is achieved by the ability to set and configure different threshold limits for parameters of interest. The tool then automatically monitors the said thresholds and raises the alarm once these thresholds are violated.
- ENGAGE will also provide an interface as it will be part of the common operational picture tool. However, it mainly targets the needs of hospital workers and first responders, and it will provide information in a more geographically narrow aspect. ENGAGE is a software suite that integrates into a single user interface all the functionalities needed by public/private safety agencies to handle their everyday procedures and respond to emergency situations. Due to its modular architecture, ENGAGE is highly configurable and extendable, assuring that it will provide the appropriate subset of functionalities that are required for its intended use.
- The CMT will provide an interface that supports senior political personnel and decision makers in the strategic tasks of sense-making, decision-making, resource management and coordination. CrisisHub’s purpose is to assist the end-user in translating operational data (such as scarcity of resources) into a strategically relevant situation reporting and strategic decision-making agenda (including advice) that supports the strategic level (e.g., regional or national health authorities) in their decision-making efforts.
- The WSMA will provide an interface that will allow users to alter the parameters for capturing web data and will provide additional information about the analysis of web resources. This interface will be implemented in order to provide the detailed information of WSMA, while a summary of this analysis will be provided also by EMT. This decision is taken because the incorporation of the diverse information provided by WSMA adversely affects user friendliness and usability.
- The POCT interface will address the needs of first responders and on-site doctors. It will not be used to visualize data from other tools, but rather to provide the ability to quickly input information about POCT devices and test site diagnosis in the system.
5. Conclusions and Future Steps
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Triboli, L.; Fornasari, E.; Ray, F.; Giorgi, F.M. Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations. J. Med. Virol. 2021, 93, 3238–3245. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhu, R.; Gill, H.K.; Sood, S. Smart monitoring and controlling of Pandemic Influenza A (H1N1) using Social Network Analysis and cloud computing. J. Comput. Sci. 2016, 12, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Neher, R.; Bedford, T. Nextflu: Real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics 2015, 31, 3546–3548. [Google Scholar] [CrossRef] [PubMed]
- Tradigo, G.; Vizza, P.; Gabriele, G.; Mazzitelli, M.; Torti, C.; Prosperi, M.; Guzzi, P.H.; Veltri, P. On the use of clinical based infection data for pandemic case studies. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020. [Google Scholar]
- Li, E.Y.; Tung, C.-Y.; Chang, S.-H. The wisdom of crowds in action: Forecasting epidemic diseases with a web-based prediction market system. Int. J. Med. Inform. 2016, 92, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Abbasimehr, H.; Paki, R.; Bahrini, A. A novel approach based on combining deep learning models with statistical methods for COVID-19 time series forecasting. Neural Comput. Appl. 2021, 34, 3135–3149. [Google Scholar] [CrossRef] [PubMed]
- Rauf, H.T.; Lali, M.I.U.; Khan, M.A.; Kadry, S.; Alolaiyan, H.; Razaq, A.; Irfan, R. Time series forecasting of COVID-19 transmission in Asia Pacific countries using deep neural networks. Pers. Ubiquitous Comput. 2021, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ayoobi, N.; Sharifrazi, D.; Alizadehsani, R.; Shoeibi, A.; Gorriz, J.M.; Moosaei, H.; Khosravi, A.; Nahavandi, S.; Chofreh, A.G.; Goni, F.A.; et al. Time series forecasting of new cases and new deaths rate for COVID-19 using deep learning methods. Results Phys. 2021, 27, 104495. [Google Scholar] [CrossRef] [PubMed]
- Bodapati, S.; Bandarupally, H.; Trupthi, M. COVID-19 Time Series Forecasting of Daily Cases, Deaths Caused and Recovered Cases using Long Short Term Memory Networks. In Proceedings of the 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA), Greater Noida, India, 30–31 October 2020; pp. 525–530. [Google Scholar] [CrossRef]
- Ketu, S.; Mishra, P.K. India perspective: CNN-LSTM hybrid deep learning model-based COVID-19 prediction and current status of medical resource availability. Soft Comput. 2021, 26, 645–664. [Google Scholar] [CrossRef] [PubMed]
- Borges, V.; Pinheiro, M.; Pechirra, P.; Guiomar, R.; Gomes, J.P. INSaFLU: An automated open web-based bioinformatics suite “from-reads” for influenza whole-genome-sequencing-based surveillance. Genome Med. 2018, 10, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argimón, S.; AbuDahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singer, J.B.; Thomson, E.C.; McLauchlan, J.; Hughes, J.; Gifford, R.J. GLUE: A flexible software system for virus sequence data. BMC Bioinform. 2018, 19, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Broeck, W.; Gioannini, C.; Gonçalves, B.; Quaggiotto, M.; Colizza, V.; Vespignani, A. The GLEaMviz computational tool, a publicly available software to explore realistic epidemic spreading scenarios at the global scale. BMC Infect. Dis. 2011, 11, 37. [Google Scholar]




| Component | Component Acronym | Brief Description |
|---|---|---|
| Web and Social Media Analytics tool | WSMA | It monitors different types of content shared on social media and produces analytics and insights (Natural Language classification, Sentiment Analysis, descriptive statistics and indicators). |
| Point-Of-Care Tools Interface | POCTI | Enters results from POCT and SmartKo devices in the system. |
| Data Management and Harmonization | DMHT | It aggregates various data sets, stores them, harmonizes them in a common schema and provides query mechanisms to other components to retrieve them. |
| Information Protection and Secure Data Sharing tool | IPT | It provides a unified and efficient access-control policy management framework. |
| Preparedness Pandemic Training tool | PPT | It provides the capability of creating realistic scenarios to perform exercises of preparedness. |
| Predictive Modelling Tool—Global Epidemic and Mobility model | GLEAM | Global epidemic and mobility model. |
| Predictive Model—Antimicrobial Resistance Model | AIR | It is a Deep Neural Network (DNN) model used for predicting excess mortality due to antimicrobial resistance. Additionally, it incorporates patient’s contextual epidemiological and infectious information. |
| Predictive Modelling Tool | FLEE | It is an agent-based modelling toolkit used for modelling the movements of refugees and internally displaced persons (IDPs). |
| Predictive Modelling Tool | BIMS | The tool aims to simulate pandemic evolution at the regional level, considering population and car movements. |
| Predictive Modelling Tool—Physical Activity Lifelong Modelling and Simulation | PALMS | It is an agent-based micro-simulation that predicts the lifelong physical activity behavior of individuals of a population and its effect on their quality of life. It is designed to perform policy-relevant cost-effectiveness analysis of cross-sectoral interventions that increase physical activity. |
| Predictive Modelling Tool—Flu and Coronavirus Simulator | FACS | It models the transmission of COVID-19 (or other viruses as defined in the input) and simulates the COVID-19 outbreak in each region using computational disease dynamics at a local (e.g., city or borough) scale. |
| Predictive Modelling Tool—dynamiC Hospital wARd Management | CHARM | It is a discrete-event simulation that models dynamic reconfiguration of hospital wards for bed-capacity planning facilitating continuation of normal ICU operations and COVID-19 outbreaks. |
| Early Warning System | EWS | EWS (with ML) will be responsible for processing real and simulated data from multiple sources to generate alerts, warnings. |
| Crisis-Management Tool | CMT | Strategic information management and decision-making support tool. It supports senior political leaders (and other actors on the strategic level) in pandemic response planning, sense-making, decision-making (agenda setting), task monitoring and resource monitoring. The tool supports collecting and organizing capacity and resource data from distributed providers. Data may be collected manually or via data exchange (API, etc.). |
| Emergency Map Tool | EMT | It offers different visualizations for geo-referenced data. |
| ENGAGE IMS/CAD | ENGAGE | It is an incident management and computer-aided dispatch system. In addition to the typical C2 functionalities, it supports hospital availability and patient-tracking functionalities |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakalos, N.; Kaselimi, M.; Doulamis, N.; Doulamis, A.; Kalogeras, D.; Bimpas, M.; Davradou, A.; Vlachostergiou, A.; Fotopoulos, A.; Plakia, M.; et al. STAMINA: Bioinformatics Platform for Monitoring and Mitigating Pandemic Outbreaks. Technologies 2022, 10, 63. https://doi.org/10.3390/technologies10030063
Bakalos N, Kaselimi M, Doulamis N, Doulamis A, Kalogeras D, Bimpas M, Davradou A, Vlachostergiou A, Fotopoulos A, Plakia M, et al. STAMINA: Bioinformatics Platform for Monitoring and Mitigating Pandemic Outbreaks. Technologies. 2022; 10(3):63. https://doi.org/10.3390/technologies10030063
Chicago/Turabian StyleBakalos, Nikolaos, Maria Kaselimi, Nikolaos Doulamis, Anastasios Doulamis, Dimitrios Kalogeras, Mathaios Bimpas, Agapi Davradou, Aggeliki Vlachostergiou, Anaxagoras Fotopoulos, Maria Plakia, and et al. 2022. "STAMINA: Bioinformatics Platform for Monitoring and Mitigating Pandemic Outbreaks" Technologies 10, no. 3: 63. https://doi.org/10.3390/technologies10030063
APA StyleBakalos, N., Kaselimi, M., Doulamis, N., Doulamis, A., Kalogeras, D., Bimpas, M., Davradou, A., Vlachostergiou, A., Fotopoulos, A., Plakia, M., Karalis, A., Tsekeridou, S., Anagnostopoulos, T., Despotopoulou, A. M., Bonavita, I., Petersen, K., Pelepes, L., Voumvourakis, L., Anagnostou, A., ... Rossi, L. (2022). STAMINA: Bioinformatics Platform for Monitoring and Mitigating Pandemic Outbreaks. Technologies, 10(3), 63. https://doi.org/10.3390/technologies10030063










