Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador
Abstract
1. Introduction
2. Results
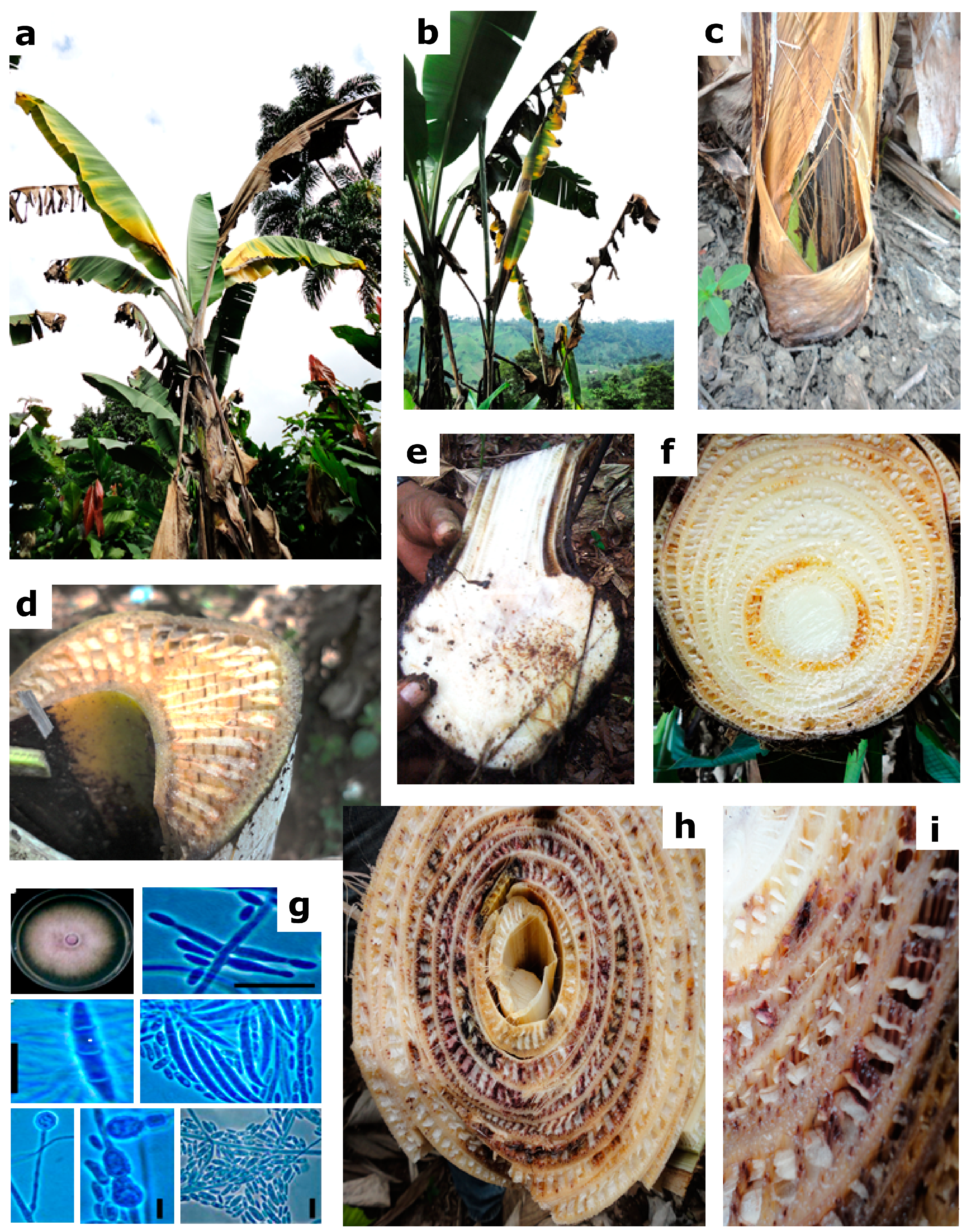
2.1. Distribution of Fusarium Wilt in Ecuador and Fungal Sampling
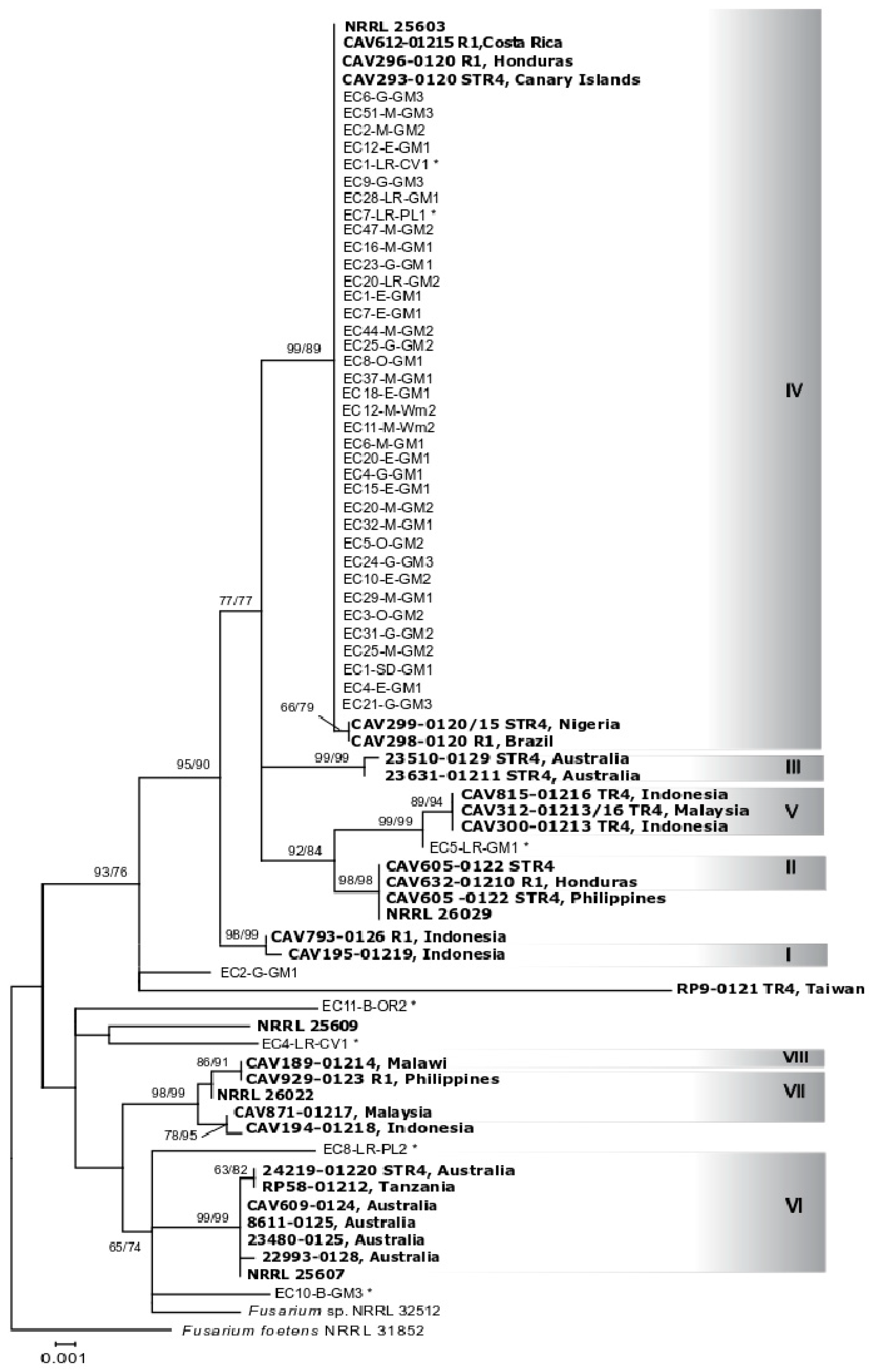
2.2. Identification of Fusarium oxysporum and Phylogenetic Analyses
2.3. Identification of Mating Types and VCG Determination
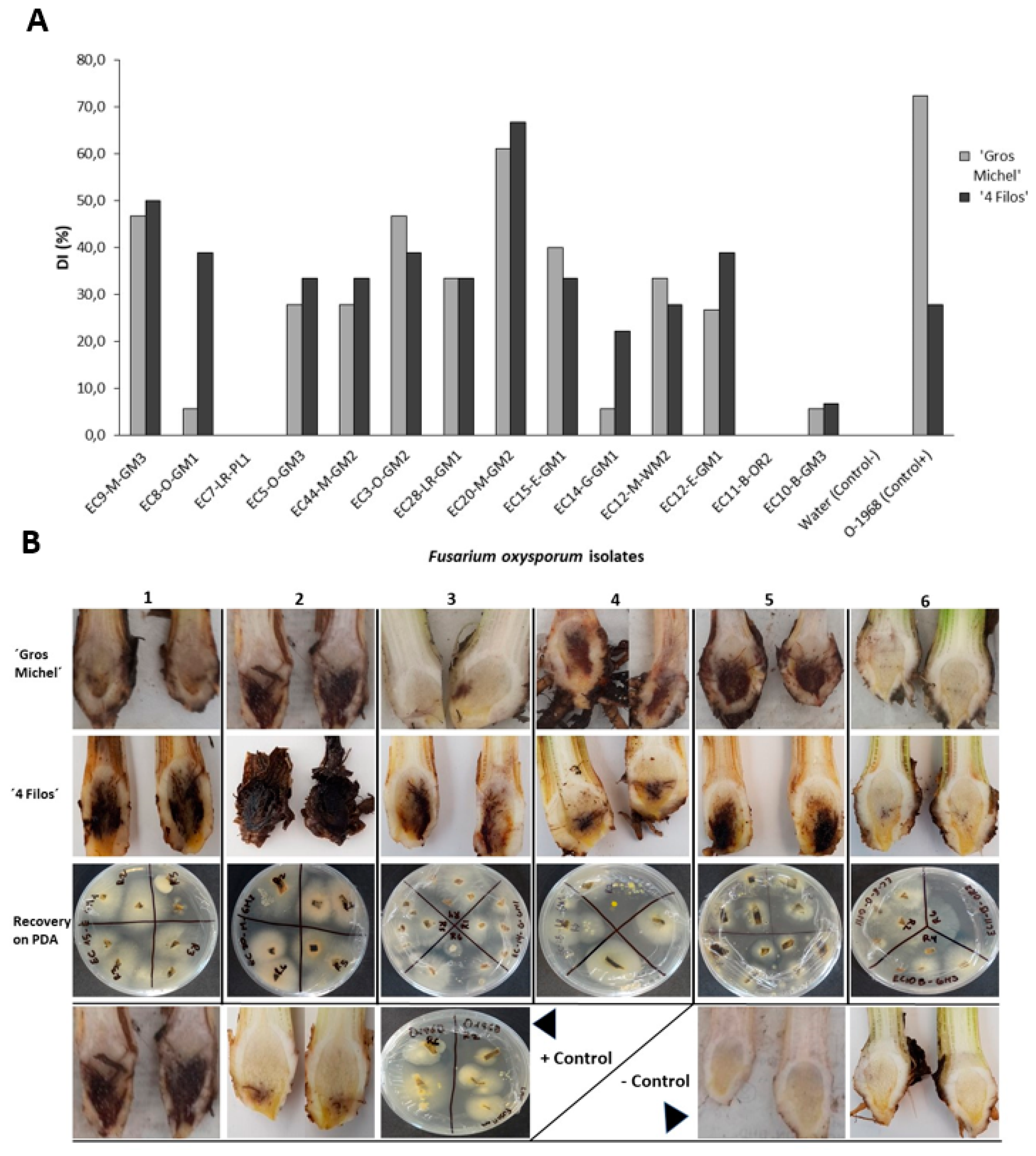
2.4. Pathogenicity Test and Race Determination
3. Discussion
4. Materials and Methods
4.1. Sampling and Fungal Isolation
4.2. DNA Extraction
4.3. PCR Amplification, Sequencing, and Phylogenetic Analyses
4.4. Identification of Mating-Type Idiomorphs
4.5. Vegetative Compatibility Group Analyses
4.6. Pathogenicity Tests
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, A.; Tumuhimbise, R.; Amah, D.; Uwimana, B.; Nyine, M.; Mduma, H.; Talengera, D.; Karamura, D.; Kuriba, J.; Swennen, R.L. Bananas and Plantains (Musa spp.). In Genetic Improvement of Tropical Crops; Campos, H., Caligari, P.D.S., Eds.; Springer: Cham, Switzerland, 2017; pp. 219–240. [Google Scholar] [CrossRef]
- Pareek, S. Nutritional and Biochemical composition of banana (Musa spp.). In Nutritional Composition of Fruit Cultivars; Simmonds, M.S.J., Preedy, V.R., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 49–81. ISBN 9780124081178. [Google Scholar]
- Aurore, G.; Parfait, B.; Fahrasmane, L. Bananas, raw materials for making processed food products. Trends Food Sci. Tech. 2009, 20, 78–91. [Google Scholar] [CrossRef]
- Banana Facts and Figures. Available online: http://www.fao.org/economic/est/est-commodities/bananas/bananafacts/en/#.Xe6UOPZFyM8 (accessed on 9 December 2019).
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 9 December 2019).
- Ploetz, R.C.; Kema, G.H.J.; Ma, L.-J. Impact of diseases on export and smallholder production of banana. Annu. Rev. Phytopathol. 2015, 53, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Lescot, T.B. Diversité génétique. Fruitrop, 15 May 2017; Volume 248. [Google Scholar]
- Ploetz, R.C.; Evans, E.A. The Future of Global Banana Production. In Horticultural Reviews, 1st ed.; Jules, J., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2015; Volume 43, pp. 311–351. ISBN 9781119107781. [Google Scholar]
- Ordonez, N.; Seidl, M.F.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.P.; Ploetz, R.C.; Kema, G.H. Worse Comes to Worst: Bananas and Panama Disease--When Plant and Pathogen Clones Meet. PLoS Pathog. 2015, 19, e1005197. [Google Scholar] [CrossRef] [PubMed]
- Fourie, G.; Steenkamp, E.T.; Gordon, T.R.; Viljoen, A. Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Appl. Environ. Microbiol. 2009, 75, 4770–4781. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F. Genetic exchange within sexual and asexual populations of the genus Fusarium. In Fusarium Wilt of Banana, 2nd ed.; Ploetz, R., Ed.; APS Press: St. Paul, MN, USA, 1990; pp. 37–48. ISBN 0890541124. [Google Scholar]
- Perrier, X.; Bakry, F.; Carreel, F.; Jenny, C.; Horry, J.-P.; Lebot, V.; Hippolyte, I. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Appl. 2009, 7, 199–216. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Zheng, S.-J.; García-Bastidas, F.A.; Li, X.; Zeng, L.; Bai, T.; Xu, S.; Yin, K.; Li, H.; Fu, G.; Yu, Y.; et al. New Geographical Insights of the Latest Expansion of Fusarium oxysporum f. sp. cubense Tropical Race 4 Into the Greater Mekong Subregion. Front. Plant Sci. 2018, 9, 457. [Google Scholar] [CrossRef]
- Damodaran, T.; Rajan, S.; Mishra, V.K.; Jha, S.K.; Ahmad, I.; Gopal, R. First report of Fusarium wilt in banana caused by Fusarium oxysporum f. sp. cubense Tropical race 4 in India. Plant. Dis. Notes. 2018, 103, 1022. [Google Scholar] [CrossRef]
- Maymon, M.; Shpatz, U.; Harel, Y.M.; Levy, E.; Elkind, G.; Teverovsky, E.; Gofman, R.; Haberman, A.; Zemorski, R.; Ezra, N.; et al. First report of Fusarium oxysporum f. sp. cubense tropical race 4 causing Fusarium wilt of Cavendish bananas in Israel. Plant Dis. Notes. 2018, 102, 2655. [Google Scholar] [CrossRef]
- TR4 Present in UK. Available online: http://www.promusa.org/blogpost580-TR4-present-in-the-UK (accessed on 24 July 2020).
- García-Bastidas, F.A.; Quintero-Vargas, J.-C.; Ayala-Vasquez, M.; Schermer, T.; Seidl, M.F.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Hofstede, R.; et al. First Report of Fusarium Wilt Tropical Race 4 in Cavendish Bananas Caused by Fusarium odoratissimum in Colombia. Plant Dis. Notes 2019. [Google Scholar] [CrossRef]
- Özarslandan, M.; Akgül, D.S. First Report of Fusarium oxysporum f. sp. cubense Race 4 Causing Fusarium Wilt Disease of Banana in Turkey. Plant Dis. Notes 2020. [Google Scholar] [CrossRef]
- First report of Fusarium oxysporum f. sp. cubense Tropical race 4 in Mayotte. Available online: https://gd.eppo.int/reporting/article-6694 (accessed on 27 July 2020).
- R&D and Policy Directions for effective Control of Fusarium Wilt Disease of Cavendish Banana in the Asia-Pacific Region. Available online: http://ap.fftc.agnet.org/ap_db.php?id=163#_ftn1 (accessed on 14 January 2020).
- Scheerer, L.; Pemsl, D.; Dita, M.; Perez Vicente, L.; Staver, C. A quantified approach to projecting losses caused by Fusarium wilt Tropical race 4. Acta Hortic. 2016, 1196, 211–218. [Google Scholar] [CrossRef]
- Cook, D.C.; Taylor, A.S.; Meldrum, R.A.; Drenth, A. Potential economic impact of Panama disease. (Tropical race 4) on the Australian banana industry. J. Plant Dis. Prot. 2015, 122, 229–237. [Google Scholar] [CrossRef]
- SIPA. Available online: http://sipa.agricultura.gob.ec/index.php/indicador-agroecomercio (accessed on 14 January 2020).
- Parsons, J.J. Bananas in Ecuador: A new chapter in the history of tropical agriculture. Econ. Geogr. 1957, 33, 201–216. [Google Scholar] [CrossRef]
- Ploetz, R.C. Population biology of Fusarium oxysporum f. sp. cubense. In Fusarium Wilt of Banana; Ploetz, R.C., Ed.; APS Press: St. Paul, MN, USA, 1990; pp. 63–76. ISBN 0-89054-112-4. [Google Scholar]
- Ploetz, R.C. Variability in Fusarium oxysporum f. sp. cubense. Can. J. Bot. 1990, 68, 1357–1363. [Google Scholar] [CrossRef]
- Perez Vicente, L. Fusarium wilt (Panama Disease) of bananas: An updating review of the current knowledge on the disease and its causal agent. In Proceedings of the XVI International ACORBAT Meeting, Anaís, Oaxaca, México, 26 October 2004; Orozco-Santos, M., Orozco-Romero, J., Robles-González, M., Velázquez-Monreal, J., Medina-Urrutia, V., Hernández-Bautista, J.A., Eds.; ACORBAT: Oaxaca, Mexico; pp. 1–15. [Google Scholar]
- Guedez, C.; Rodríguez, D. Compatibilidad vegetativa y raza patogénica de Fusarium oxysporum f. sp. cubense del estado de Trujillo, Venezuela. Fitopatol. Venez. 2004, 17, 30–32. [Google Scholar]
- Ploetz, R.C. Panama disease: An old nemesis rears its ugly head. Part 2. The Cavendish era and beyond. Plant Health Prog. 2005. [Google Scholar] [CrossRef]
- Batlle-Viera, A.; Pérez-Vicente, L. Variabilidad genética de las poblaciones de Fusarium oxysporum f. sp. cubense en bananos y plátanos en Cuba. Fitosanidad 2009, 13, 169–186. [Google Scholar]
- Costa, S.N.; Bragança, C.A.D.; Ribeiro, L.R.; Amorim, E.P.; Oliveira, S.A.S.; Dita, M.A.; Laranjeira, F.F.; Haddad, F. Genetic structure of Fusarium oxysporum f. sp. cubense in different regions from Brazil. Plant Pathol. 2015, 64, 137–146. [Google Scholar]
- Cunha, C.M.S.; Hinz, R.H.; Pereira, A.; Tcacenco, F.A.; Stadnik, M.J. Aggressiveness and genetic diversity of Fusarium oxysporum f. sp. cubense from Santa Catarina, southern Brazil. Trop. Plant Pathol. 2015, 40, 326–334. [Google Scholar]
- Fiusa Araujo, N.A.; Pasqual, M.; Salles Pio, L.A.; Alves, E.; de Matos Moura, N.; da Silva Costa, S. Identification and aggressiveness of four isolates of Fusarium oxysporum f. sp. cubense from Latundan banana in Brazil. J. Phytopathol. 2017, 165, 257–264. [Google Scholar]
- Garcia, R.O.; Rivera-Vargas, L.I.; Ploetz, R.; Correll, J.C.; Irish, B.M. Characterization of Fusarium spp. isolates recovered from bananas (Musa spp.) affected by Fusarium wilt in Puerto Rico. Eur. J. Plant. Pathol. 2018, 152, 599–611. [Google Scholar] [CrossRef]
- Roman, C.H. Consideraciones Epidemiológicas para el Manejo de la Marchitez por Fusarium (Fusarium oxysporum f. sp. cubense) del Banano en la Región Central del Perú. Master’s Thesis, Centro Agronómico Tropical de Investigación y Enseñanza, Turrialba, Costa Rica, 2012. [Google Scholar]
- Zhan, J.; Thrall, P.H.; Papaïx, J.; Xie, L.; Burdon, J.J. Playing on a pathogen’s weakness: Using evolution to guide sustainable plant disease control strategies. Annu. Rev. Phytopathol. 2015, 53, 19–43. [Google Scholar] [CrossRef] [PubMed]
- Burdon, J.J.; Thrall, P.H. Pathogen evolution across the agro-ecological interface: Implications for disease management. Evol. Appl. 2008, 1, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Stukenbrock, E.H.; McDonald, B.A. The origins of plant pathogens in agroecosystems. Annu. Rev. Phytopathol. 2008, 46, 75–100. [Google Scholar] [CrossRef]
- McKinney, H. A new system of grading plant diseases. J. Agric. Res. 1923, 26, 195–218. [Google Scholar]
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 2019, 10, 1395. [Google Scholar] [CrossRef]
- Baayen, R.P.; O’Donnell, K.; Bonants, P.J.; Cigelnik, E.; Kroon, L.P.; Roebroeck, E.J.; Waalwijk, C. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 2000, 90, 891–900. [Google Scholar] [CrossRef]
- Geiser, D.M.; Jiménez-Gasco, M.M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- O’Donnell, K.; Gueidan, C.; Sink, S.; Johnston, P.R.; Crous, P.W.; Glenn, A.; Riley, R.; Zitomer, N.C.; Colyer, P.; Waalwijk, C.; et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009, 46, 936–948. [Google Scholar] [CrossRef]
- Laurence, M.H.; Burgess, L.W.; Summerell, B.A.; Liew, E.C.Y. High levels of diversity in Fusarium oxysporum from non-cultivated ecosystems in Australia. Fungal Biol. 2012, 116, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Laurence, M.H.; Summerell, B.A.; Burgess, L.W.; Liew, E.C.Y. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biol. 2014, 118, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Lievens, B.; van Baarlen, P.; Verreth, C.; van Kerckhove, S.; Rep, M.; Thomma, B.P. Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici isolates inferred from mating type, elongation factor-1alpha and exopolygalacturonase sequences. Mycol. Res. 2009, 113, 1181–1191. [Google Scholar]
- Mostert, D.; Molina, A.B.; Daniells, J.; Fourie, G.; Hermanto, C.; Chao, C.P.; Fabregar, E.; Sinohin, V.G.; Masdek, N.; Thangavelu, R.; et al. The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE 2017, 12, e0181630. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A. The status of Fusarium wilt (Panama disease) of Banana in South Africa. S. Afr. J. Sci. 2002, 98, 341–344. [Google Scholar]
- Magdama, F.; Monserrate-Maggi, L.; Serrano, L.; Sosa, D.; Geiser, D.M.; Jiménez-Gasco, M.d.M. Comparative analysis uncovers the limitations of current molecular detection methods for Fusarium oxysporum f. sp. cubense race 4 strains. PLoS ONE 2019, 14, e0222727. [Google Scholar] [CrossRef]
- Demers, J.E.; Gugino, B.K.; Jimenez-Gasco, M.M. Highly diverse endophytic and soil Fusarium oxysporum populations associated with field-grown tomato plants. Appl. Environ. Microbiol. 2015, 81, 81–90. [Google Scholar] [CrossRef]
- Balmas, V.; Migheli, Q.; Scherm, B.; Garau, P.; O’Donnell, K.; Ceccherelli, G.; Kang, S.; Geiser, D.M. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 2010, 102, 803–812. [Google Scholar] [CrossRef]
- Buddenhagen, I. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of Tropical race 4 to better manage banana production. Acta Hortic. 2009, 828, 193–204. [Google Scholar] [CrossRef]
- Ordoñez, N. A Global Genetic Diversity Analysis of Fusarium oxysporum f. sp. cubense the Panama Disease Pathogen of Banana. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 16 October 2018. [Google Scholar]
- Julian, R. The International Banana Trade, 1st ed.; Woodhead Publishing Limited: Cambridge, UK, 1998; p. 288. ISBN 9781855734050. [Google Scholar]
- Langdon, R. The banana as a key to early American and Polynesian history. J. Pacific. Hist. 1993, 28, 15–35. [Google Scholar] [CrossRef]
- Nayar, N.M. The Bananas: Botany, Origin, Dispersal. In Horticultural Reviews, 1st ed.; Janick, J., Ed.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2010; Volume 36, pp. 117–164. [Google Scholar]
- Marin, D.H.; Sutton, T.B.; Barker, K.R. Dissemination of bananas in Latin America and the Caribbean and its relationship to the occurrence of Radopholus similis. Plant Dis. 1998, 82, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Striffer, S.; Moberg, M. Banana Wars: Power, Production and History in the Americas; Striffer, S., Moberg, M., Eds.; Duke University Press: Durham, NC, USA, 2003; p. 360. ISBN 0822331594. [Google Scholar]
- SIPA. Available online: http://sipa.agricultura.gob.ec/index.php/comercio-exterior (accessed on 10 February 2020).
- Instituto Colombiano Agropecuario (ICA). Available online: https://www.ica.gov.co/noticias/ica-amplia-y-refuerza-las-medidas-que-ya-venia-im (accessed on 10 February 2020).
- Nash, S.M.; Snyder, W.C. Quantitative estimations by plate counts of propagules of bean root rot Fusarium in fields soils. Phytopathology 1962, 52, 567–572. [Google Scholar]
- Cenis, J.L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids. Res. 1992, 20, 2380. [Google Scholar] [CrossRef] [PubMed]
- Appel, D.J.; Gordon, T.R. Relationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Mol. Plant. Microbe. Interact. 1996, 9, 125–138. [Google Scholar] [CrossRef]
- Edel, V.; Steinberg, C.; Gautheron, N.; Recorbet, G.; Alabouvette, C. Genetic diversity of Fusarium oxysporum populations isolated from different soils in France. FEMS Microbiol. Ecol. 2001, 36, 61–71. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Edel, V.; Steinberg, C.; Avelange, I. Comparison of three molecular methods for the characterization of Fusarium oxysporum strains. Phytopathology 1995, 85, 579–585. [Google Scholar] [CrossRef]
- Zwickl, D.J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. Ph.D. Thesis, The University of Texas at Austin, Austin, TX, USA, 2006. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Arie, T.; Kaneko, I.; Yoshida, T.; Noguchi, M.; Nomura, Y.; Yamaguchi, I. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant. Microbe. Interact. 2000, 13, 1330–1339. [Google Scholar] [CrossRef]
- Steenkamp, E.T.; Wingfield, B.D.; Coutinho, T.A.; Zeller, K.A.; Wingfield, M.J.; Marasas, W.F.O.; Leslie, J.F. PCR-Based Identification of MAT-1 and MAT-2 in the Gibberella fujikuroi Species Complex. Appl. Environ. Microbiol. 2000, 66, 4378–4382. [Google Scholar] [CrossRef]
- Puhalla, J. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Can. J. Bot. 1985, 63, 179–183. [Google Scholar] [CrossRef]
- Correll, J.C.; Klittich, C.J.R.; Leslie, J.F. Nitrate non-utilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 1987, 77, 1640–1646. [Google Scholar] [CrossRef]
- Jiménez-Gasco, M.M.; Pérez-Artés, E.; Jiménez-Díaz, R.M. Identification of pathogenic races 0, 1B/C, 5, and 6 of Fusarium oxysporum f. sp. ciceris with random amplified polymorphic DNA (RAPD). Eur. J. Plant Pathol. 2001, 107, 237–248. [Google Scholar]
- Li, W.M.; Dita, M.; Wu, W.; Hu, G.B.; Xie, J.H.; Ge, X.J. Resistance sources to Fusarium oxysporum f. sp. cubense tropical race 4 in banana wild relatives. Plant Pathol. 2015, 64, 1061–1067. [Google Scholar]


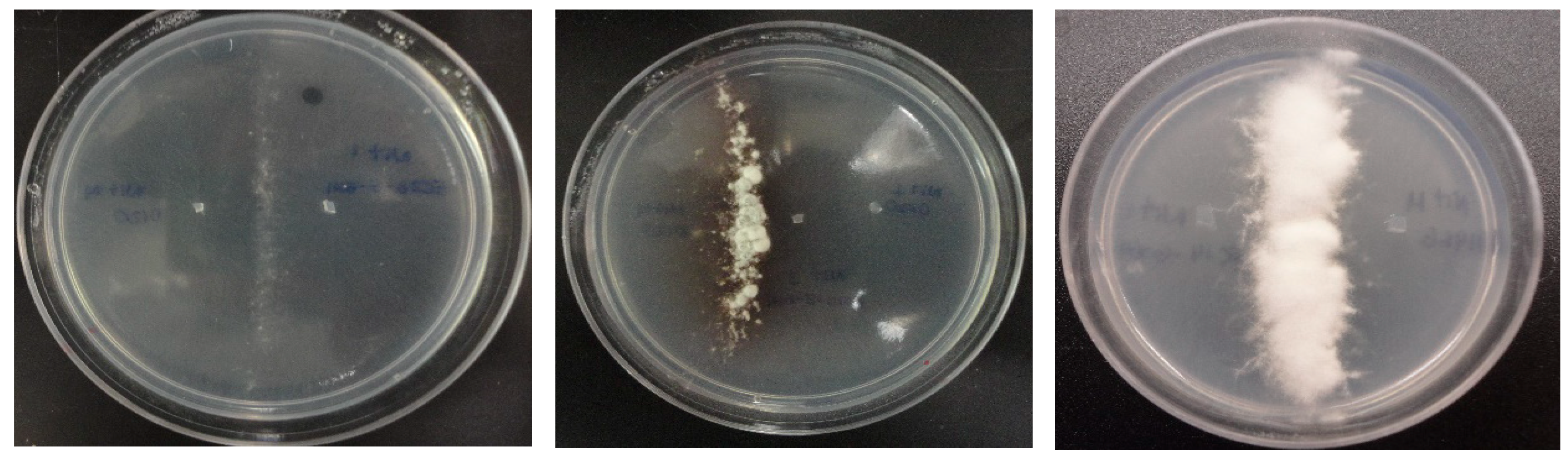
| Isolate Code | VCG 0120 | VCG 0123 | VCG 01213 |
|---|---|---|---|
| EC4-E-GM1 | ++ | − | − |
| EC10-E-GM2 | ++ | − | − |
| EC12-E-GM1 | +++ | − | − |
| EC15-E-GM1 | +++ | − | − |
| EC18-E-GM1 | +++ | − | − |
| EC2-M-GM2 | − | − | − |
| EC11-M-GM2 | − | − | − |
| EC18-M-GM2 | +++ | − | − |
| EC29-M-GM1 | ++ | − | − |
| EC32-M-GM2 | +++ | − | − |
| EC44-M-GM2 | +++ | − | − |
| EC51-M-GM3 | + | − | − |
| EC8-G-GM1 | +++ | − | − |
| EC9-G-GM3 | − | − | − |
| EC14-G-GM1 | +++ | − | − |
| EC21-G-GM3 | − | − | − |
| EC23-G-GM1 | +++ | − | − |
| EC24-G-GM3 | ++ | − | − |
| EC25-G-GM1 | ++ | − | − |
| EC32-G-GM1 | + | − | − |
| EC28-LR-GM1 | + | − | − |
| EC3-O-GM2 | +++ | − | − |
| EC5-O-GM2 | − | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magdama, F.; Monserrate-Maggi, L.; Serrano, L.; García Onofre, J.; Jiménez-Gasco, M.d.M. Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador. Plants 2020, 9, 1133. https://doi.org/10.3390/plants9091133
Magdama F, Monserrate-Maggi L, Serrano L, García Onofre J, Jiménez-Gasco MdM. Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador. Plants. 2020; 9(9):1133. https://doi.org/10.3390/plants9091133
Chicago/Turabian StyleMagdama, Freddy, Lorena Monserrate-Maggi, Lizette Serrano, José García Onofre, and María del Mar Jiménez-Gasco. 2020. "Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador" Plants 9, no. 9: 1133. https://doi.org/10.3390/plants9091133
APA StyleMagdama, F., Monserrate-Maggi, L., Serrano, L., García Onofre, J., & Jiménez-Gasco, M. d. M. (2020). Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador. Plants, 9(9), 1133. https://doi.org/10.3390/plants9091133








