Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium
Abstract
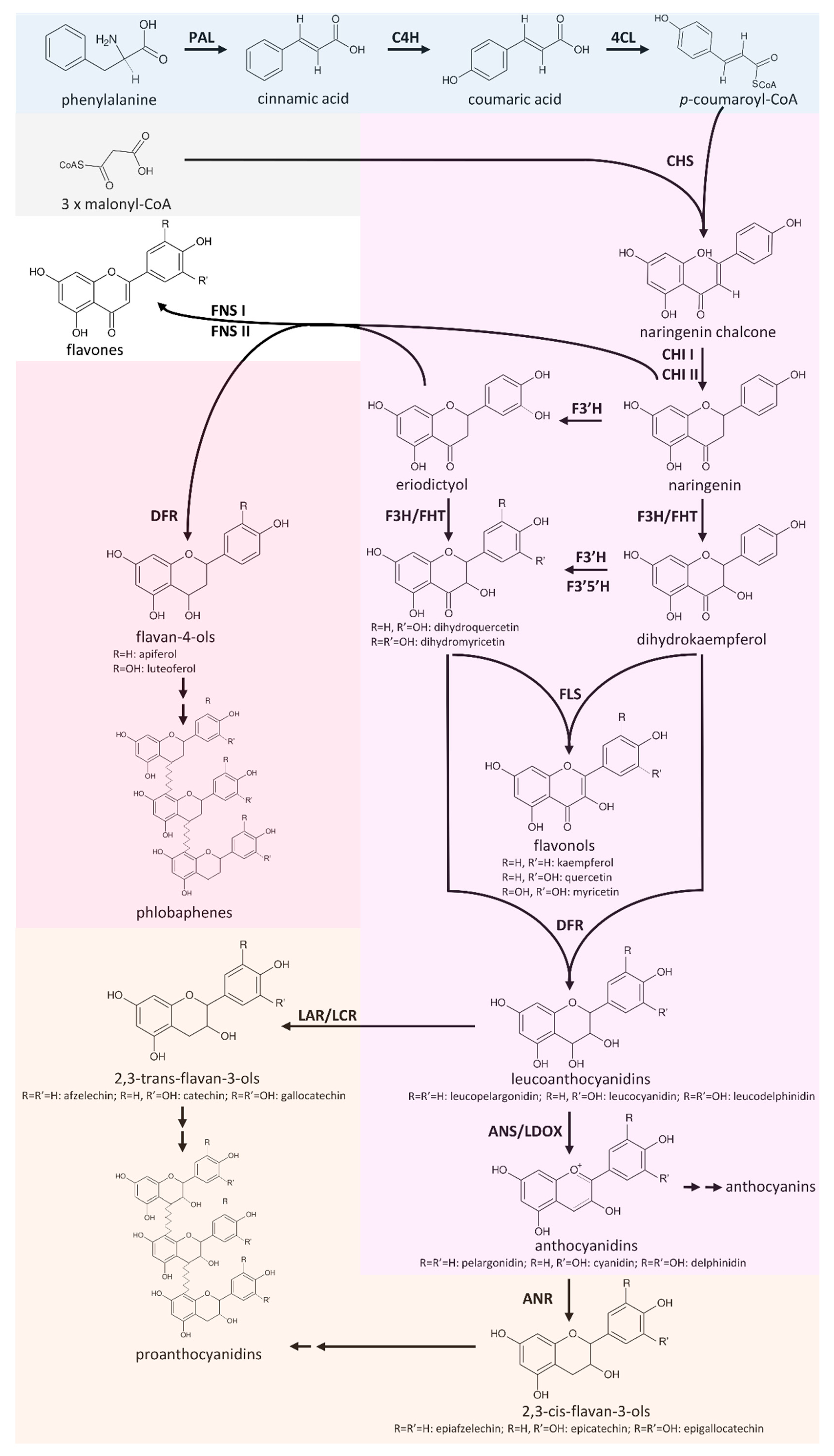
:1. Introduction
2. Results
2.1. Concept and Components of Knowledge-Based Identification of Pathway Enzymes (KIPEs)
2.1.1. General Concept
2.1.2. Three Modes
2.1.3. Final Filtering
2.2. Technical Validation of KIPEs
2.3. The Flavonoid Biosynthesis Enzymes in Croton tiglium
2.4. Transcriptional Regulators of the Flavonoid Biosynthesis in Croton tiglium
3. Discussion
4. Materials and Methods
4.1. Retrieval of Bait and Reference Sequences
4.2. Collection of Information about Important Amino Acid Residues
4.3. Implementation and Availability of KIPEs
4.4. Phylogenetic Analysis
4.5. Transcript Abundance Quantification
4.6. Application of KIPEs for the Identification of Transcription Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Williams, C.A.; Grayer, R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Kandaswami, C.; Kanadaswami, C.; Lee, L.-T.; Lee, P.-P.H.; Hwang, J.-J.; Ke, F.-C.; Huang, Y.-T.; Lee, M.-T. The antitumor activities of flavonoids. In Vivo 2005, 19, 895–909. [Google Scholar] [PubMed]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Marais, J.P.J.; Deavours, B.; Dixon, R.A.; Ferreira, D. The Stereochemistry of Flavonoids; Grotewold, E., Ed.; Springer: New York, NY, USA, 2006; pp. 47–69. [Google Scholar]
- Pourcel, L.; Routaboul, J.-M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant. Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef]
- Murphy, A.; Peer, W.A.; Taiz, L. Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 2000, 211, 315–324. [Google Scholar] [CrossRef]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Harborne, J.B. Recent advances in chemical ecology. Nat. Prod. Rep. 1999, 16, 509–523. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef]
- Kozłowska, A.; Szostak-Wegierek, D. Flavonoids--food sources and health benefits. Rocz. Panstw. Zakl. Hig. 2014, 65, 79–85. [Google Scholar] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Rice-Evans, C.; Miller, N.; Paganga, G. Antioxidant properties of phenolic compounds. Trends Plant. Sci. 1997, 2, 152–159. [Google Scholar] [CrossRef]
- Chen, A.Y.; Chen, Y.C. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013, 138, 2099–2107. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Furusaki, S. Production of anthocyanins by plant cell cultures. Biotechnol. Bioprocess. Eng. 1999, 4, 231–252. [Google Scholar] [CrossRef]
- Appelhagen, I.; Wulff-Vester, A.K.; Wendell, M.; Hvoslef-Eide, A.-K.; Russell, J.; Oertel, A.; Martens, S.; Mock, H.-P.; Martin, C.; Matros, A. Colour bio-factories: Towards scale-up production of anthocyanins in plant cell cultures. Metab. Eng. 2018, 48, 218–232. [Google Scholar] [CrossRef]
- Forkmann, G. Flavonoids as Flower Pigments: The Formation of the Natural Spectrum and its Extension by Genetic Engineering. Plant Breed. 1991, 106, 1–26. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Koornneef, M. Mutations affecting the testa color in Arabidopsis. Arab. Inf. Serv. 1990, 28, 1–4. [Google Scholar]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Casas, M.I.; Questa, J.I.; Herrera, A.L.; Deblasio, S.; Wang, J.; Jackson, D.; Grotewold, E.; Casati, P. Evolution and expression of tandem duplicated maize flavonol synthase genes. Front. Plant. Sci. 2012, 3, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer, J.-L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar] [CrossRef]
- Van der Krol, A.R.; Mur, L.A.; Beld, M.; Mol, J.N.; Stuitje, A.R. Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 1990, 2, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Schröder, G.; Schröder, J. A single change of histidine to glutamine alters the substrate preference of a stilbene synthase. J. Biol. Chem. 1992, 267, 20558–20560. [Google Scholar]
- Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 2000, 7, 786–791. [Google Scholar] [CrossRef]
- Jez, J.M.; Noel, J.P. Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences. J. Biol. Chem. 2002, 277, 1361–1369. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.-X.; Zhang, X.; Han, X.-J.; Zhang, Y.-Y.; Gao, S.; Liu, C.-J.; Lou, H.-X. Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for synthesis of flavonoids. New Phytol. 2018, 217, 909–924. [Google Scholar] [CrossRef] [Green Version]
- Kaltenbach, M.; Burke, J.R.; Dindo, M.; Pabis, A.; Munsberg, F.S.; Rabin, A.; Kamerlin, S.C.L.; Noel, J.P.; Tawfik, D.S. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol. 2018, 14, 548–555. [Google Scholar] [CrossRef] [Green Version]
- Forkmann, G.; Heller, W.; Grisebach, H. Anthocyanin Biosynthesis in Flowers of Matthiola incana Flavanone 3-and Flavonoid 3′-Hydroxylases. Z. Nat. C 1980, 35, 691–695. [Google Scholar] [CrossRef]
- Prescott, A.G.; John, P. DIOXYGENASES: Molecular Structure and Role in Plant Metabolism. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1996, 47, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.-X.; Han, X.-J.; Wu, Y.-F.; Lou, H.-X. The Function and Catalysis of 2-Oxoglutarate-Dependent Oxygenases Involved in Plant Flavonoid Biosynthesis. Int J. Mol. Sci 2014, 15, 1080–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, D.K.; Alerding, A.B.; Crosby, K.C.; Bandara, A.B.; Westwood, J.H.; Winkel, B.S.J. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008, 147, 1046–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Kim, D.-H.; Park, B.-R.; Lee, J.-Y.; Lim, S.-H. Molecular and Functional Characterization of Oryza sativa Flavonol Synthase (OsFLS), a Bifunctional Dioxygenase. J. Agric. Food Chem. 2019, 67, 7399–7409. [Google Scholar] [CrossRef]
- Xu, F.; Li, L.; Zhang, W.; Cheng, H.; Sun, N.; Cheng, S.; Wang, Y. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 2012, 39, 2285–2296. [Google Scholar] [CrossRef]
- Turnbull, J.J.; Nagle, M.J.; Seibel, J.F.; Welford, R.W.D.; Grant, G.H.; Schofield, C.J. The C-4 stereochemistry of leucocyanidin substrates for anthocyanidin synthase affects product selectivity. Bioorganic Med. Chem. Lett. 2003, 13, 3853–3857. [Google Scholar] [CrossRef]
- Welford, R.W.D.; Turnbull, J.J.; Claridge, T.D.W.; Prescott, A.G.; Schofield, C.J. Evidence for oxidation at C-3 of the flavonoid C-ring during anthocyanin biosynthesis. Chem. Commun. 2001, 1828–1829. [Google Scholar] [CrossRef]
- Yan, Y.; Chemler, J.; Huang, L.; Martens, S.; Koffas, M.A.G. Metabolic Engineering of Anthocyanin Biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 3617–3623. [Google Scholar] [CrossRef] [Green Version]
- Almeida, J.R.M.; D’Amico, E.; Preuss, A.; Carbone, F.; de Vos, C.H.R.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; et al. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria xananassa). Arch. Biochem. Biophys. 2007, 465, 61–71. [Google Scholar] [CrossRef]
- Brugliera, F.; Barri-Rewell, G.; Holton, T.A.; Mason, J.G. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant. J. 1999, 19, 441–451. [Google Scholar] [CrossRef] [PubMed]
- de Vetten, N.; ter Horst, J.; van Schaik, H.P.; de Boer, A.; Mol, J.; Koes, R. A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 1999, 96, 778–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, K.M.; Hehn, A.; Jugdé, H.; Slimestad, R.; Larbat, R.; Bourgaud, F.; Lillo, C. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010, 10, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, C.; Ameres, S.; Forkmann, G. Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett. 2007, 581, 3429–3434. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant. J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Forkmann, G.; Vlaming, P.; de Spribille, R.; Wiering, H.; Schram, A.W. Genetic and Biochemical Studies on the Conversion of Dihydroflavonols to Flavonols in Flowers of Petunia hybrida. Z. Nat. C 1985, 41, 179–186. [Google Scholar] [CrossRef]
- Britsch, L.; Heller, W.; Grisebach, H. Conversion of Flavanone to Flavone, Dihydroflavonol and Flavonol with an Enzyme System from Cell Cultures of Parsley. Z. Nat. C 1981, 36, 742–750. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of Flavonol Synthase and Leucoanthocyanidin Dioxygenase Genes in Arabidopsis (Further Evidence for Differential Regulation of “Early” and “Late” Genes). Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Hostetler, G.L.; Ralston, R.A.; Schwartz, S.J. Flavones: Food Sources, Bioavailability, Metabolism, and Bioactivity. Adv. Nutr. 2017, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.; Forkmann, G.; Britsch, L.; Wellmann, F.; Matern, U.; Lukacin, R. Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley. FEBS Lett. 2003, 544, 93–98. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.; Forkmann, G. Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J. 1999, 20, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, Y.H.; Witte, S.; Steuber, H.; Matern, U.; Martens, S. Evolution of Flavone Synthase I from Parsley Flavanone 3β-Hydroxylase by Site-Directed Mutagenesis. Plant Physiol. 2007, 144, 1442–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Lewis, D.H.; Bloor, S.J.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant. J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Miosic, S.; Thill, J.; Milosevic, M.; Gosch, C.; Pober, S.; Molitor, C.; Ejaz, S.; Rompel, A.; Stich, K.; Halbwirth, H. Dihydroflavonol 4-Reductase Genes Encode Enzymes with Contrasting Substrate Specificity and Show Divergent Gene Expression Profiles in Fragaria Species. PLoS ONE 2014, 9, e112707. [Google Scholar] [CrossRef] [Green Version]
- Katsu, K.; Suzuki, R.; Tsuchiya, W.; Inagaki, N.; Yamazaki, T.; Hisano, T.; Yasui, Y.; Komori, T.; Koshio, M.; Kubota, S.; et al. A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 2017, 17, 239. [Google Scholar] [CrossRef]
- Gang, D.R.; Kasahara, H.; Xia, Z.Q.; Vander Mijnsbrugge, K.; Bauw, G.; Boerjan, W.; Van Montagu, M.; Davin, L.B.; Lewis, N.G. Evolution of plant defense mechanisms. Relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J. Biol. Chem. 1999, 274, 7516–7527. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Shen, L.; Yuan, J.; Zheng, H.; Su, Q.; Yang, W.; Zhang, L.; Nnaemeka, V.E.; Sun, J.; Ke, L.; et al. Functional analysis of GhCHS, GhANR and GhLAR in colored fiber formation of Gossypium hirsutum L. BMC Plant Biol. 2019, 19, 455. [Google Scholar] [CrossRef] [Green Version]
- Timberlake, C.F.; Bridle, P. Spectral Studies of Anthocyanin and Anthocyanidin Equilibria in Aqueous Solution. Nature 1966, 212, 158–159. [Google Scholar] [CrossRef]
- Kovinich, N.; Saleem, A.; Rintoul, T.L.; Brown, D.C.W.; Arnason, J.T.; Miki, B. Coloring genetically modified soybean grains with anthocyanins by suppression of the proanthocyanidin genes ANR1 and ANR2. Transgenic Res. 2012, 21, 757–771. [Google Scholar] [CrossRef]
- Xie, D.-Y.; Sharma, S.B.; Dixon, R.A. Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch. Biochem. Biophys. 2004, 422, 91–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisshaar, B.; Jenkins, G.I. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant. Biol. 1998, 1, 251–257. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.-G.; Tran, L.-S.P.; Lu, K.; Huang, Y.-B.; Li, J.-N. The Evolutionary History of R2R3-MYB Proteins Across 50 Eukaryotes: New Insights Into Subfamily Classification and Expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Pillet, J.; Yu, H.-W.; Chambers, A.H.; Whitaker, V.M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef]
- Pandey, A.; Alok, A.; Lakhwani, D.; Singh, J.; Asif, M.H.; Trivedi, P.K. Genome-wide Expression Analysis and Metabolite Profiling Elucidate Transcriptional Regulation of Flavonoid Biosynthesis and Modulation under Abiotic Stresses in Banana. Sci. Rep. 2016, 6, 31361. [Google Scholar] [CrossRef] [Green Version]
- Otani, M.; Kanemaki, Y.; Oba, F.; Shibuya, M.; Funayama, Y.; Nakano, M. Comprehensive isolation and expression analysis of the flavonoid biosynthesis-related genes in Tricyrtis spp. Biol Plant 2018, 62, 684–692. [Google Scholar] [CrossRef]
- Qu, C.; Zhao, H.; Fu, F.; Wang, Z.; Zhang, K.; Zhou, Y.; Wang, X.; Wang, R.; Xu, X.; Tang, Z.; et al. Genome-Wide Survey of Flavonoid Biosynthesis Genes and Gene Expression Analysis between Black- and Yellow-Seeded Brassica napus. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.; Xia, H.; Qiu, J.; Li, T.; Zhang, Y.; Wang, X.; et al. Genome-Wide Analysis of Gene Expression Provides New Insights into Cold Responses in Thellungiella salsuginea. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Firouzabadi, F.N.; Shobbar, Z.-S. Transcriptome response of roots to salt stress in a salinity-tolerant bread wheat cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef] [PubMed]
- Sicilia, A.; Testa, G.; Santoro, D.F.; Cosentino, S.L.; Lo Piero, A.R. RNASeq analysis of giant cane reveals the leaf transcriptome dynamics under long-term salt stress. BMC Plant Biol 2019, 19, 355. [Google Scholar] [CrossRef] [Green Version]
- Pucker, B.; Schilbert, H.M. Genomics and Transcriptomics Advance in Plant Sciences. In Molecular Approaches in Plant Biology and Environmental Challenges; Energy, Environment, and Sustainability; Singh, S.P., Upadhyay, S.K., Pandey, A., Kumar, S., Eds.; Springer: Singapore, 2019; pp. 419–448. ISBN 9789811506901. [Google Scholar]
- Kalwij, J.M. Review of ‘The Plant List, a working list of all plant species. J. Veg. Sci. 2012, 23, 998–1002. [Google Scholar] [CrossRef]
- Pope, J. On a New Preparation of Croton Tiglium. Med. Chir. Trans. 1827, 13, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Gläser, S.; Sorg, B.; Hecker, E. A Method for Quantitative Determination of Polyfunctional Diterpene Esters of the Tigliane Type in Croton tiglium. Planta Med. 1988, 54, 580. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.J.; Han, Y.B.; Moon, J.J.; Kim, J.B. Isolation of isoguanosine from Croton tiglium and its antitumor activity. Arch. Pharm. Res. 1994, 17, 115–118. [Google Scholar] [CrossRef]
- El-Mekkawy, S.; Meselhy, M.R.; Nakamura, N.; Hattori, M.; Kawahata, T.; Otake, T. Anti-HIV-1 phorbol esters from the seeds of Croton tiglium. Phytochemistry 2000, 53, 457–464. [Google Scholar] [CrossRef]
- Abbas, M.; Shahid, M.; Sheikh, M.A.; Muhammad, G. Phytochemical Screening of Plants Used in Folkloric Medicine: Effect of Extraction Method and Solvent. Asian J. Chem. 2014, 26, 6194–6198. [Google Scholar] [CrossRef]
- Palmeira, S.F., Jr.; Conserva, L.M.; Silveira, E.R. Two clerodane diterpenes and flavonoids from Croton brasiliensis. J. Braz. Chem. Soc. 2005, 16, 1420–1424. [Google Scholar] [CrossRef] [Green Version]
- Kostova, I.; Iossifova, T.; Rostan, J.; Vogler, B.; Kraus, W.; Navas, H. Chemical and biological studies on Croton panamensis latex (Dragon’s Blood). Pharm. Pharmacol. Lett. 1999, 9, 34–36. [Google Scholar]
- Haak, M.; Vinke, S.; Keller, W.; Droste, J.; Rückert, C.; Kalinowski, J.; Pucker, B. High Quality de Novo Transcriptome Assembly of Croton tiglium. Front. Mol. Biosci. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, X.; Sun, R.; Zhao, P.; Liu, F.; Zhang, C. Croton Tiglium Extract Induces Apoptosis via Bax/Bcl-2 Pathways in Human Lung Cancer A549 Cells. Asian Pac. J. Cancer Prev. 2016, 17, 4893–4898. [Google Scholar] [CrossRef] [PubMed]
- Salatino, A.; Salatino, M.L.F.; Negri, G. Traditional uses, chemistry and pharmacology of Croton species (Euphorbiaceae). J. Braz. Chem. Soc. 2007, 18, 11–33. [Google Scholar] [CrossRef]
- Tsacheva, I.; Rostan, J.; Iossifova, T.; Vogler, B.; Odjakova, M.; Navas, H.; Kostova, I.; Kojouharova, M.; Kraus, W. Complement inhibiting properties of dragon’s blood from Croton draco. Z. Nat. C J. Biosci. 2004, 59, 528–532. [Google Scholar] [CrossRef]
- Maciel, M.A.; Pinto, A.C.; Arruda, A.C.; Pamplona, S.G.; Vanderlinde, F.A.; Lapa, A.J.; Echevarria, A.; Grynberg, N.F.; Côlus, I.M.; Farias, R.A.; et al. Ethnopharmacology, phytochemistry and pharmacology: A successful combination in the study of Croton cajucara. J. Ethnopharmacol. 2000, 70, 41–55. [Google Scholar] [CrossRef]
- Guerrero, M.F.; Puebla, P.; Carrón, R.; Martín, M.L.; Román, L.S. Quercetin 3,7-dimethyl ether: A vasorelaxant flavonoid isolated from Croton schiedeanus Schlecht. J. Pharm. Pharmacol. 2002, 54, 1373–1378. [Google Scholar] [CrossRef]
- Krebs, H.C.; Ramiarantsoa, H. Clerodane diterpenes of Croton hovarum. Phytochemistry 1997, 45, 379–381. [Google Scholar] [CrossRef]
- Shimada, S.; Otsuki, H.; Sakuta, M. Transcriptional control of anthocyanin biosynthetic genes in the Caryophyllales. J. Exp. Bot. 2007, 58, 957–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 Gene Encodes a Basic Helix-Loop-Helix Domain Protein Required for Expression of DFR and BAN Genes in Arabidopsis Siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, R.; Zhu, T.-T.; Zhang, X.-S.; Wang, P.-Y.; Sun, C.-J.; Qiao, Y.-N.; Lou, H.-X.; Cheng, A.-X. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J. Exp. Bot. 2020, 71, 290–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Evans, F.J.; Roberts, M.F.; Phillipson, J.D.; Zenk, M.H.; Gleba, Y.Y. Polyphenolic compounds from Croton lechleri. Phytochemistry 1991, 30, 2033–2040. [Google Scholar] [CrossRef]
- Furlan, C.M.; Santos, K.P.; Sedano-Partida, M.D.; Motta, L.B.; da Santos, D.Y.A.C.; Salatino, M.L.F.; Negri, G.; Berry, P.E.; van Ee, B.W.; Salatino, A. Flavonoids and antioxidant potential of nine Argentinian species of Croton (Euphorbiaceae). Braz. J. Bot. 2015, 38, 693–702. [Google Scholar] [CrossRef]
- de Sampaio e Spohr, T.C.L.; Stipursky, J.; Sasaki, A.C.; Barbosa, P.R.; Martins, V.; Benjamim, C.F.; Roque, N.F.; Costa, S.L.; Gomes, F.C.A. Effects of the flavonoid casticin from Brazilian Croton betulaster in cerebral cortical progenitors in vitro: Direct and indirect action through astrocytes. J. Neurosci. Res. 2010, 88, 530–541. [Google Scholar] [CrossRef]
- Biscaro, F.; Parisotto, E.B.; Zanette, V.C.; Günther, T.M.F.; Ferreira, E.A.; Gris, E.F.; Correia, J.F.G.; Pich, C.T.; Mattivi, F.; Filho, D.W.; et al. Anticancer activity of flavonol and flavan-3-ol rich extracts from Croton celtidifolius latex. Pharm. Biol. 2013, 51, 737–743. [Google Scholar] [CrossRef] [Green Version]
- Aderogba, M.A.; Ndhlala, A.R.; Rengasamy, K.R.R.; Van Staden, J. Antimicrobial and selected in vitro enzyme inhibitory effects of leaf extracts, flavonols and indole alkaloids isolated from Croton menyharthii. Molecules 2013, 18, 12633–12644. [Google Scholar] [CrossRef]
- Cruz, B.G.; Dos Santos, H.S.; Bandeira, P.N.; Rodrigues, T.H.S.; Matos, M.G.C.; Nascimento, M.F.; de Carvalho, G.G.C.; Braz-Filho, R.; Teixeira, A.M.R.; Tintino, S.R.; et al. Evaluation of antibacterial and enhancement of antibiotic action by the flavonoid kaempferol 7-O-β-D-(6″-O-cumaroyl)-glucopyranoside isolated from Croton piauhiensis müll. Microb. Pathog. 2020, 143, 104144. [Google Scholar] [CrossRef]
- Nascimento, A.M.; Maria-Ferreira, D.; Dal Lin, F.T.; Kimura, A.; de Santana-Filho, A.P.; Werner, M.F.D.P.; Iacomini, M.; Sassaki, G.L.; Cipriani, T.R.; de Souza, L.M. Phytochemical analysis and anti-inflammatory evaluation of compounds from an aqueous extract of Croton cajucara Benth. J. Pharm. Biomed. Anal. 2017, 145, 821–830. [Google Scholar] [CrossRef]
- Prescott, A.G.; Stamford, N.P.J.; Wheeler, G.; Firmin, J.L. In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 2002, 60, 589–593. [Google Scholar] [CrossRef]
- Lukacin, R.; Wellmann, F.; Britsch, L.; Martens, S.; Matern, U. Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry 2003, 62, 287–292. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keilwagen, J.; Hartung, F.; Paulini, M.; Twardziok, S.O.; Grau, J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinform. 2018, 19, 189. [Google Scholar] [CrossRef] [Green Version]
- Pucker, B.; Brockington, S.F. Genome-wide analyses supported by RNA-Seq reveal non-canonical splice sites in plant genomes. BMC Genom. 2018, 19, 980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Li, B.; Zhang, Y.; Zhang, X.; Xu, Y.; Li, C. Evolutionary dynamic analyses on monocot flavonoid 3′-hydroxylase gene family reveal evidence of plant-environment interaction. BMC Plant Biol. 2019, 19, 347. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Brown, J.W.; Walker, J.F.; Smith, S.A. Phyx: Phylogenetic tools for unix. Bioinformatics 2017, 33, 1886–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Canedo-Téxon, A.; Ramón-Farias, F.; Monribot-Villanueva, J.L.; Villafán, E.; Alonso-Sánchez, A.; Pérez-Torres, C.A.; Ángeles, G.; Guerrero-Analco, J.A.; Ibarra-Laclette, E. Novel findings to the biosynthetic pathway of magnoflorine and taspine through transcriptomic and metabolomic analysis of Croton draco (Euphorbiaceae). BMC Plant Biol. 2019, 19, 560. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [Green Version]
- Pucker, B.; Pandey, A.; Weisshaar, B.; Stracke, R. The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. bioRxiv 2020. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix–Loop–Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, T.-Y.; Liu, Y.-Y.; Zhu, H.-H.; Zhang, J.; Yang, J.-X.; Fu, Q.; Wang, N.; Wang, Z. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ 2019, 7, e7153. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Qiu, J.-Y.; Hui, Q.-L.; Xu, Y.-Y.; He, Y.-Z.; Peng, L.-Z.; Fu, X.-Z. Systematic analysis of the basic/helix-loop-helix (bHLH) transcription factor family in pummelo (Citrus grandis) and identification of the key members involved in the response to iron deficiency. BMC Genom. 2020, 21, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Jia, C.; Wang, J.-Y.; Miao, H.-X.; Liu, J.-H.; Chen, C.; Yang, H.-X.; Xu, B.; Jin, Z. Genome-Wide Analysis of Basic Helix-Loop-Helix Transcription Factors to Elucidate Candidate Genes Related to Fruit Ripening and Stress in Banana (Musa acuminata L. AAA Group, cv. Cavendish). Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Li, L.; Wei, M.; Yang, F. Genome-wide analysis of basic helix–loop–helix superfamily members related to anthocyanin biosynthesis in eggplant (Solanum melongena L.). PeerJ 2019, 7, e7768. [Google Scholar] [CrossRef]
- van Nocker, S.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, R.; Xiao, J.; Gu, T.; Yu, X.; Zhang, Y.; Chang, J.; Yang, G.; He, G. Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genom. 2018, 19, 803. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.K.; Muthamilarasan, M.; Khan, Y.; Parida, S.K.; Prasad, M. Genome-Wide Investigation and Expression Analyses of WD40 Protein Family in the Model Plant Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e86852. [Google Scholar] [CrossRef] [Green Version]

| Sequence ID | Function | Leaf | Stem | Inflorescence | Root | Seed |
|---|---|---|---|---|---|---|
| DN23351_c0_g1_i2 | CtPALa | 18 | 10 | 0 | 1 | 29 |
| DN32981_c5_g1_i1 | CtPALb | 0 | 0 | 0 | 0 | 0 |
| DN32981_c5_g1_i16 | CtPALc | 4 | 3 | 0 | 0 | 0 |
| DN32981_c5_g1_i9 | CtPALd | 0 | 0 | 0 | 0 | 0 |
| DN32981_c5_g1_i17 | CtPALe | 0 | 25 | 0 | 5 | 10 |
| DN32981_c5_g1_i12 | CtPALf | 5 | 233 | 0 | 18 | 48 |
| DN32981_c5_g1_i5 | CtPALg | 0 | 318 | 0 | 3 | 3 |
| DN32981_c5_g1_i13 | CtPALh | 0 | 3 | 0 | 0 | 0 |
| DN32981_c5_g1_i14 | CtPALi | 0 | 0 | 0 | 0 | 0 |
| DN23351_c0_g2_i1 | CtPALj | 18 | 1 | 0 | 9 | 12 |
| DN32464_c6_g3_i2 | CtC4Ha | 122 | 110 | 2 | 77 | 233 |
| DN15593_c0_g1_i1 | CtC4Hb | 0 | 0 | 0 | 0 | 3 |
| DN32164_c5_g1_i2 | Ct4CLa | 46 | 19 | 1 | 2 | 113 |
| DN50385_c0_g1_i1 | CtCHSa | 3 | 6 | 0 | 1 | 588 |
| DN27125_c0_g1_i1 | CtCHI Ia | 11 | 2 | 19 | 3 | 88 |
| DN33424_c3_g3_i1 | CtF3Ha | 4 | 21 | 1 | 2 | 342 |
| DN33407_c7_g7_i2 1 | CtFNS IIa | 1 | 7 | 0 | 95 | 1 |
| DN33407_c7_g7_i1 1 | CtFNS IIb | 0 | 3 | 1 | 4 | 2 |
| DN27999_c0_g1_i2 1 | CtFNS IIc | 0 | 2 | 0 | 75 | 0 |
| DN33407_c7_g6_i4 1 | CtFNS IId | 0 | 0 | 0 | 22 | 0 |
| DN252_c0_g1_i1 | CtF3′Ha | 111 | 62 | 0 | 9 | 165 |
| DN32466_c16_g7_i1 | CtF3′5′Ha | 0 | 0 | 0 | 7 | 266 |
| DN32466_c16_g7_i3 | CtF3′5′Hb | 0 | 0 | 0 | 0 | 3 |
| DN25915_c0_g1_i3 | CtFLSa | 18 | 19 | 0 | 0 | 84 |
| DN25915_c0_g2_i1 | CtFLSb | 0 | 1 | 0 | 1 | 2 |
| DN27402_c0_g1_i3 | CtDFRa | 0 | 0 | 0 | 0 | 51 |
| DN32893_c8_g1_i1 | CtANSa | 0 | 0 | 0 | 0 | 25 |
| DN33042_c3_g1_i3 | CtLARa | 3 | 2 | 0 | 1 | 101 |
| DN30161_c9_g1_i2 | CtANRa | 0 | 1 | 0 | 1 | 375 |
| DN30161_c9_g1_i3 | CtANRb | 0 | 0 | 0 | 0 | 3 |
| Sequence ID | Group | Leaf | Stem | Inflorescence | Root | Seed |
|---|---|---|---|---|---|---|
| DN30455_c10_g1_i1 | Subgroup 7 | 0 | 1 | 0 | 0 | 4 |
| DN21046_c0_g1_i3 | Subgroup 7 | 0 | 0 | 0 | 0 | 0 |
| DN21046_c0_g1_i2 | Subgroup 7 | 0 | 1 | 0 | 0 | 9 |
| DN28041_c1_g1_i4 | Subgroup 6 | 0 | 0 | 0 | 0 | 7 |
| DN28041_c1_g1_i2 | Subgroup 6 | 0 | 0 | 0 | 0 | 0 |
| DN33356_c3_g1_i2 | Subgroup 6 | 0 | 0 | 0 | 0 | 4 |
| DN31144_c5_g1_i2 | MYB123 | 0 | 0 | 0 | 10 | 29 |
| DN33314_c5_g2_i2 | MYB123 | 3 | 8 | 0 | 1 | 14 |
| DN33314_c5_g2_i3 | MYB123 | 0 | 0 | 0 | 0 | 1 |
| DN33314_c5_g2_i4 | MYB123 | 1 | 2 | 0 | 0 | 6 |
| DN31260_c4_g2_i2 | MYB123 | 0 | 0 | 0 | 0 | 5 |
| DN30681_c1_g1_i1 | bHLH2 | 0 | 0 | 0 | 0 | 0 |
| DN30681_c1_g1_i2 | bHLH2 | 0 | 2 | 0 | 2 | 4 |
| DN30681_c1_g1_i3 | bHLH2 | 0 | 0 | 0 | 0 | 0 |
| DN30681_c1_g1_i6 | bHLH2 | 0 | 0 | 0 | 0 | 0 |
| DN30681_c1_g1_i7 | bHLH2 | 2 | 13 | 0 | 16 | 16 |
| DN30681_c1_g1_i8 | bHLH2 | 2 | 9 | 0 | 20 | 18 |
| DN30681_c1_g1_i9 | bHLH2 | 0 | 0 | 0 | 0 | 0 |
| DN32219_c4_g2_i2 | bHLH42 | 0 | 0 | 0 | 0 | 0 |
| DN32219_c4_g2_i5 | bHLH42 | 0 | 0 | 0 | 0 | 2 |
| DN32219_c4_g2_i12 | bHLH42 | 0 | 0 | 0 | 0 | 2 |
| DN32219_c4_g2_i11 | bHLH42 | 0 | 0 | 0 | 0 | 3 |
| DN32219_c4_g2_i4 | bHLH42 | 0 | 1 | 0 | 0 | 25 |
| DN32219_c4_g2_i7 | bHLH42 | 0 | 0 | 0 | 0 | 4 |
| DN32219_c4_g2_i8 | bHLH42 | 0 | 0 | 0 | 0 | 5 |
| DN32272_c1_g1_i1 | TTG1 | 0 | 0 | 0 | 0 | 0 |
| DN32272_c1_g2_i2 | TTG1 | 12 | 8 | 4 | 10 | 9 |
| DN32604_c4_g1_i2 | TTG1 | 0 | 1 | 0 | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pucker, B.; Reiher, F.; Schilbert, H.M. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants 2020, 9, 1103. https://doi.org/10.3390/plants9091103
Pucker B, Reiher F, Schilbert HM. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants. 2020; 9(9):1103. https://doi.org/10.3390/plants9091103
Chicago/Turabian StylePucker, Boas, Franziska Reiher, and Hanna Marie Schilbert. 2020. "Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium" Plants 9, no. 9: 1103. https://doi.org/10.3390/plants9091103
APA StylePucker, B., Reiher, F., & Schilbert, H. M. (2020). Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants, 9(9), 1103. https://doi.org/10.3390/plants9091103







