Intra- and Inter-Specific Crosses among Centaurea aspera L. (Asteraceae) Polyploid Relatives—Influences on Distribution and Polyploid Establishment
Abstract
:1. Introduction
2. Results
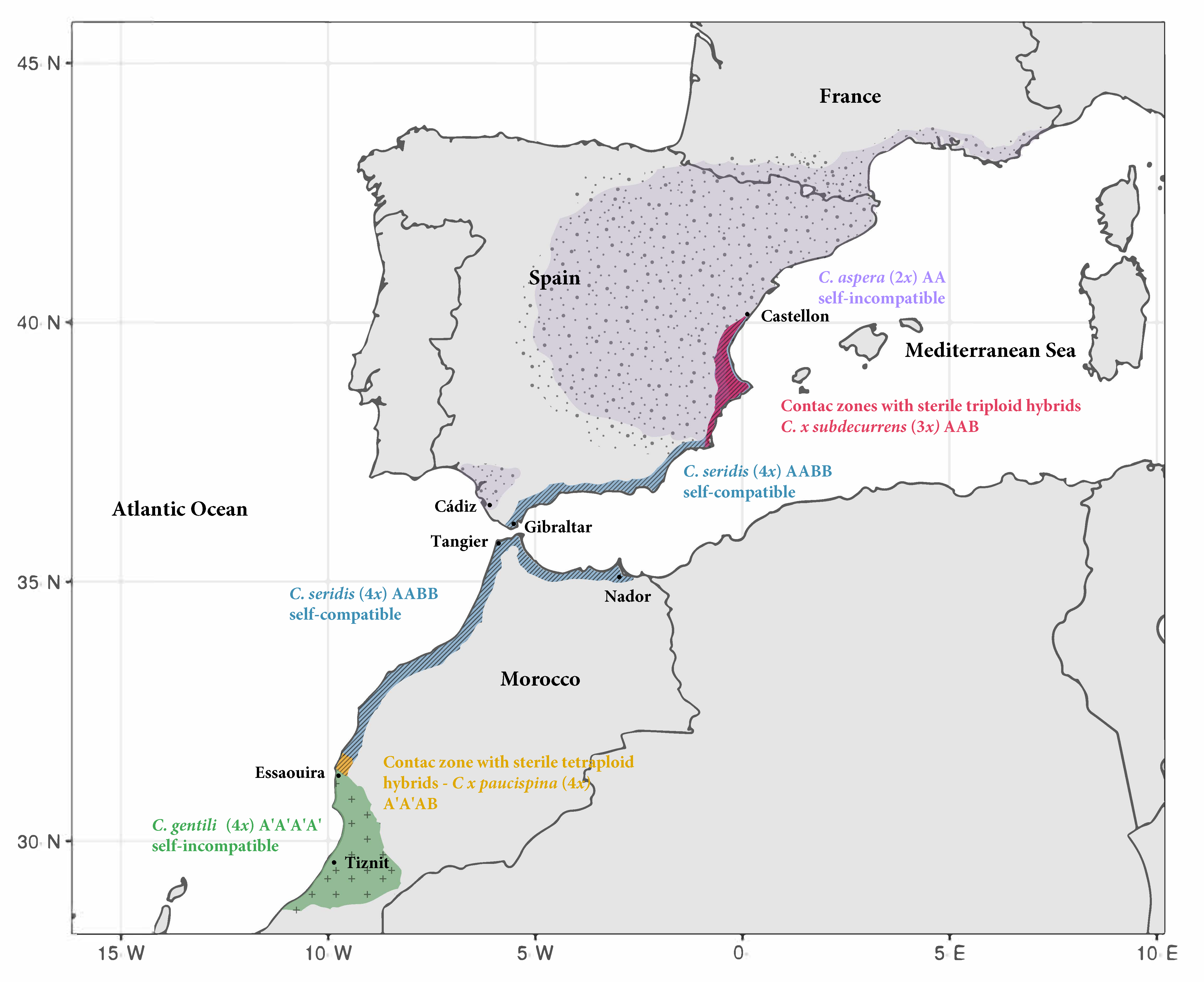
2.1. Centaurea gentilii and C. seridis Geographical Distribution and Ploidy Level in Morocco
2.2. Cypselae Production in Intra-Specific Crosses
2.3. Intra-Specific Cypselae Production between and within Populations in C. aspera and C. gentilii
2.4. Inter-Specific Crosses between C. aspera and C. gentilii
2.5. Offspring Analysis
2.6. Intra- vs. Inter-Specific Treatments
3. Discussion
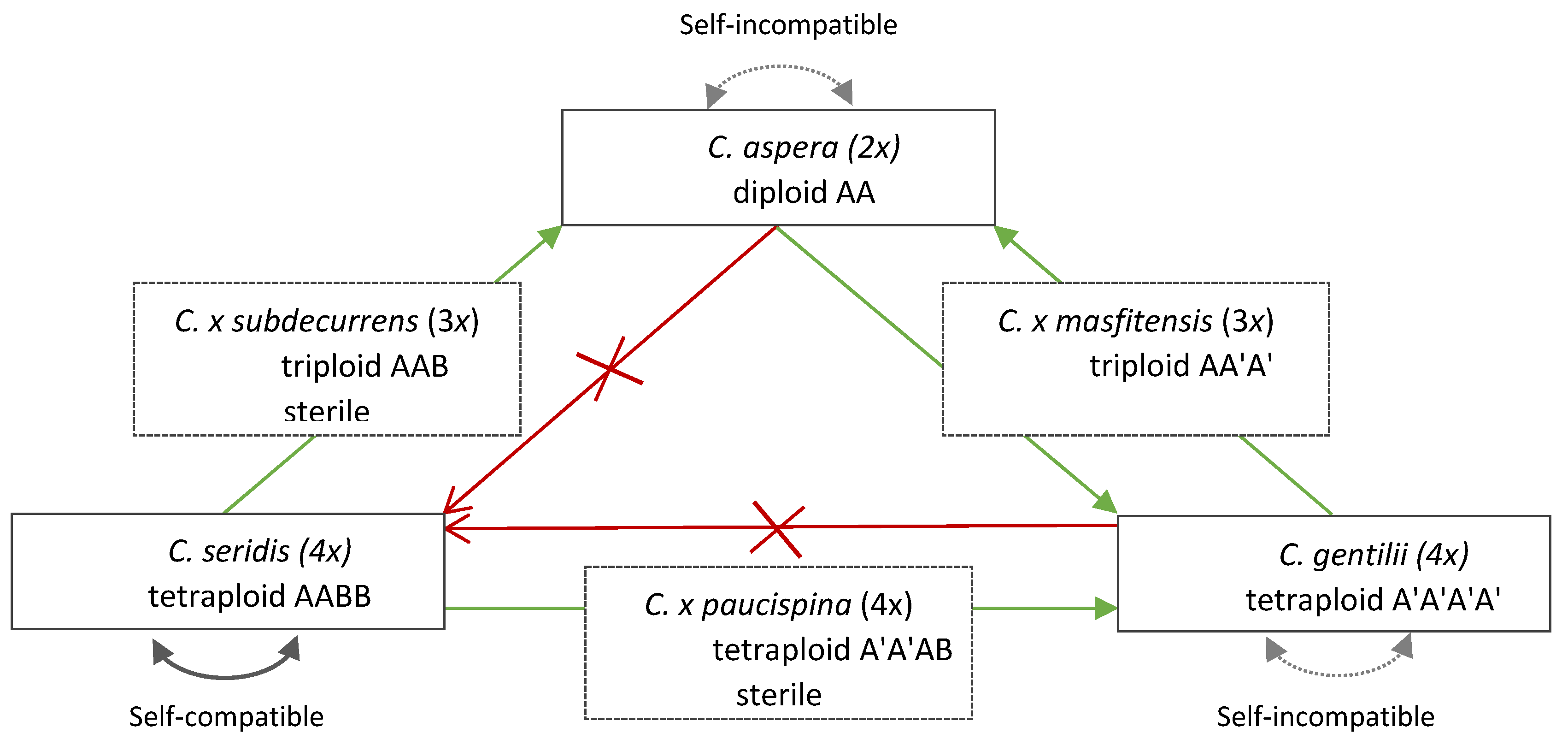
3.1. Biogeography of Centaurea aspera, C. gentilii, and C. seridis
3.2. Mating System of Centaurea aspera Polyploid Complex
3.3. Influence on Polyploid Establishment
4. Materials and Methods
4.1. Geographical Distribution and Ploidy Level of Moroccan Populations
4.2. Controlled Pollinations
4.3. Progeny Analysis
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Ann. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.A. Plant speciation in the age of climate change. Ann. Bot. 2019, 124, 769–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stebbins, G.L., Jr. Types of polyploids: Their classification and significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar] [PubMed]
- Chester, M.; Gallagher, J.P.; Symonds, V.V.; da Silva, A.V.C.; Mavrodiev, E.V.; Leitch, A.R.; Soltis, P.S.; Soltis, D.E. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 2012, 109, 1176–1181. [Google Scholar] [CrossRef] [Green Version]
- Arrigo, N.; Barker, M.S. Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 2012, 15, 140–146. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Pegoraro, L.; De Vos, J.M.; Cozzolino, S.; Scopece, G. Shift in flowering time allows diploid and autotetraploid Anacamptis pyramidalis (Orchidaceae) to coexist by reducing competition for pollinators. Bot. J. Linn. Soc. 2019, 191, 274–284. [Google Scholar] [CrossRef]
- Clausen, J.; Keck, D.D.; Hiesey, W.M. Experimental Studies on the Nature of Species. II. Plant Evolution through Amphidiploidy and Autoploidy, with Examples from the Madiinae; Carnegie Institute of Washington: Washington, DC, USA, 1945. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold Ltd.: London, UK, 1971. [Google Scholar]
- Molina-Henao, Y.F.; Hopkins, R. Autopolyploid lineage shows climatic niche expansion but not divergence in Arabidopsis arenosa. Am. J. Bot. 2019, 106, 61–70. [Google Scholar] [CrossRef]
- Spoelhof, J.P.; Soltis, P.S.; Soltis, D.E. Pure polyploidy: Closing the gaps in autopolyploid research. J. Syst. Evol. 2017, 55, 340–352. [Google Scholar] [CrossRef] [Green Version]
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.A.; Feldman, M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. 2004, 82, 607–613. [Google Scholar] [CrossRef] [Green Version]
- Mavrodiev, E.V.; Chester, M.; Sußrez-Santiago, V.N.; Visger, C.J.; Rodriguez, R.; Susanna, A.; Baldini, R.M.; Soltis, P.S.; Soltis, D.E. Multiple origins and chromosomal novelty in the allotetraploid Tragopogon castellanus (Asteraceae). New Phytol. 2015, 206, 1172–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Bian, Y.; Gou, X.; Zhu, B.; Xu, C.; Qi, B.; Li, N.; Rustgi, S.; Zhou, H.; Han, F. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. Natl. Acad. Sci. USA 2013, 110, 3447–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozkan, H.; Tuna, M.; Galbraith, D.W. No DNA loss in autotetraploids of Arabidopsis thaliana. Plant Breed. 2006, 125, 288–291. [Google Scholar] [CrossRef]
- Eilam, T.; Anikster, Y.; Millet, E.; Manisterski, J.; Feldman, M. Genome size in natural and synthetic autopolyploids and in a natural segmental allopolyploid of several Triticeae species. Genome 2009, 52, 275–285. [Google Scholar] [CrossRef]
- Buggs, R.J.; Zhang, L.; Miles, N.; Tate, J.A.; Gao, L.; Wei, W.; Schnable, P.S.; Barbazuk, W.B.; Soltis, P.S.; Soltis, D.E. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 2011, 21, 551–556. [Google Scholar] [CrossRef] [Green Version]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Grandbastien, M.-A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010, 186, 37–45. [Google Scholar] [CrossRef]
- Stupar, R.M.; Bhaskar, P.B.; Yandell, B.S.; Rensink, W.A.; Hart, A.L.; Ouyang, S.; Veilleux, R.E.; Busse, J.S.; Erhardt, R.J.; Buell, C.R. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 2007, 176, 2055–2067. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J.; Coate, J.E. Autopolyploidy: An epigenetic macromutation. Am. J. Bot. 2020. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Barker, G.L.; Wilson, I.D.; Abbott, R.J.; Edwards, K.J.; Hiscock, S.J. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 2006, 16, 1652–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsey, J.; Schemske, D.W. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 2002, 33, 589–639. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, A.; Bomblies, K. Meiosis in autopolyploid and allopolyploid Arabidopsis. Curr. Opin. Plant Biol. 2016, 30, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Barke, B.H.; Daubert, M.; Hörandl, E. Establishment of apomixis in diploid F2 hybrids and inheritance of apospory from F1 to F2 hybrids of the Ranunculus auricomus complex. Front. Plant Sci. 2018, 9, 1111. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Nybom, H.; Esselink, G.D.; Werlemark, G.; Vosman, B. Microsatellite DNA marker inheritance indicates preferential pairing between two highly homologous genomes in polyploid and hemisexual dog-roses, Rosa L. Sect. Caninae DC. Heredity 2004, 92, 139–150. [Google Scholar] [CrossRef]
- Harlan, J.R.; DeWet, J.M. On Ö. Winge and a prayer: The origins of polyploidy. Bot. Rev. 1975, 41, 361–390. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef] [Green Version]
- Ferriol, M.; Garmendia, A.; Gonzalez, A.; Merle, H. Allogamy-autogamy switch enhance assortative mating in the allotetraploid Centaurea seridis L. coexisting with the diploid Centaurea aspera L. and triggers the asymmetrical formation of triploid hybrids. PLoS ONE 2015, 10, e0140465. [Google Scholar] [CrossRef] [Green Version]
- Norden, E.H.; Lyrene, P.M.; Chaparro, J.X. Ploidy, fertility, and phenotypes of F1 hybrids between tetraploid highbush blueberry cultivars and diploid Vaccinium elliottii. HortScience 2020, 1, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Schinkel, C.C.; Kirchheimer, B.; Dullinger, S.; Geelen, D.; De Storme, N.; Hörandl, E. Pathways to polyploidy: Indications of a female triploid bridge in the alpine species Ranunculus kuepferi (Ranunculaceae). Plant Syst. Evol. 2017, 303, 1093–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crismani, W.; Girard, C.; Mercier, R. Tinkering with meiosis. J. Exp. Bot. 2013, 64, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, A.S.; Pires, J.C. Unreduced gametes: Meiotic mishap or evolutionary mechanism? Trends Genet. 2015, 31, 5–10. [Google Scholar] [CrossRef]
- Decanter, L.; Colling, G.; Elvinger, N.; Heiðmarsson, S.; Matthies, D. Ecological niche differences between two polyploid cytotypes of Saxifraga rosacea. Am. J. Bot. 2020, 107, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Garmendia, A.; Merle, H.; Ruiz, P.; Ferriol, M. Distribution and ecological segregation on regional and microgeographic scales of the diploid Centaurea aspera L., the tetraploid C. seridis L., and their triploid hybrids (Compositae). PeerJ 2018, 6, e5209. [Google Scholar] [CrossRef]
- Pandit, M.K.; Pocock, M.J.; Kunin, W.E. Ploidy influences rarity and invasiveness in plants. J. Ecol. 2011, 99, 1108–1115. [Google Scholar] [CrossRef]
- Ehrendorfer, F. Polyploidy and distribution. In Polyploidy: Biological Relevance; Lewis, H., Walter, H., Eds.; Plenum Press: London, UK, 1980; pp. 45–60. [Google Scholar]
- Linder, H.P.; Barker, N.P. Does polyploidy facilitate long-distance dispersal? Ann. Bot. 2014, 113, 1175–1183. [Google Scholar] [CrossRef]
- Hellwig, F.H. Centaureinae (Asteraceae) in the Mediterranean-History of ecogeographical radiation. Plant Syst. Evol. 2003, 246, 137–162. [Google Scholar] [CrossRef]
- Romaschenko, K.; Ertuǧrul, K.; Susanna, A.; Garcia-Jacas, N.; Uysal, T.; Arslan, E. New chromosome counts in the Centaurea Jacea group (Asteraceae, Cardueae) and some related taxa. Bot. J. Linn. Soc. 2004, 145, 345–352. [Google Scholar] [CrossRef]
- Consortium, M.E.R.P.D.; Austin, J.D.; Bertin, A.; Bórquez, J.P.; Cárdenas, L.; Cardoza, T.B.; Chapman, F.; De Sousa, A.C.B.; De Souza, A.P.; Douglas, K.C. Permanent genetic resources added to molecular ecology resources database 1 February 2011–31 March 2011. Mol. Ecol. Resour. 2011, 11, 757–758. [Google Scholar]
- Ferrer-Gallego, P.P.; Merle, H.; Laguna, E. Typification of Centaurea gentilii (Asteraceae). Phytotaxa 2018, 334, 83–86. [Google Scholar] [CrossRef]
- Ferrer-Gallego, P.P.; Merle Farinós, H.B.; Ferriol Molina, M.; Garmendia, A. A new combination and change in Rank for a Moroccan hybrid in Centaurea (Asteraceae). Flora Montiberica. 2018, 71, 35–37. [Google Scholar]
- Ferriol, M.; Merle, H.; Garmendia, A. Microsatellite evidence for low genetic diversity and reproductive isolation in tetraploid Centaurea seridis (Asteraceae) coexisting with diploid Centaurea aspera and triploid hybrids in contact zones. Bot. J. Linn. Soc. 2014, 176, 82–98. [Google Scholar] [CrossRef] [Green Version]
- Ferriol, M.; Garmendia, A.; Ruiz, J.J.; Merle, H.; Boira, H. Morphological and molecular analysis of natural hybrids between the diploid Centaurea aspera L. and the tetraploid C. seridis L. (Compositae). Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2012, 146, 86–100. [Google Scholar]
- Garmendia, A.; Ferriol, M.; Juarez, J.; Zając, A.; Kałużny, K.; Merle, H. A rare case of a natural contact zone in Morocco between an autopolyploid and an allopolyploid of Centaurea aspera with sterile tetraploid hybrids. Plant Biol. 2015, 17, 746–757. [Google Scholar] [CrossRef]
- Garmendia, A.; Farinós, H.M.; Segura, I.; Ferriol, M. Biogeografía de Centaurea x subdecurrens Pau nothosubsp. subdecurrens como indicador del estado de degradación de las dunas litorales del levante español. In Proceedings of the Biogeografía: Una ciencia para la conservación del medio: (VI Congreso Español de Biogeografía), Alicante, Spain, 7–11 September 2010; pp. 463–469. [Google Scholar]
- Merle Farinós, H.B.; Garmendia Salvador, A.; Ferriol Molina, M. Nuevo híbrido del género Centaurea L. (Compositae) sección Seridia (Juss.) Czerep. Flora Montiberica 2009, 44, 66–71. [Google Scholar]
- Ferriol, M.; Garmendia, A.; Benavent, D.; Ferrer-Gallego, P.P.; Merle, H. Mating system of Centaurea aspera L. (Asteraceae) polyploid relatives—Short Communication. Plant Biosyst. 2020, in press. [Google Scholar]
- Ferrer-Gallego, P.P.; Benavent, D.; Ferriol, M.; Garmendia, A.; Merle, H. Centaurea × masfitensis, Nothosp. nov. (sect. Seridia (Juss.) DC., Asteraceae). Flora Montiberica 2020, in press. [Google Scholar]
- Levin, D.A. Minoritycytotype exclusion in local plant populations. TAXON 1975, 24, 35–43. [Google Scholar] [CrossRef]
- Nielsen, L.R.; Siegismund, H.R.; Hansen, T. Inbreeding depression in the partially self-incompatible endemic plant species Scalesia affinis (Asteraceae) from Galápagos islands. Evol. Ecol. 2007, 21, 1–12. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S. Polyploidy: Recurrent formation and genome evolution. Trends Ecol. Evol. 1999, 14, 348–352. [Google Scholar] [CrossRef]
- Parisod, C.; Besnard, G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae). Mol. Ecol. 2007, 16, 2755–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segraves, K.A.; Thompson, J.N.; Soltis, P.S.; Soltis, D.E. Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia. Mol. Ecol. 1999, 8, 253–262. [Google Scholar] [CrossRef] [Green Version]
- Mráz, P.; Garcia-Jacas, N.; Gex-Fabry, E.; Susanna, A.; Barres, L.; Müller-Schärer, H. Allopolyploid origin of highly invasive Centaurea stoebe s.l. (Asteraceae). Mol. Phylogenetics Evol. 2012, 62, 612–623. [Google Scholar] [CrossRef] [Green Version]
- Čertner, M.; Sudová, R.; Weiser, M.; Suda, J.; Kolář, F. Ploidy-altered phenotype interacts with local environment and may enhance polyploid establishment in Knautia serpentinicola (Caprifoliaceae). New Phytol. 2019, 221, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Ronfort, J. The mutation load under tetrasomic inheritance and its consequences for the evolution of the selfing rate in autotetraploid species. Genet. Res. 1999, 74, 31–42. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 2000, 97, 7051–7057. [Google Scholar] [CrossRef] [Green Version]
- Pannell, J.R.; Obbard, D.J.; Buggs, R.J.A. Polyploidy and the sexual system: What can we learn from Mercurialis annua? Biol. J. Linn. Soc. 2004, 82, 547–560. [Google Scholar] [CrossRef] [Green Version]
- Husband, B.C.; Schemske, D.W. The effect of inbreeding in diploid and tetraploid populations of Epilobium angustifolium (Onagraceae): Implications for the genetic basis of inbreeding depression. Evolution 1997, 51, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Rosquist, G. Reproductive biology in diploid Anthericum ramosum and tetraploid A. liliago (Anthericaceae). Oikos 2001, 92, 143–152. [Google Scholar] [CrossRef]
- Johnston, M.O.; Schoen, D.J. Correlated evolution of self-fertilization and inbreeding depression: An experimental study of nine populations of Amsinckia (Boraginaceae). Evolution 1996, 50, 1478–1491. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. Polyploidy and the distribution of the arctic-alpine flora: New evidence and a new approach. Bot. Helv. 1984, 94, 1–13. [Google Scholar]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2018. [Google Scholar]
- RStudio Team. RStudio: Integrated Development Environment for r; RStudio Team: Boston, MA, USA, 2015. [Google Scholar]
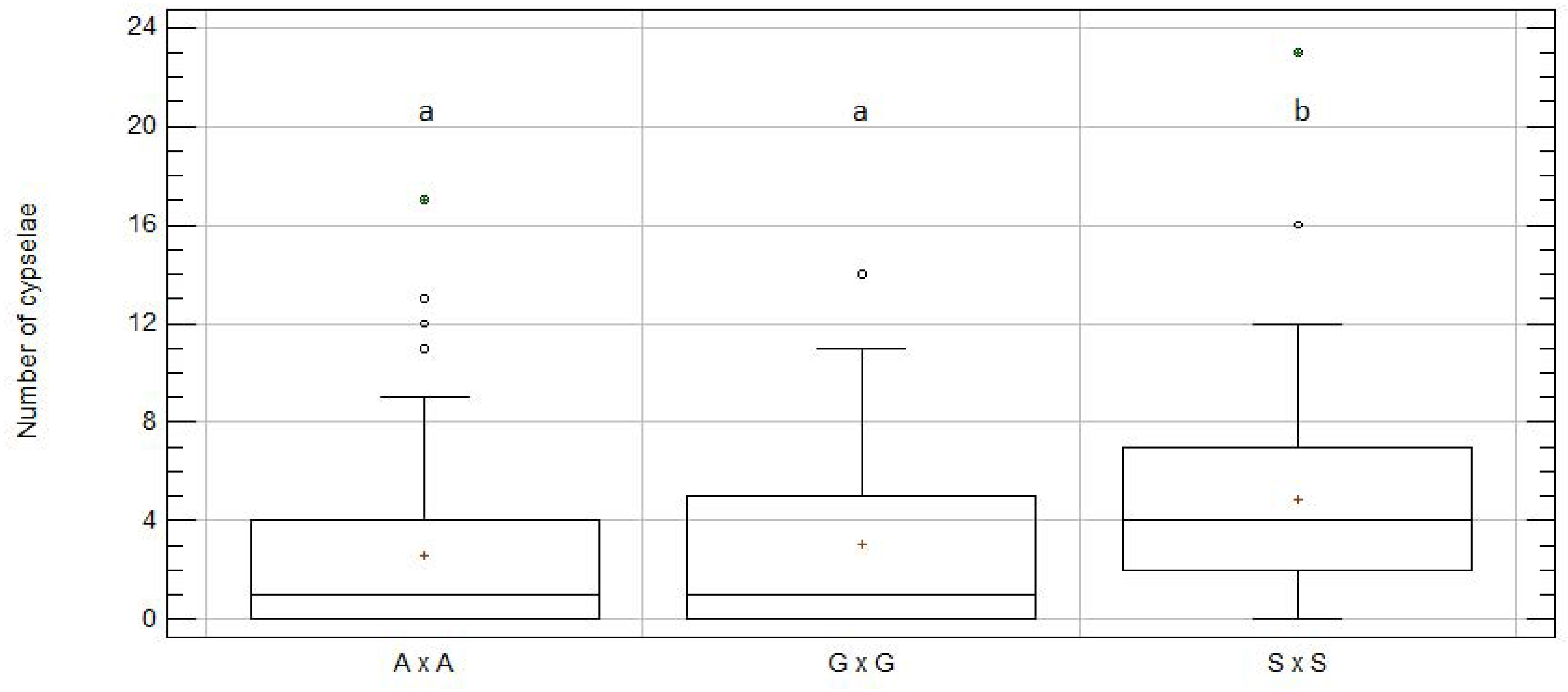
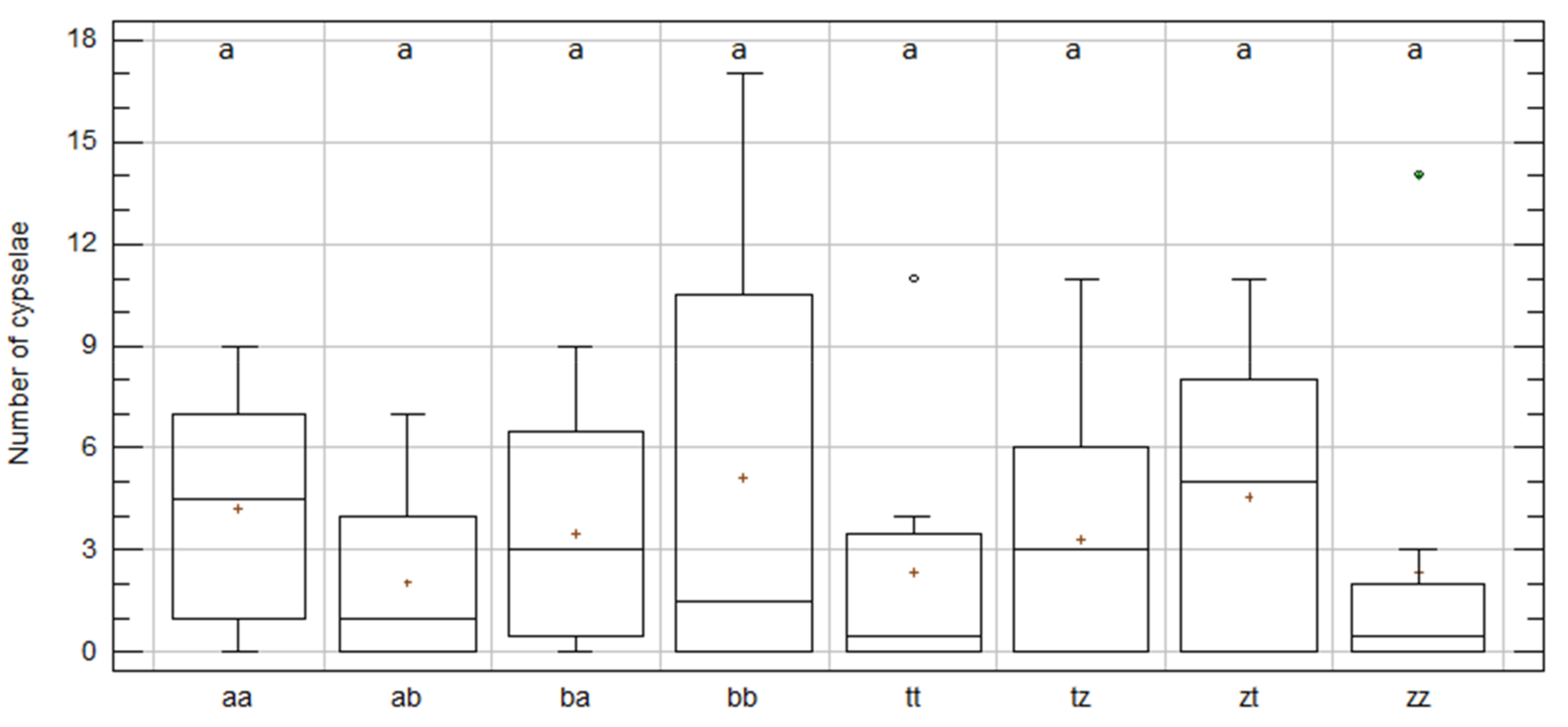
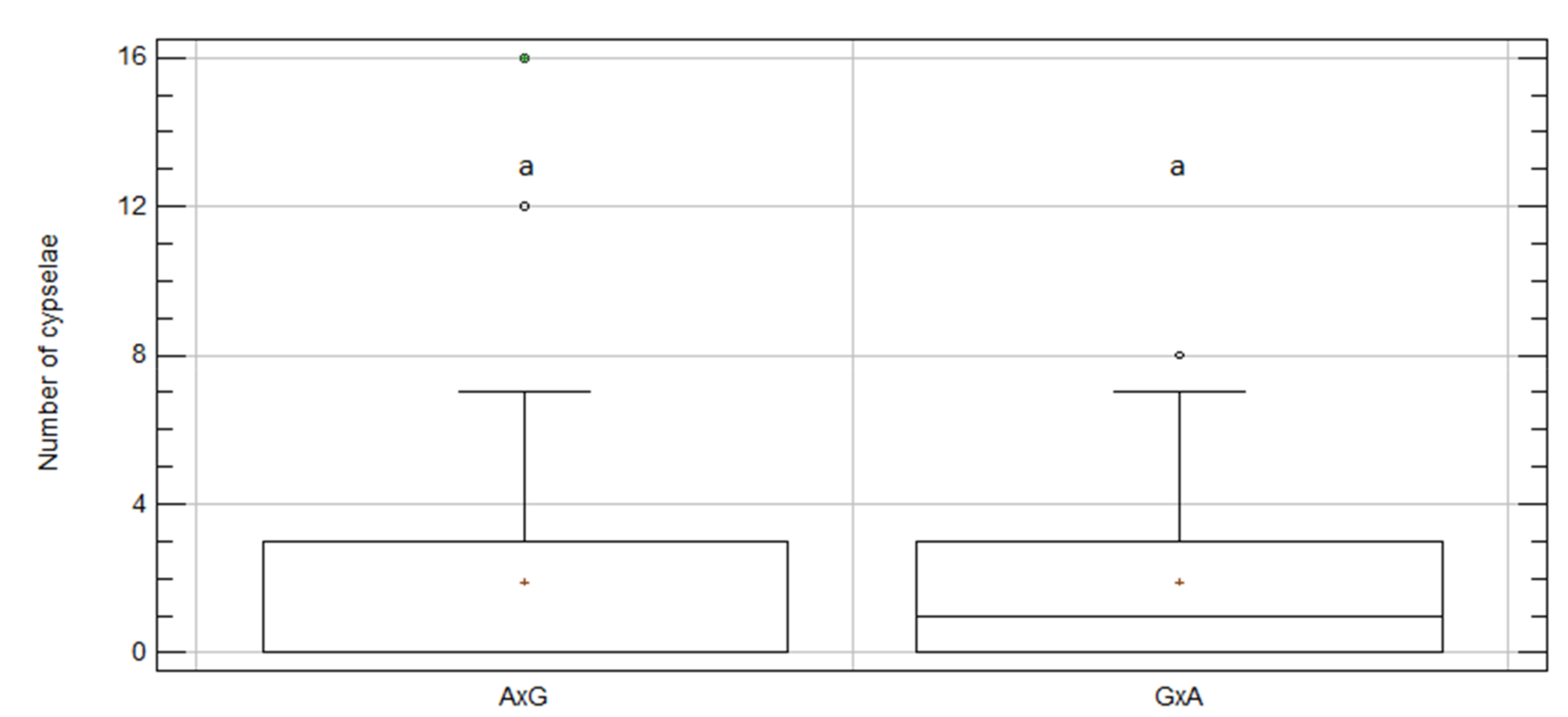

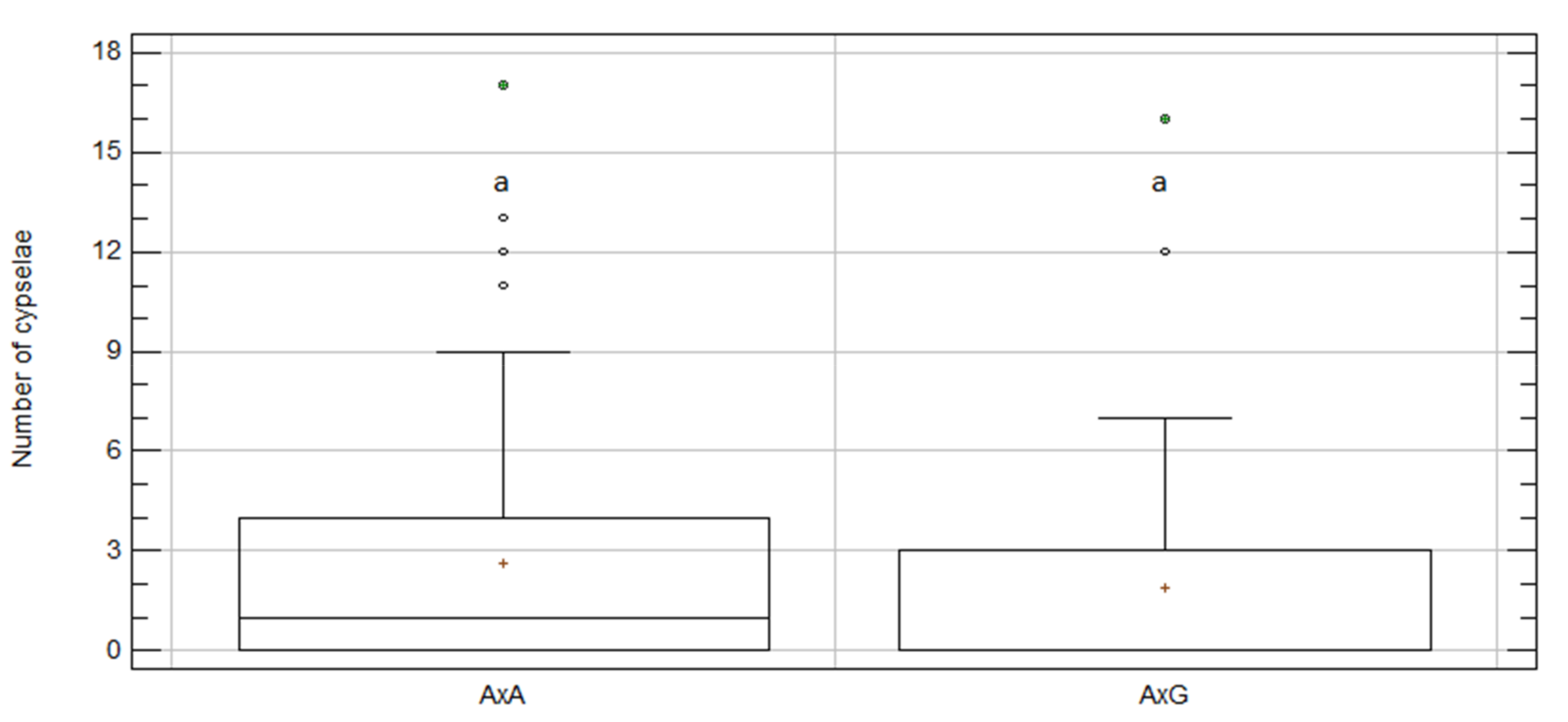
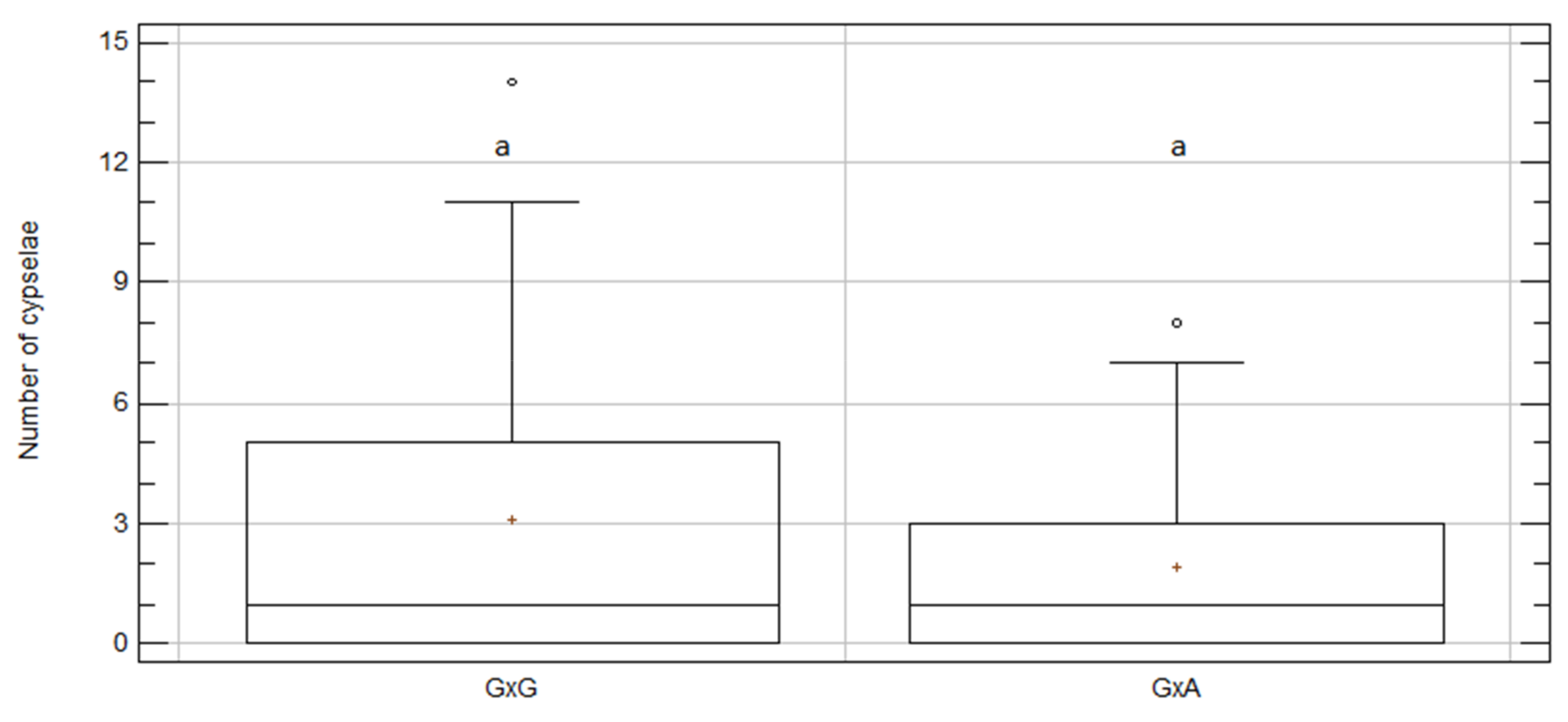
| Taxon | N | Mean | SE | KW | Skew | Kurtosis | Cypselae_sum |
|---|---|---|---|---|---|---|---|
| A × A | 98 | 2.62 | 0.35 | a | 6.59 | 5.50 | 257 |
| G × G | 58 | 3.09 | 0.51 | a | 3.59 | 0.33 | 179 |
| S × S | 48 | 4.88 | 0.64 | b | 4.94 | 7.01 | 234 |
| Total | 204 | 3.28 | 0.28 | - | 9.10 | 9.31 | 670 |
| Location | Sp. | N | Mean | SE | KW | Skew | Kurtosis | Cypselae_sum |
|---|---|---|---|---|---|---|---|---|
| aa | C. aspera | 8 | 4.25 | 1.28 | a | −0.03 | −1.23 | 34 |
| ab | C. aspera | 16 | 2.06 | 0.59 | a | 1.52 | −0.36 | 41 |
| ba | C. aspera | 16 | 3.50 | 0.79 | a | 0.60 | −1.11 | 33 |
| bb | C. aspera | 8 | 5.13 | 2.39 | a | 1.16 | −0.33 | 56 |
| tt | C. gentilii | 8 | 2.38 | 1.35 | a | 2.34 | 2.47 | 19 |
| tz | C. gentilii | 17 | 3.35 | 0.84 | a | 1.49 | −0.19 | 19 |
| zt | C. gentilii | 17 | 4.59 | 1.03 | a | 0.42 | −1.34 | 57 |
| zz | C. gentilii | 8 | 2.38 | 1.70 | a | 2.99 | 3.98 | 78 |
| Total | 98 | 3.44 | 0.40 | - | 4.57 | 1.39 | 337 |
| Mother Taxa | N | Mean | SE | KW | Skew | Kurtosis | Cypselae_sum |
|---|---|---|---|---|---|---|---|
| A × G | 44 | 1.91 | 0.51 | a | 6.67 | 9.41 | 84 |
| G × A | 44 | 1.93 | 0.36 | a | 2.89 | 0.10 | 85 |
| Total | 88 | 1.92 | 0.31 | - | 8.31 | 12.01 | 169 |
| Treatment | N | Mean | SE | KW | Skew | Kurtosis | Cypselae_sum |
|---|---|---|---|---|---|---|---|
| A × A | 98 | 2.62 | 0.35 | a | 6.59 | 5.50 | 257 |
| A × G | 44 | 1.91 | 0.51 | a | 6.67 | 9.41 | 84 |
| Total | 142 | 2.40 | 0.29 | - | 8.92 | 8.60 | 341 |
| Treatment | N | Mean | SE | KW | Skew | Kurtosis | Cypselae_sum |
|---|---|---|---|---|---|---|---|
| G × G | 58 | 3.09 | 0.51 | a | 3.59 | 0.33 | 179 |
| G × A | 44 | 1.93 | 0.36 | a | 2.89 | 0.10 | 85 |
| Total | 102 | 2.59 | 0.33 | - | 5.65 | 2.38 | 264 |
| Population | Species | Code | UTM Coordinates | No. Ind | Cypselae | Country |
|---|---|---|---|---|---|---|
| Takat | C. gentilii | TK | 29 R 440900 3347409 | 66 | no | Morocco |
| Sous Massa | C. gentilii | SM | 29 R 422360 3300019 | 64 | no | Morocco |
| Cap Beddouza | C. seridis | CB | 29 S 492206 3617401 | 66 | no | Morocco |
| Zaouiat Kourati | C. gentilii | ZA/z | 29 R 439299 3508884 | 65 | yes | Morocco |
| Tamri | C. gentilii | TM/t | 29 R 422640 3403933 | 114 | yes | Morocco |
| Bouznika | C. seridis | b | 29 R 670580 3740367 | 15 | yes | Morocco |
| Axdir | C. seridis | a | 29 R 416555 3895607 | 15 | yes | Morocco |
| El Saler | C. aspera | s | 30 R 729751 4362619 | 15 | yes | Spain |
| Chulilla | C. aspera | c | 30 R 680542 4391645 | 15 | yes | Spain |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garmendia, A.; Ferriol, M.; Benavent, D.; Ferrer-Gallego, P.P.; Merle, H. Intra- and Inter-Specific Crosses among Centaurea aspera L. (Asteraceae) Polyploid Relatives—Influences on Distribution and Polyploid Establishment. Plants 2020, 9, 1142. https://doi.org/10.3390/plants9091142
Garmendia A, Ferriol M, Benavent D, Ferrer-Gallego PP, Merle H. Intra- and Inter-Specific Crosses among Centaurea aspera L. (Asteraceae) Polyploid Relatives—Influences on Distribution and Polyploid Establishment. Plants. 2020; 9(9):1142. https://doi.org/10.3390/plants9091142
Chicago/Turabian StyleGarmendia, Alfonso, María Ferriol, David Benavent, P. Pablo Ferrer-Gallego, and Hugo Merle. 2020. "Intra- and Inter-Specific Crosses among Centaurea aspera L. (Asteraceae) Polyploid Relatives—Influences on Distribution and Polyploid Establishment" Plants 9, no. 9: 1142. https://doi.org/10.3390/plants9091142
APA StyleGarmendia, A., Ferriol, M., Benavent, D., Ferrer-Gallego, P. P., & Merle, H. (2020). Intra- and Inter-Specific Crosses among Centaurea aspera L. (Asteraceae) Polyploid Relatives—Influences on Distribution and Polyploid Establishment. Plants, 9(9), 1142. https://doi.org/10.3390/plants9091142








