QTL Mapping and Candidate Gene Analysis for Pod Shattering Tolerance in Soybean (Glycine max)
Abstract
1. Introduction
2. Results
2.1. Variation in Pod Shattering Ratios
2.2. Linkage Map Construction
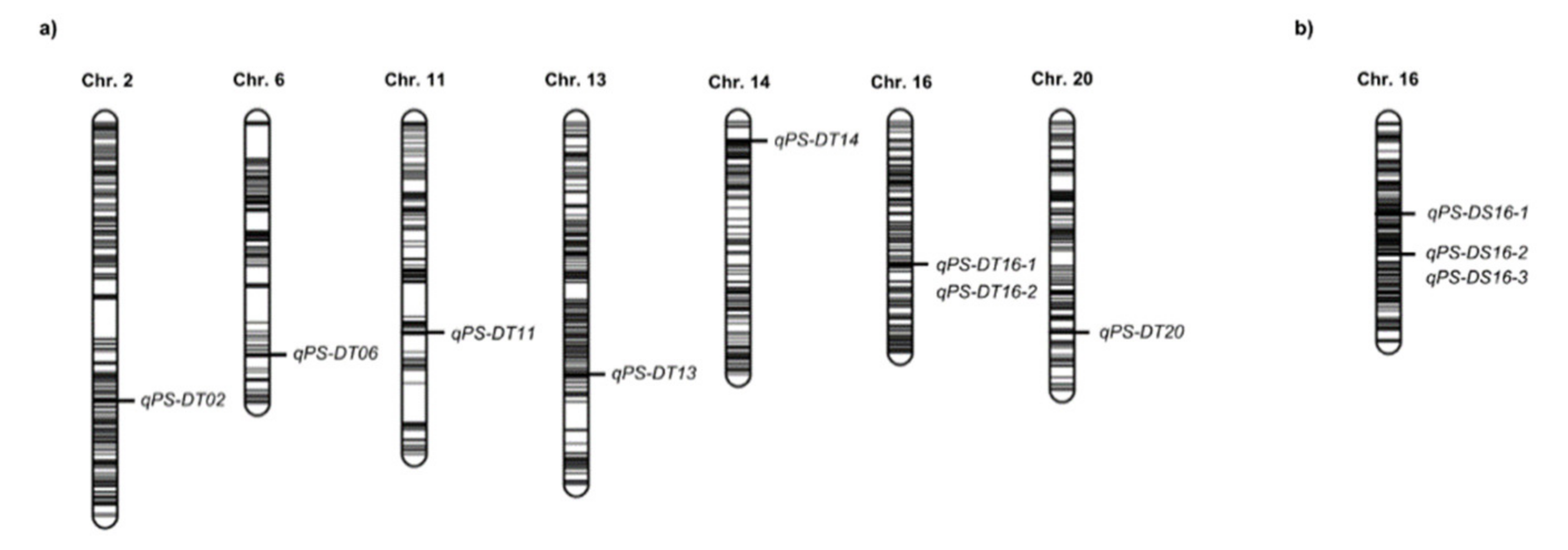
2.3. QTL Analysis
2.4. Phenotypic Variation According to the Allele Patterns
2.5. Identification of Candidate Genes within and Near QTL
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Evaluation of Pod Shattering Tolerance
4.3. Genomic DNA Extraction and Genotyping
4.4. Linkage Map Construction and QTL Mapping
4.5. Prediction of Novel Candidate QTL and Genes
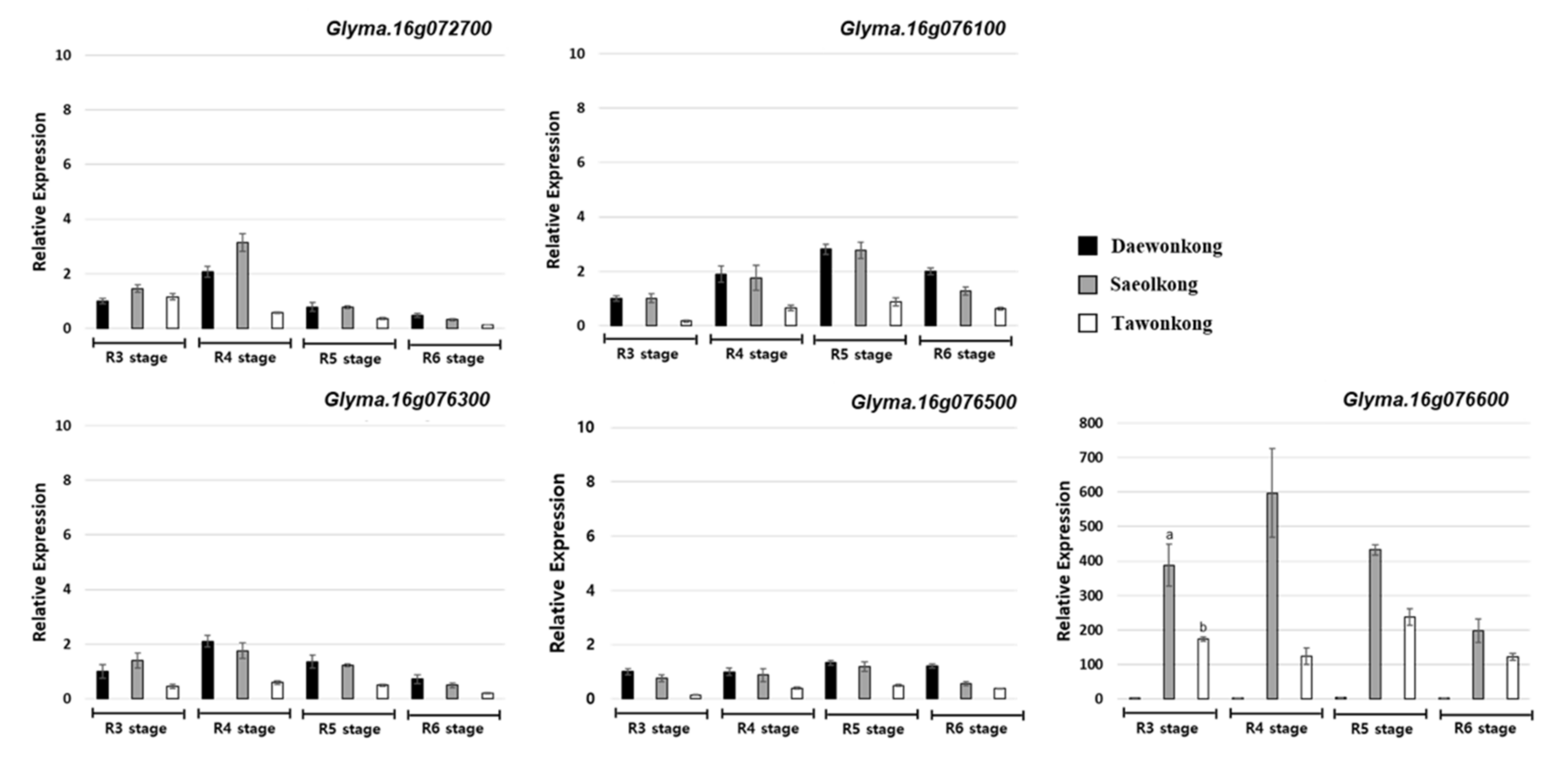
4.6. Gene Expression Analysis of the Candidate Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fuller, D.Q. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. 2007, 100, 903–924. [Google Scholar] [CrossRef]
- Funatsuki, H.; Suzuki, M.; Hirose, A.; Inaba, H.; Yamada, T.; Hajika, M.; Komatsu, K.; Katayama, T.; Sayama, T.; Ishimoto, M. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. USA 2014, 111, 17797–17802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tu, B.; Liu, C.; Liu, X. Pod anatomy, morphology and dehiscing forces in pod dehiscence of soybean (Glycine max L. Merrill). Flora 2018, 248, 48–53. [Google Scholar] [CrossRef]
- Di Vittori, V.; Gioia, T.; Rodriguez, M.; Bellucci, E.; Bitocchi, E.; Nanni, L.; Attene, G.; Rau, D.; Papa, R. Convergent evolution of the seed shattering trait. Genes 2019, 10, 68. [Google Scholar] [CrossRef]
- Kang, X.; Cai, J.; Chen, Y.; Yan, Y.; Yang, S.; He, R.; Wang, D.; Zhu, Y. Pod-shattering characteristics differences between two groups of soybeans are associated with specific changes in gene expression. Funct. Integr. Genom. 2020, 20, 201–210. [Google Scholar] [CrossRef]
- Bandillo, N.B.; Anderson, J.E.; Kantar, M.B.; Stupar, R.M.; Specht, J.E.; Graef, G.L.; Lorenz, A.J. Dissecting the genetic basis of local adaptation in soybean. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Tu, B.; Liu, C.; Wang, X.; Li, Y.; Zhang, Q.; Liu, X.; Herbert, S.J. Greater Anatomical Differences of Pod Ventral Suture in Shatter-Susceptible and Shatter-Resistant Soybean Cultivars. Crop Sci. 2019, 59, 2784–2793. [Google Scholar] [CrossRef]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, D.; Li, C.; Luo, J.; Zhu, B.-F.; Zhu, J.; Shangguan, Y.; Wang, Z.; Sang, T.; Zhou, B. Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 2012, 24, 1034–1048. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, X.; Liu, J.; Wang, B.-H.; Liu, B.-L.; Wang, Y.-Z. Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Rau, D.; Murgia, M.L.; Rodriguez, M.; Bitocchi, E.; Bellucci, E.; Fois, D.; Albani, D.; Nanni, L.; Gioia, T.; Santo, D. Genomic dissection of pod shattering in common bean: Mutations at non-orthologous loci at the basis of convergent phenotypic evolution under domestication of leguminous species. Plant J. 2019, 97, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Fourquin, C.; del Cerro, C.; Victoria, F.C.; Vialette-Guiraud, A.; de Oliveira, A.C.; Ferrándiz, C. A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago genus. Plant Physiol. 2013, 162, 907–917. [Google Scholar] [CrossRef]
- Suanum, W.; Somta, P.; Kongjaimun, A.; Yimram, T.; Kaga, A.; Tomooka, N.; Takahashi, Y.; Srinives, P. Co-localization of QTLs for pod Fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 2016, 36, 80. [Google Scholar] [CrossRef]
- Lo, S.; Muñoz-Amatriaín, M.; Boukar, O.; Herniter, I.; Cisse, N.; Guo, Y.-N.; Roberts, P.A.; Xu, S.; Fatokun, C.; Close, T.J. Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bhor, T.; Chimote, V.; Deshmukh, M. Inheritance of pod shattering in soybean (Glycine max L) Merrill). Electron. J. Plant Breed. 2014, 5, 671–676. [Google Scholar]
- Kang, S.-T.; Kwak, M.; Kim, H.-K.; Choung, M.-G.; Han, W.-Y.; Baek, I.-Y.; Kim, M.Y.; Van, K.; Lee, S.-H. Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean (Glycine max L. Merr.). Euphytica 2009, 166, 15. [Google Scholar] [CrossRef]
- Funatsuki, H.; Hajika, M.; Yamada, T.; Suzuki, M.; Hagihara, S.; Tanaka, Y.; Fujita, S.; Ishimoto, M.; Fujino, K. Mapping and use of QTLs controlling pod dehiscence in soybean. Breed. Sci. 2012, 61, 554–558. [Google Scholar] [CrossRef][Green Version]
- Bailey, M.; Mian, M.; Carter, T., Jr.; Ashley, D.; Boerma, H. Pod dehiscence of soybean: Identification of quantitative trait loci. J. Hered. 1997, 88, 152–154. [Google Scholar] [CrossRef]
- Funatsuki, H.; Ishimoto, M.; Tsuji, H.; Kawaguchi, K.; Hajika, M.; Fujino, K. Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breed. 2006, 125, 195–197. [Google Scholar] [CrossRef]
- Funatsuki, H.; Hajika, M.; Hagihara, S.; Yamada, T.; Tanaka, Y.; Tsuji, H.; Ishimoto, M.; Fujino, K. Confirmation of the location and the effects of a major QTL controlling pod dehiscence, qPDH1, in soybean. Breed. Sci. 2008, 58, 63–69. [Google Scholar] [CrossRef]
- Suzuki, M.; Fujino, K.; Funatsuki, H. A major soybean QTL, qPDH1, controls pod dehiscence without marked morphological change. Plant Prod. Sci. 2009, 12, 217–223. [Google Scholar] [CrossRef]
- Yamada, T.; Funatsuki, H.; Hagihara, S.; Fujita, S.; Tanaka, Y.; Tsuji, H.; Ishimoto, M.; Fujino, K.; Hajika, M. A major QTL, qPDH1, is commonly involved in shattering resistance of soybean cultivars. Breed. Sci. 2009, 59, 435–440. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, K.R.; Ha, B.-K.; Kang, S. Identification of SNPs tightly linked to the QTL for pod shattering in soybean. Mol. Breed. 2017, 37, 54. [Google Scholar] [CrossRef]
- Miranda, C.; Culp, C.; Škrabišová, M.; Joshi, T.; Belzile, F.; Grant, D.M.; Bilyeu, K. Molecular tools for detecting Pdh1 can improve soybean breeding efficiency by reducing yield losses due to pod shatter. Mol. Breed. 2019, 39, 27. [Google Scholar] [CrossRef]
- Suzuki, M.; Fujino, K.; Nakamoto, Y.; Ishimoto, M.; Funatsuki, H. Fine mapping and development of DNA markers for the qPDH1 locus associated with pod dehiscence in soybean. Mol. Breed. 2010, 25, 407–418. [Google Scholar] [CrossRef]
- Gao, M.; Zhu, H. Fine mapping of a major quantitative trait locus that regulates pod shattering in soybean. Mol. Breed. 2013, 32, 485–491. [Google Scholar] [CrossRef]
- Hu, D.; Kan, G.; Hu, W.; Li, Y.; Hao, D.; Li, X.; Yang, H.; Yang, Z.; He, X.; Huang, F.; et al. Identification of Loci and Candidate Genes Responsible for Pod Dehiscence in Soybean via Genome-Wide Association Analysis across Multiple Environments. Front. Plant Sci. 2019, 10, 811. [Google Scholar] [CrossRef]
- Han, J.; Han, D.; Guo, Y.; Yan, H.; Wei, Z.; Tian, Y.; Qiu, L. QTL mapping pod dehiscence resistance in soybean (Glycine max L. Merr.) using specific-locus amplified fragment sequencing. Theor. Appl. Genet. 2019, 132, 2253–2272. [Google Scholar] [CrossRef]
- Kim, S.; Park, K.; Yun, H.; Lee, Y.; Lee, S.; Ryu, Y.; Hwang, C.; Kim, Y.; Kim, Y.; Seung, Y. A new soybean variety for soypaste with large seed and disease resistant “Daewonkong”. RDA J. Crop Sci. 1998, 40, 107–111. [Google Scholar]
- Seo, J.; Kang, B.; Kim, H.; Kim, H.; Choi, M.; Oh, J.; Shin, S.; Baek, I.; Kwak, D. Variation in pod shattering in a RIL population and selection for pod shattering tolerance in soybean (Glycine max (L.) Merr). Korean J. Crop Sci. 2019, 64, 414–421. [Google Scholar]
- Fehr, W.; Caviness, C.; Burmood, D.; Pennington, J. Stage of development descriptions for soybeans, Glycine max L. Merrill 1. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, G.; Jin, J.; Liu, J.; Zhang, Q.; Liu, X. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol. Biochem. 2009, 41, 919–925. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Todoroki, Y. ABA 8′-hydroxylase and its chemical inhibitors. Phytochem. Rev. 2006, 5, 385. [Google Scholar] [CrossRef]
- Takeuchi, J.; Okamoto, M.; Mega, R.; Kanno, Y.; Ohnishi, T.; Seo, M.; Todoroki, Y. Abscinazole-E3M, a practical inhibitor of abscisic acid 8′-hydroxylase for improving drought tolerance. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Jaradat, M.R.; Ruegger, M.; Bowling, A.; Butler, H.; Cutler, A.J. A comprehensive transcriptome analysis of silique development and dehiscence in Arabidopsis and Brassica integrating genotypic, interspecies and developmental comparisons. GM Crop. Food 2014, 5, 302–320. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, K.; Lee, Y.; Yun, H.; Lee, S.; Ryu, Y.; Son, Y.; Kim, Y.; Kim, Y.; Seung, Y. A black seed coat soybean variety with small seed and lodging resistant “Tawankong”. RDA J. Crop Sci. 1998, 40, 102–106. [Google Scholar]
- Baek, I.; Shin, D.; Kang, S.; Song, S.; Hur, S.; Kwack, Y.; Lim, M. A New Early Maturity, Good Seed Quality and High Yielding Soybean Variety “Saeolkong”. Korean J. Breed. Sci. 1998, 30, 397. [Google Scholar]
- Kang, S.T.; Kim, H.K.; Baek, I.Y.; Chung, M.G.; Han, W.Y.; Shin, D.C.; Lee, S.-H. Genetic analysis of pod dehiscence in soybean. Korean J. Crop Sci. 2005, 50, 281–285. [Google Scholar]
- Dhungana, S.K.; Kim, H.-S.; Kang, B.-K.; Seo, J.-H.; Kim, H.-T.; Shin, S.-O.; Park, C.-H.; Kwak, D.-Y. Evaluation of Flooding Tolerance of Soybean (Glycine max L. Merr.) in Greenhouse under Upland and Paddy Soil Conditions. J. Crop Sci. Biotechnol. 2019, 22, 283–290. [Google Scholar] [CrossRef]
- Lee, Y.G.; Jeong, N.; Kim, J.H.; Lee, K.; Kim, K.H.; Pirani, A.; Ha, B.K.; Kang, S.T.; Park, B.S.; Moon, J.K. Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J. 2015, 81, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Jeong, N.; Kim, K.-S.; Jeong, S.; Kim, J.-Y.; Park, S.-K.; Lee, J.S.; Jeong, S.-C.; Kang, S.-T.; Ha, B.-K.; Kim, D.-Y. Korean soybean core collection: Genotypic and phenotypic diversity population structure and genome-wide association study. PLoS ONE 2019, 14, e0224074. [Google Scholar] [CrossRef]
- Kim, K.H.; Kim, J.-Y.; Lim, W.-J.; Jeong, S.; Lee, H.-Y.; Cho, Y.; Moon, J.-K.; Kim, N. Genome-wide association and epistatic interactions of flowering time in soybean cultivar. PLoS ONE 2020, 15, e0228114. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Toker, C. Estimates of broad-sense heritability for seed yield and yield criteria in faba bean (Vicia faba L.). Hereditas 2004, 140, 222–225. [Google Scholar] [CrossRef] [PubMed]
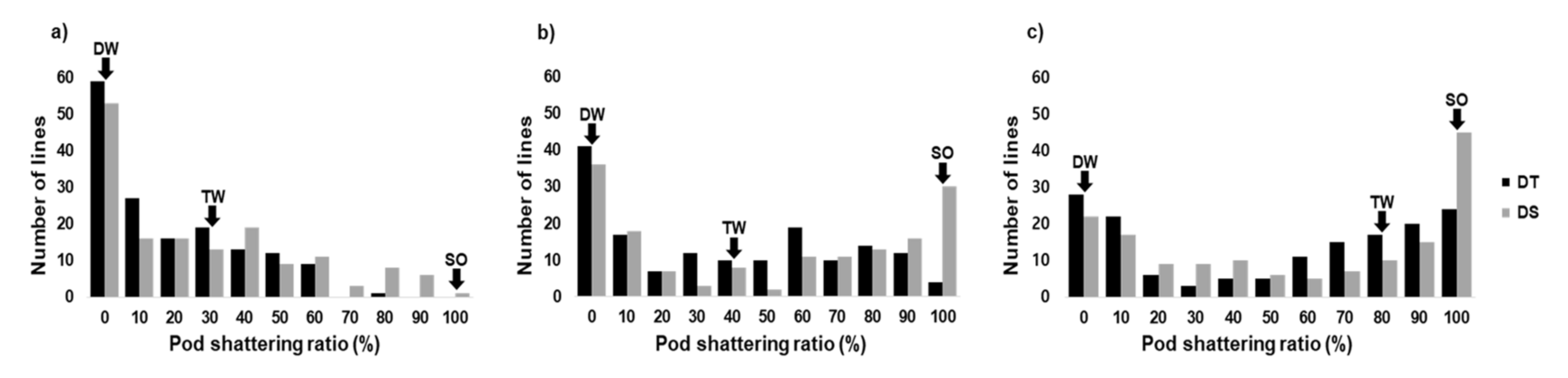
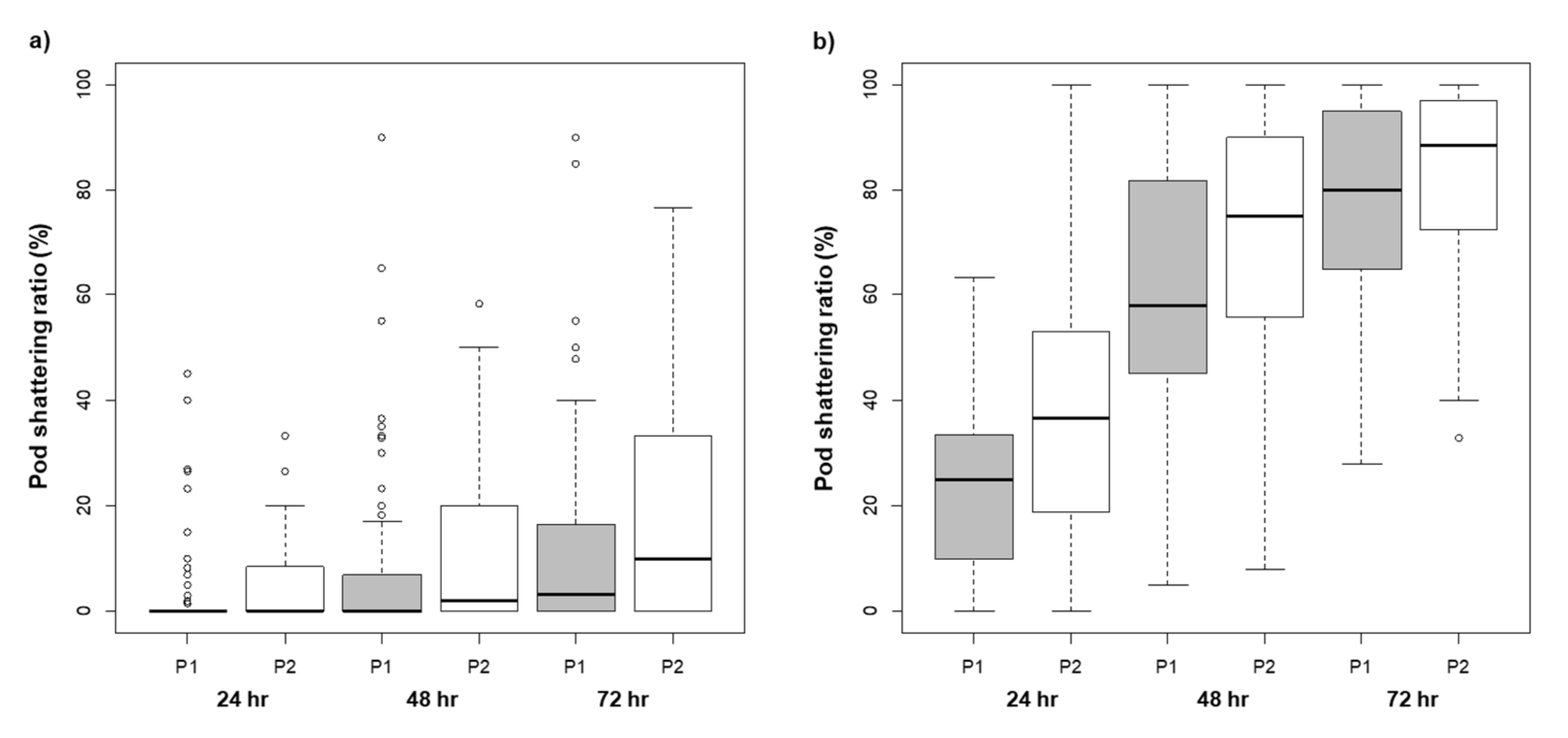
| Drying Time (Hour) | Year | Pod Shattering Ratio ± SE (%) † | Broad-Sense Heritability | |||||
|---|---|---|---|---|---|---|---|---|
| Parents | RILs | |||||||
| DW | TW | SO | DT | DS | DT | DS | ||
| 24 | 2016 | 0 | 25 | 100 | 12.1 ± 24.6 | 24.4 ± 33.5 | ||
| 2017 | 0 | 0 | 100 | 9.9 ± 34.8 | 23.3 ± 33.8 | |||
| 2018 | 0 | 50 | 100 | 27.8 ± 40.4 | 32.5 ± 32.0 | |||
| average | 0 | 25 | 100 | 16.6 ± 19.0c ‡ | 26.7 ± 26.2c | 0.38 | 0.62 | |
| 48 | 2016 | 0 | 35 | 100 | 27.8 ± 34.7 | 47.7 ± 42.9 | ||
| 2017 | 0 | 20 | 100 | 38.4 ± 39.6 | 48.0 ± 46.1 | |||
| 2018 | 0 | 50 | 100 | 44.8 ± 36.4 | 50.5 ± 38.6 | |||
| average | 0 | 35 | 100 | 37.0 ± 31.9b | 48.7 ± 39.1b | 0.78 | 0.84 | |
| 72 | 2016 | 0 | 80 | 100 | 43.6 ± 40.3 | 54.7 ± 43.6 | ||
| 2017 | 0 | 100 | 100 | 53.5 ± 43.2 | 51.1 ± 46.8 | |||
| 2018 | 0 | 60 | 100 | 52.0 ± 37.7 | 60.1 ± 37.3 | |||
| average | 0 | 80 | 100 | 49.7 ± 37.4a | 55.3 ± 39.0a | 0.88 | 0.82 | |
| Name of QTL | Chr. (LG) 1 | Year | Drying Time (Hour) | Position (cM) | Physical Interval 2 (bp) | LOD 3 | PVE 4 (%) | Add 5 | No. Candidate Genes | References |
|---|---|---|---|---|---|---|---|---|---|---|
| qPS-DT02 | 2 (D1b) | 2017 | 24 | 122 | 7,211,739–8,082,789 | 6.0 | 1.9 | −5.9 | 87 | Kang et al. 2009 |
| qPS-DT06 | 6 (C2) | 2016 | 48 | 103 | 5,498,397–6,170,213 | 3.6 | 6.1 | −9.0 | 82 | Novel |
| qPS-DT11 | 11 (B1) | 2017 | 48 | 92 | 26,717,840–30,547,921 | 17.4 | 13.3 | −20.0 | 189 | Novel |
| qPS-DT13 | 13 (F) | 2017 | 24 | 108 | 35,019,516–35,459,889 | 4.3 | 1.3 | −5.0 | 54 | Novel |
| qPS-DT14 | 14 (B2) | 2017 | 24 | 11 | 48,063,838–48,101,634 | 33.4 | 16.4 | −17.8 | 2 | Novel |
| qPS-DT16-1 | 16 (J) | 2017 | 24 | 63 | 29,532,807–29,809,243 | 7.4 | 2.3 | −6.7 | 21 | qPDH1(nearby) |
| 2018 | 72 | 29,532,807–29,809,243 | 31.0 | 67.8 | −32.0 | |||||
| qPS-DT16-2 | 16 (J) | 2016 | 48 | 64 | 29,913,393–29,972,096 | 13.4 | 26.1 | −19.0 | 5 | qPDH1 (Funatsuki et al. 2008) |
| 2016 | 72 | 29,913,393–29,972,096 | 23.9 | 48.0 | −29.2 | |||||
| 2017 | 48 | 29,913,393–29,972,096 | 30.1 | 29.6 | −30.3 | |||||
| 2017 | 72 | 29,913,393–29,972,096 | 67.1 | 82.8 | −42.0 | |||||
| Combined 6 | 24 | 29,913,393–29,972,096 | 13.4 | 33.1 | −11.2 | |||||
| Combined | 48 | 29,913,393–29,972,096 | 35.7 | 60.8 | −25.9 | |||||
| Combined | 72 | 29,913,393–29,972,096 | 55.4 | 78.7 | −34.6 | |||||
| qPS-DT20 | 20 | Combined | 48 | 93 | 2,151,689–2,392,383 | 3.7 | 3.7 | −6.3 | 17 | Novel |
| Name of QTL | Chr. (LG) 1 | Year | Drying Time (Hour) | Position (cM) | Physical Interval 2 (bp) | LOD 3 | PVE 4 (%) | Add 5 | No. Candidate Genes | References |
|---|---|---|---|---|---|---|---|---|---|---|
| qPS-DS16-1 | 16 (J) | Combined 6 | 24 | 61 | 7,325,202–7,760,393 | 5.4 | 11.2 | −7.1 | 41 | Novel |
| qPS-DS16-2 | 16 (J) | 2016 | 48 | 45 | 29,857,920–30,146,414 | 19.5 | 44.8 | −28.7 | 20 | qPDH1 (Funatsuki et al. 2008) |
| 2016 | 72 | 29,857,920–30,146,414 | 18.0 | 42.4 | −28.3 | |||||
| 2017 | 24 | 29,857,920–30,146,414 | 14.6 | 36.0 | −20.2 | |||||
| 2017 | 48 | 29,857,920–30,146,414 | 56.0 | 61.2 | −40.9 | |||||
| 2017 | 72 | 29,857,920–30,146,414 | 60.0 | 63.0 | −41.9 | |||||
| Combined | 24 | 29,857,920–30,146,414 | 19.2 | 48.7 | −14.8 | |||||
| Combined | 48 | 29,857,920–30,146,414 | 40.9 | 70.3 | −26.8 | |||||
| Combined | 72 | 29,857,920–30,146,414 | 40.8 | 71.8 | −26.9 | |||||
| qPS-DS16-3 | 16 (J) | 2018 | 24 | 43 | 30,144,267–30,580,464 | 15.2 | 56.8 | −24.4 | 25 | qPDH1 (nearby) |
| 2018 | 48 | 30,144,267–30,580,464 | 31.8 | 80.1 | −37.0 | |||||
| 2018 | 72 | 30,144,267–30,580,464 | 27.2 | 76.4 | −34.3 |
| Genotype 1 | qPS-DS16-1 | qPS-DT11 | qPS-DT14 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h |
| P1 | P1 | 2.5 | 6.5 | 10.9 | 2.6 | 6.5 | 10.7 | 3.5 | 8.1 | 12.7 |
| P2 | 6.2 | 12.5 | 18.6 | 4.3 | 10.7 | 15.6 | 3.8 | 9.0 | 13.6 | |
| t-value 2 | −1.68 * | −1.50 ns | −1.61 ns | −1.15 ns | −1.45 ns | −1.44 ns | −0.15 ns | −0.29 ns | −0.24 ns | |
| P2 | P1 | 24.3 | 60.5 | 76.1 | 37.2 | 70.3 | 82.8 | 32.0 | 65.6 | 81.7 |
| P2 | 37.6 | 69.9 | 83.5 | 31.9 | 65.9 | 80.8 | 36.8 | 69.8 | 81.6 | |
| t-value | −3.81 ** | −2.07 * | −2.00 * | 1.43 ns | 1.12 ns | 0.73 ns | −1.32 ns | −1.09 ns | 0.04 ns | |
| Gene Name | Position (bp) | Reference (Williams82) | DW | TW | SO | AA Change | Description |
|---|---|---|---|---|---|---|---|
| Glyma.16g072700 | 7,335,363 | - | - | 12 bp deletion | 12 bp deletion | Deletion | Myb-like DNA-binding domain, DNA binding |
| Glyma.16g073700 | 7,446,096 | A | A | T | T | Tyr/Asn | Unknown |
| Glyma.16g076100 | 7,665,996 | C | T | C | C | Glu/Lys | Rho guanyl-nucleotide exchange factor activity |
| Glyma.16g076300 | 7,725,602 | A | C | A | A | Met/Leu | GMC oxidoreductase |
| Glyma.16g076500 | 7,760,100 | T | C | T | T | Leu/Ser | Adaptin N terminal region |
| 7,760,393 | A | C | A | A | Ile/Leu | ||
| Glyma.16g076600 | 7,775,892 | C | T | C | C | Glu/Lys | Cytochrome P450, |
| 7,775,945 | T | A | T | T | Lys/Met | (+)-abscisic acid 8′-hydroxylase activity | |
| 7,775,948 | A | G | A | A | Ile/Thr | ||
| 7,775,970 | - | 18 bp insertion | - | - | Insertion | ||
| 7,776,045 | C | T | C | C | Met/Ile | ||
| 7,777,575 | - | 3 bp deletion | - | - | Asn/- |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, J.-H.; Kang, B.-K.; Dhungana, S.K.; Oh, J.-H.; Choi, M.-S.; Park, J.-H.; Shin, S.-O.; Kim, H.-S.; Baek, I.-Y.; Sung, J.-S.; et al. QTL Mapping and Candidate Gene Analysis for Pod Shattering Tolerance in Soybean (Glycine max). Plants 2020, 9, 1163. https://doi.org/10.3390/plants9091163
Seo J-H, Kang B-K, Dhungana SK, Oh J-H, Choi M-S, Park J-H, Shin S-O, Kim H-S, Baek I-Y, Sung J-S, et al. QTL Mapping and Candidate Gene Analysis for Pod Shattering Tolerance in Soybean (Glycine max). Plants. 2020; 9(9):1163. https://doi.org/10.3390/plants9091163
Chicago/Turabian StyleSeo, Jeong-Hyun, Beom-Kyu Kang, Sanjeev K. Dhungana, Jae-Hyeon Oh, Man-Soo Choi, Ji-Hee Park, Sang-Ouk Shin, Hong-Sik Kim, In-Youl Baek, Jung-Sook Sung, and et al. 2020. "QTL Mapping and Candidate Gene Analysis for Pod Shattering Tolerance in Soybean (Glycine max)" Plants 9, no. 9: 1163. https://doi.org/10.3390/plants9091163
APA StyleSeo, J.-H., Kang, B.-K., Dhungana, S. K., Oh, J.-H., Choi, M.-S., Park, J.-H., Shin, S.-O., Kim, H.-S., Baek, I.-Y., Sung, J.-S., Jung, C.-S., Kim, K.-S., & Jun, T.-H. (2020). QTL Mapping and Candidate Gene Analysis for Pod Shattering Tolerance in Soybean (Glycine max). Plants, 9(9), 1163. https://doi.org/10.3390/plants9091163






