1. Introduction
According to the U.S. Centers for Disease Control and Prevention (CDC), almost 30% of antibiotics prescribed in acute care hospitals are unnecessary or suboptimal. Inappropriate use of antibiotics increases the risk of resistance, adverse drug events, and the emergence of secondary infections [
1]. Additionally, evidence shows that patients are often discharged from hospitals with excess antibiotics, leading to unnecessary use of antibiotics and the emergence of antibiotic resistance [
2].
Research indicates that antibiotic usage is correlated with the emergence of antibiotic resistance [
3,
4,
5,
6,
7,
8,
9,
10,
11]. For example, a statistically significant correlation was shown between consumption of antibiotics and resistance rates of Pseudomonas [
9]. Ryu et al. [
12] showed that the use of beta-lactam/beta-lactamase inhibitor antibiotics such as ampicillin/sulbactam and piperacillin/tazobactam is significantly correlated with increased rates of piperacillin/tazobactam-resistant Klebsiella pneumoniae [
12]. Additionally, broad spectrum antibiotics have been shown to correlate with the emergence of multi-drug resistant infections [
6,
13].
Resistant infections pose not only clinical challenges, but a financial burden. Hospital operational costs are increased by patients’ prolonged hospital stays, which are frequently required to treat resistant diseases. More specifically, a resistant
E. coli infection on average increases the LOS for a patient in the United States by 5 days [
9]; this observation can be extrapolated to other resistant Gram-negative infections such as Klebsiella and Pseudomonas [
9]. Since overnight hospital stays cost an average of
$2883 per day in the U.S. [
14], a resistant Gram-negative infection can cost approximately
$14,415, just for the extended LOS alone.
In addition to reporting correlation coefficients between antibiotic usage and antibiotic resistance, scientists have used time series models such as the Box–Jenkins method to document the development of antibiotic resistance over time as a result of antibiotic usage [
11,
15,
16,
17,
18]. At the same time, machine learning techniques such as artificial neural networks (ANNs) and random forest algorithms are becoming an integral forecasting tool in pharmaceutical and healthcare-related research [
19,
20]. Those ANNs are being applied in the pharmaceutical arena to predict how novel drug molecules would behave in the human body [
21] and to predict antibiotic resistance [
22,
23,
24]. Michael Kane, a researcher and a professor at Yale University, showed that the performance of random forest time series algorithms can outperform existing time series models for predicting infectious disease outbreaks [
19].
Statistical and machine learning models can be used to better understand the relationship between antibiotic usage and the development of antibiotic resistance over time, and to predict the clinical and financial cost of using culpable antibiotics. The CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC) recommend implementing ASP programs and guidelines for antibiotic use to ensure appropriate selection, dose, route of administration, and duration of therapy [
25]. This study was devised and executed to kickstart and encourage the application of statistical and machine learning approaches to region-specific data. The aim is to guide and support local hospital ASPs in both acute care and discharge settings.
4. Discussion
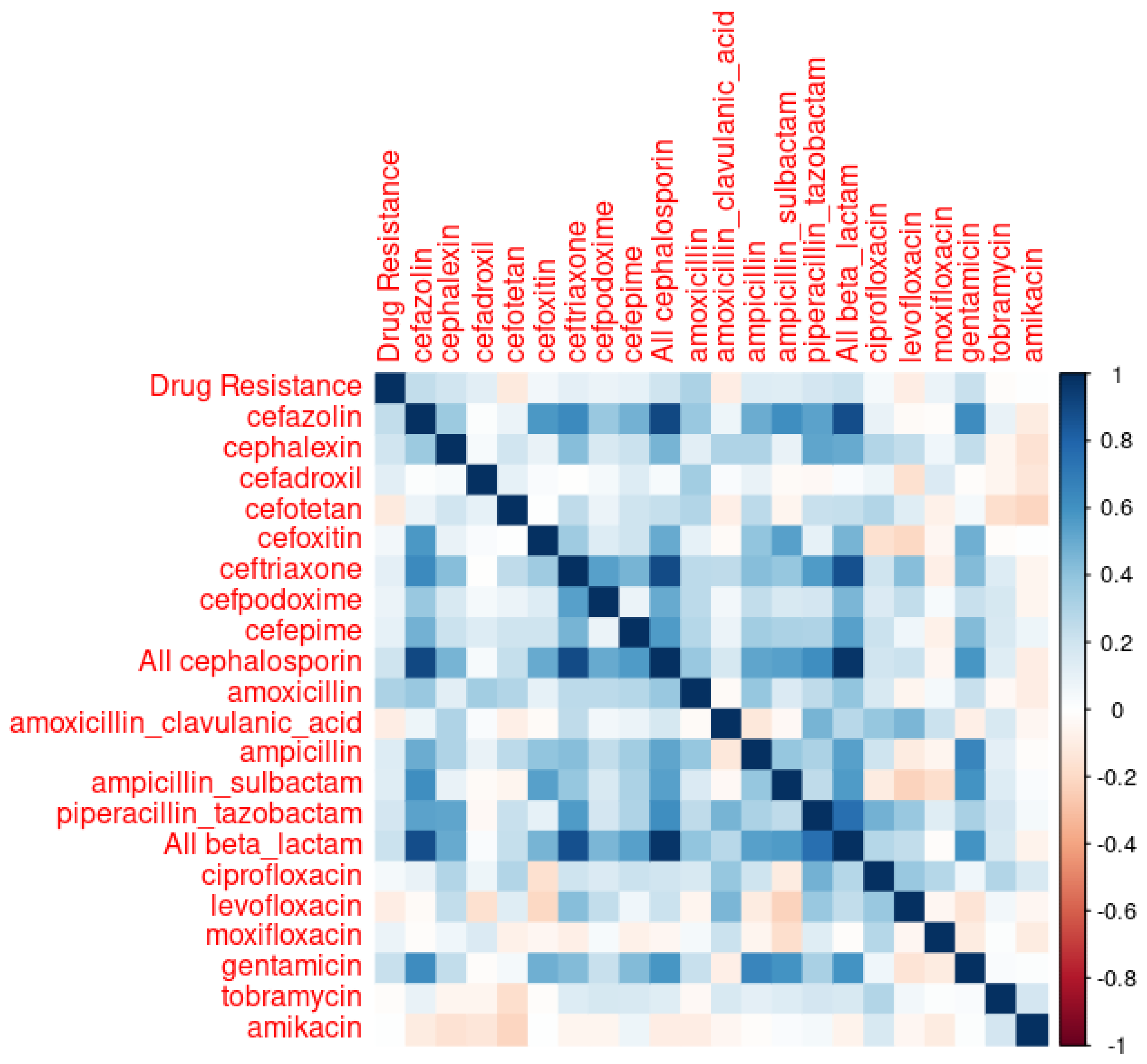
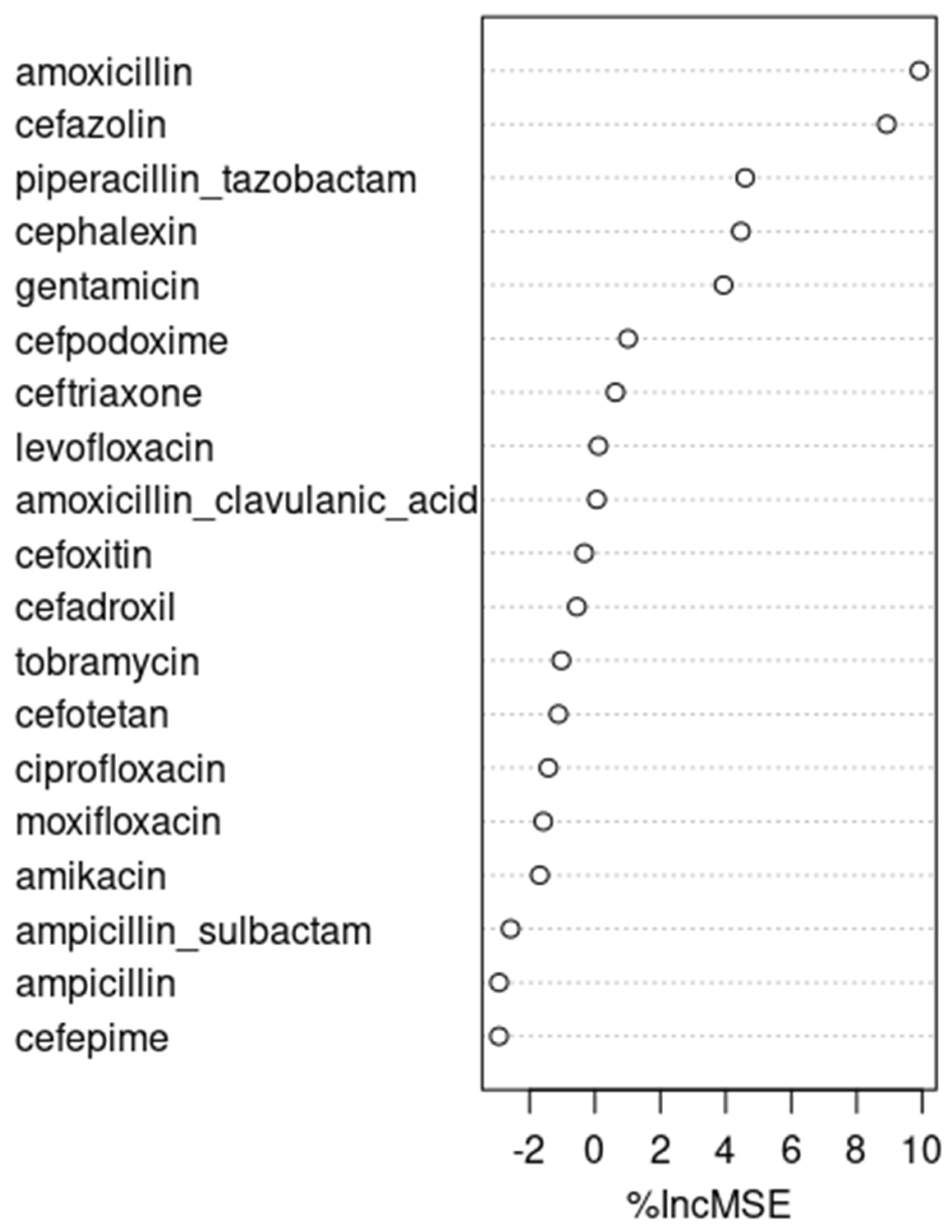
It is interesting to note that cefazolin and cephalexin, both first-generation cephalosporins with similar spectrums of activity, were found to be closely associated with the resistant E. coli infections we observed. Cefazolin is frequently used in perioperative settings and in long-term bacteremia treatments, representing 11.2% of all antibiotics usage observed in our data set. On the other hand, cephalexin is usually used in outpatient settings, and represents only 0.57% of the observed inpatient antibiotics usage. Given the relatively narrower spectrums of activity of cefazolin and cephalexin compared to other antibiotics, targeting these two antibiotics for ASP interventions is often challenging. We encounter this scenario because antibiotics stewardship programs try to limit the use of unnecessarily broad-spectrum antibiotics under the assumption that they exert Darwinian pressure, generating antibiotics-resistant microorganisms.
At the same time, although piperacillin/tazobactam, a broad-spectrum antibiotic, did not perform as well as cefazolin and cephalexin, it performed fairly well in the ARIMA and neural network time series models, ranking right after cefazolin and cephalexin. Had we picked piperacillin/tazobactam to make our predictions, the predicted rate of resistant E. coli in 2020, according to the ARIMA model, and assuming the same rate of piperacillin/tazobactam usage as in 2019, was 5.52%, with its 95% prediction interval being 0.14% to 10.9%. When a 50% reduction is made in piperacillin/tazobactam usage, the predicted rate of resistant E. coli in 2020 was 4.31%, with its 95% prediction interval being −1.1% to 9.7%. With a 25% reduction in piperacillin/tazobactam usage, the predicted rate of resistant E. coli was 4.91%, with its 95% prediction interval being −0.47% to 10.3%.
While one might be tempted to build a time series model with both cefazolin and piperacillin/tazobactam—excluding cephalexin because it is rarely used in inpatient settings—such a model does not perform better than cefazolin alone, as shown in
Table 9:
Although the variance inflation factor (VIF)—a measure to quantify the severity of multicollinearity in an ordinary least squares regression analysis—value for cefazolin and piperacillin/tazobactam is 1.39, which is not particularly concerning for collinearity, the performance of a linear model combining these two factors is less than satisfactory. Specifically, the adjusted R-value for a model with piperacillin/tazobactam and cefazolin together is 0.03477, which is lower than the 0.0446 adjusted R-value for a model with cefazolin alone. This lower adjusted R-value suggests that the combined model does not explain the variance in the dependent variable as well as the cefazolin-alone model does. Moreover, the moderate increase in the adjusted R-value from a piperacillin/tazobactam-only model (which is 0.02) to a piperacillin/tazobactam and cefazolin combination model (which is 0.03477) does not seem sufficient to justify the time series model that includes both piperacillin/tazobactam and cefazolin as regressors.
An interesting analysis can be made with amoxicillin and cephalexin, whose inpatient use is negligible at 0.15% and 0.57%, respectively. Those figures might underestimate the actual use of amoxicillin and cephalexin, as those two oral antibiotics are frequently prescribed just before patient discharge. Although patients might receive those medications for only one or two days during their inpatient stay, the therapy usually extends much longer post-discharge. Those two antibiotics became notable because of the stepwise linear regression shown in
Table 3, in which the usage of four antibiotics amoxicillin, cefotetan, cephalexin, and amoxicillin/clavulanic acid explain a non-negligible amount—more specifically, 16.4%—of variations associated with resistant
E. coli. Of those, the use of amoxicillin, cefotetan, and cephalexin demonstrated significant correlations with the emergence of antibiotic resistance, with respective
p-values of 0.00289, 0.01918, and 0.03301. After excluding cefotetan, an intravenous antibiotic that is rarely (i.e., 0.032%) used, and then primarily in perioperative settings, we built a linear model of the two oral antibiotics, amoxicillin and cephalexin. That linear model explains 9.5% of variations associated with observed resistant
E. coli cases, with a
p-value of 0.01369. Therefore, it might be worthwhile to consider those two antibiotics as external regressors when building time-series models.
The performance of those models, tabulated in
Table 10 (with amoxicillin at lag 5 and cephalexin at lag 0), is not particularly outstanding. However, the best of them, the ARIMA model, was used to make predictions in recognition of the potential significance of those two antibiotics, as indicated by the strength of their linear model.
Using the ARIMA model with amoxicillin at lag 5 and cephalexin at lag 0 as external regressors, we concluded that the predicted rate of resistant E. coli infections in 2020, assuming the same rate of amoxicillin and cephalexin usage as in 2019, was 5.5%, with a 95% prediction interval of 0.31% to 10.7%. When a 50% reduction is made in amoxicillin and cephalexin usage, the predicted rate of resistant E. coli in 2020 was 4.56%, with a 95% prediction interval of −0.61% to 9.72%. With a 25% reduction in amoxicillin and cephalexin usage, the predicted rate of resistant E. coli in 2020 was 5.0%, with a 95% prediction interval of −0.15% to 10.2%.
Overall, our analyses suggest that while making interventions for cefazolin might not be practical in light of its relatively narrow spectrum, and the focus of many ASP programs that primarily target the use of broad-spectrum antibiotics, judiciously reducing the use of piperacillin/tazobactam in inpatient settings and the use of amoxicillin and cephalexin at discharge could lower
E. coli-resistant infection rates [
6,
13]. Both time series models with either piperacillin/tazobactam or amoxicillin and cephalexin showed about a 0.5% drop in antibiotic resistance rates, from approximately 5.5 to 5%, when the use of either group of antibiotics was reduced by 25%. In 2019, there were 1009 total
E. coli infection cases at the Kaiser Vacaville facility in the U.S., and this number on average increased by 81 cases every year. Extrapolating, we can expect 1090 cases in 2020. Therefore, a 0.5% drop in antibiotic resistance rates achieved via an ASP program could translate to approximately
$80,000 per year in savings for
E. coli infections alone, because each Gram-negative resistant infection extends the LOS of a patient by 5 days on average [
9] and the average cost of inpatient stay per day is
$2883 [
14]. Considering many other Gram-negative infections caused by other organisms, such as Klebsiella, Pseudomonas, and Acinetobacter, we could anticipate multiples of
$80,000 in savings per year per hospital.
It is noteworthy that the ARIMA model outperformed both the neural network and the random forest models. This is in line with a systematic review published in 2019, which found a lack of evidence to support the superiority of machine learning algorithms over logistic regression in clinical prediction models [
32]. At the same time, one of the reasons the neural network model did not perform as well as we initially expected might be attributed to the limitations associated with the rather simple nnetar() function in R that we used.
The results of this study are based on longitudinal data collected from one Kaiser Permanente facility located in a somewhat rural region with relatively low population migration. In the future, a broader study could be conducted using data from multiple facilities. Additionally, while the performance of the statistical and machine learning models was evaluated against the test dataset from the final year in our data set, 2019, a follow-up study could be carried out to validate the predictions made for 2020 using actual data from that year. However, conducting such a study is currently beyond our scope, as this study was approved by Kaiser Permanente for a pharmacist resident project in 2021, utilizing data spanning from 2013 to 2019.
Finally, while this study identifies significant correlations between antibiotic usage and the emergence of resistant E. coli infections, it is crucial to recognize that these findings do not establish direct causation. The inherent limitations of observational data in confirming causal relationships necessitate caution in interpretation. Consequently, our results highlight the need for further research, including experimental or more comprehensive longitudinal studies, to rigorously explore causality.