Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops
Abstract
1. Introduction
2. Results
2.1. Transcriptomic Responses to Drought and Salinity
2.2. Gene Set and Pathway Enrichment Analysis
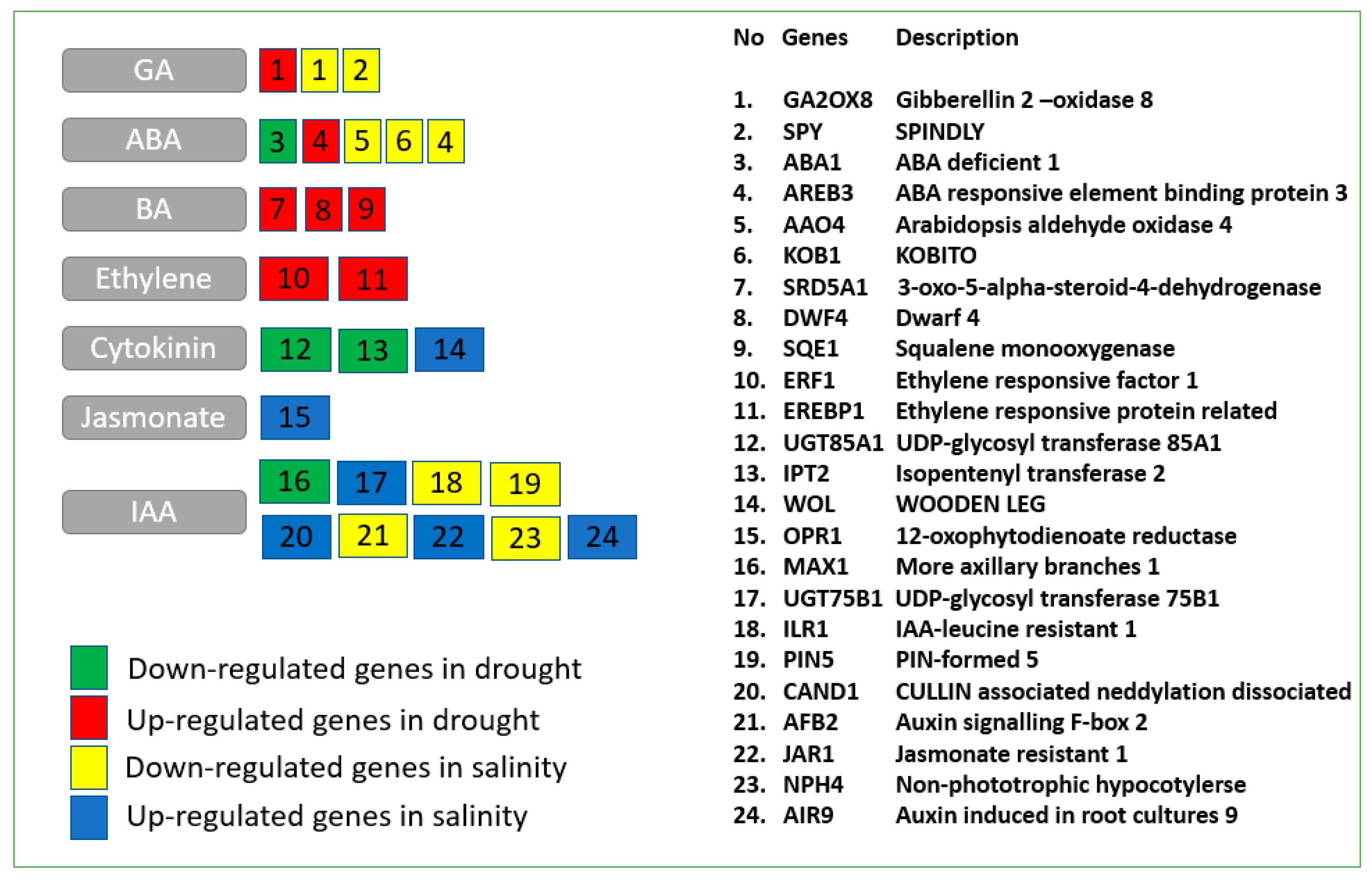
2.3. Transcriptomic Responses Related to Hormone Metabolism
2.4. Transcription Factors (TFs)
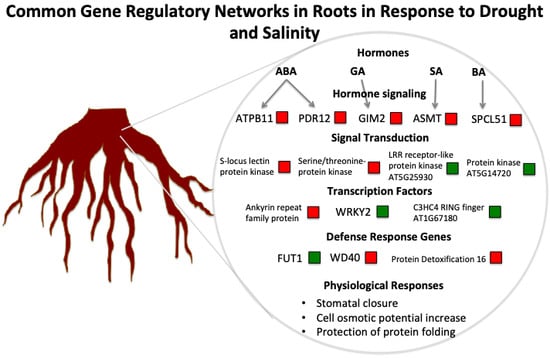
2.5. Stress-Related Genes Involved in Both Drought and Salinity
2.6. Protein–Protein Interaction Network Analysis in Response to Abiotic Stresses
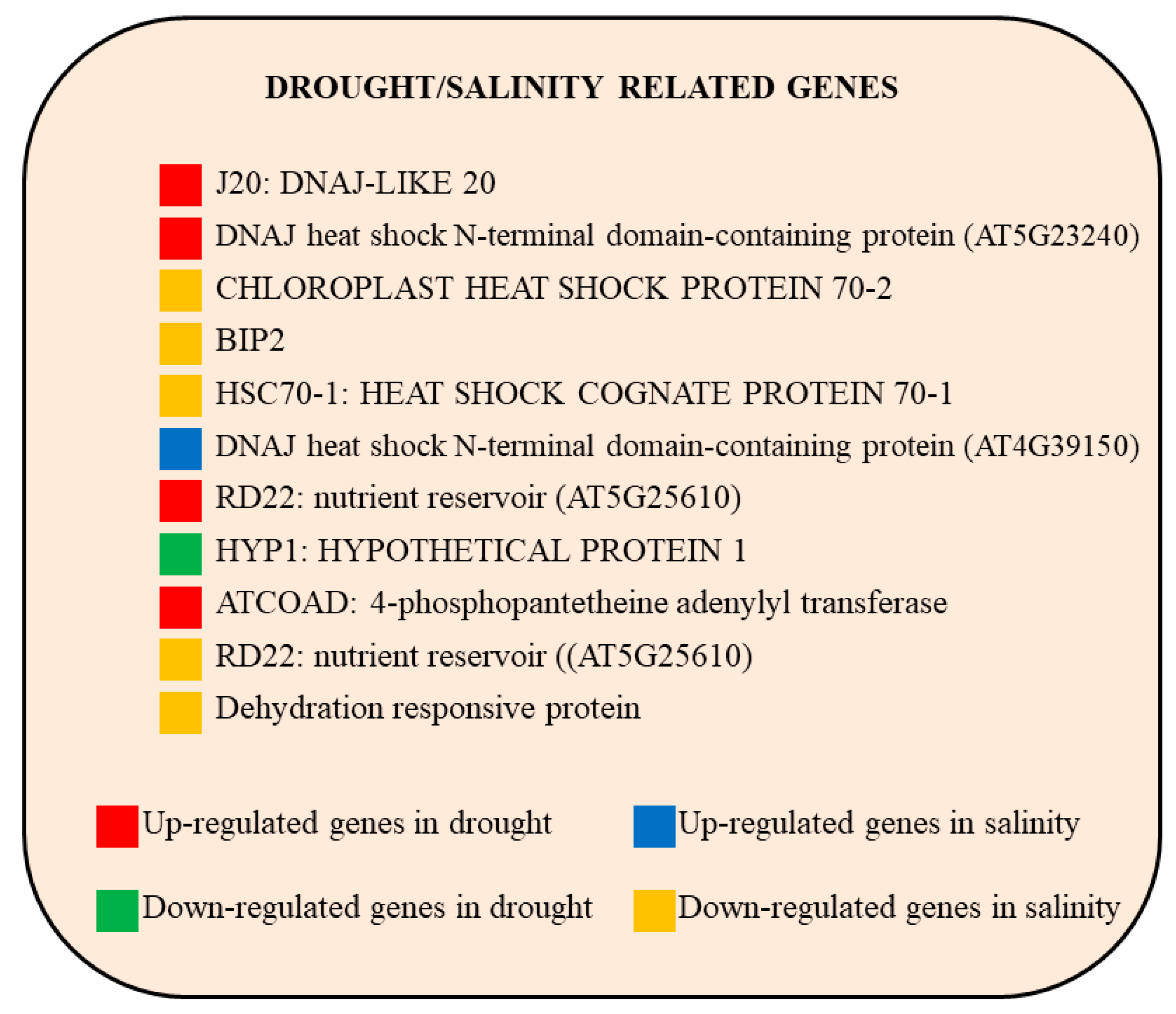
2.7. Genes Involved in General Dehydration Stresses
2.8. Leave-One-Out Cross-Validation (LOOCV) of Meta-Analysis
3. Discussion
3.1. The Role of Hormones in Drought and Salinity Responses
3.2. Key Genes and Chromosome Regions in Abiotic Stress Tolerance/Resistance
3.3. A Hypothetical Transductional Signal in Response to Osmotic Stresses
4. Materials and Methods
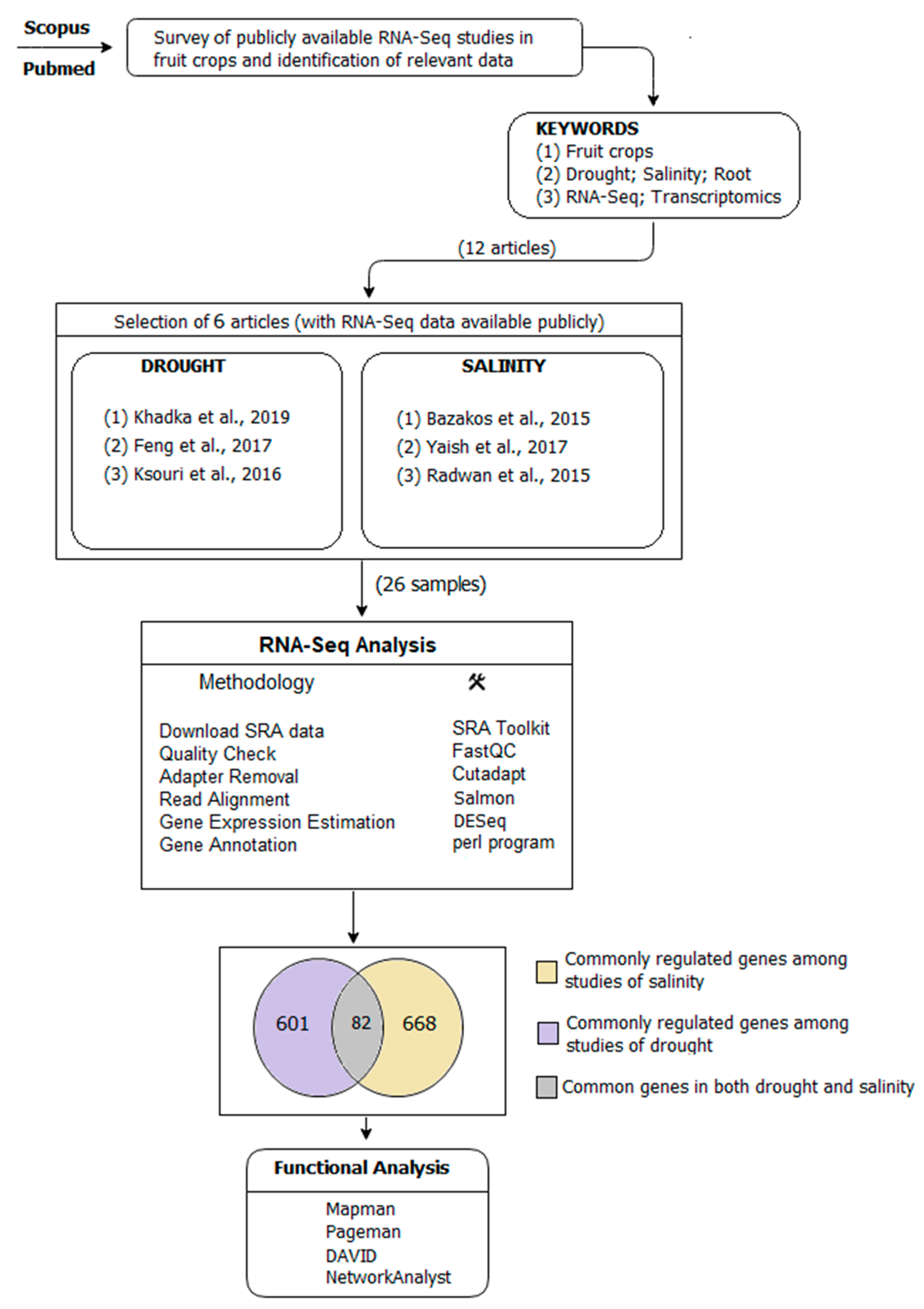
4.1. Search Strategy for Selection of RNA-Seq Studies
- (A)
- Commonly regulated genes among three articles in drought.
- (B)
- Commonly regulated genes among three articles in salinity.
- (C)
- Commonly regulated genes among both (A) and (B).
4.2. Read Alignment, Gene Differential Expression, and Annotation
4.3. Statistical and Cluster Analysis
4.4. Gene Set and Pathway Enrichment Analysis
4.5. Mapping of Genes to Corresponding Chromosomes
4.6. Protein–Protein Interaction Network
4.7. Validation Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lynch, J.P. Rightsizing root phenotypes for drought resistance. J. Exp. Bot. 2018, 69, 3279–3292. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Ortiz, S.M.; Arrieta-Montiel, M.D.L.P.; Acosta-Gallegos, J.; Covarrubias, A.A. Relationship between carbohydrate partitioning and drought resistance in common bean. Plant Cell Environ. 2008, 31, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Benny, J.; Pisciotta, A.; Caruso, T.; Martinelli, F. Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 2019, 19, 194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.L.; Zhang, W.; Zhang, G.H.; Wang, J.J.; Meng, Z.G.; Long, G.Q.; Yang, S.C. Unigene-based RNA-seq provides insights on drought stress responses in Marsdenia tenacissima. PLoS ONE 2018, 13, e0202848. [Google Scholar] [CrossRef]
- Alexandersson, E.; Danielson, J.A.; Rade, J.; Moparthi, V.K.; Fontes, M.; Kjellbom, P.; Johanson, U. Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 2010, 61, 650–660. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Maldonado, J.; Biondi, S.; Silva, H. RNA-seq Analysis of Salt-Stressed Versus Non Salt-Stressed Transcriptomes of Chenopodium quinoa Landrace R49. Genes 2019, 10, 1042. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. ABA control of plant macro element membrane transport systems in response to water deficit and high salinity. New Phytol. 2013, 202, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, D.; Liu, B.; Luo, W.; Lu, J.; Ma, T.; Wan, D. Transcriptome dynamics of a desert poplar (Populus pruinosa) in response to continuous salinity stress. Plant Cell Rep. 2014, 33, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.S.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, S.; Liu, Q.; Huang, J.; Zhang, W.; Lin, J.; Wang, Y.; Ke, Y.; He, H. Expression and interaction of small heat shock proteins (sHsps) in rice in response to heat stress. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2014, 1844, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Benny, J.; Perrone, A.; Marra, F.P.; Pisciotta, A.; Caruso, T.; Martinelli, F. Deciphering transcriptional regulation mechanisms underlining fruit development and ripening in Vitis vinifera. J. Berry Res. 2019, 9, 641–664. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS: Int. J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Kwasniewski, M.; Daszkowska-Golec, A.; Janiak, A.; Chwialkowska, K.; Nowakowska, U.; Sablok, G.; Szarejko, I. Transcriptome analysis reveals the role of the root hairs as environmental sensors to maintain plant functions under water-deficiency conditions. J. Exp. Bot. 2015, 67, 1079–1094. [Google Scholar] [CrossRef]
- Polania, J.A.; Poschenrieder, C.; Beebe, S.; Rao, I.M. Effective Use of Water and Increased Dry Matter Partitioned to Grain Contribute to Yield of Common Bean Improved for Drought Resistance. Front. Plant Sci. 2016, 7, 660. [Google Scholar] [CrossRef]
- Yaish, M.W.; Patankar, H.V.; Assaha, D.V.M.; Zheng, Y.; Al-Yahyai, R.; Sunkar, R. Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genom. 2017, 18, 246. [Google Scholar] [CrossRef]
- Radwan, O.; Arro, J.; Keller, C.; Korban, S.S. RNA-Seq Transcriptome Analysis in Date Palm Suggests Multi-Dimensional Responses to Salinity Stress. Trop. Plant Biol. 2015, 8, 74–86. [Google Scholar] [CrossRef]
- Rozhon, W.; Akter, S.; Fernandez, A.; Poppenberger, B. Inhibitors of Brassinosteroid Biosynthesis and Signal Transduction. Molecules 2019, 24, 4372. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Huang, S.; Wang, S.; Cheng, D.; Liu, J.; Lv, S.; Li, Q.; Wang, X. Enhancing Brassinosteroid Signaling via Overexpression of Tomato (Solanum lycopersicum) SlBRI1 Improves Major Agronomic Traits. Front. Plant Sci. 2017, 8, 1386. [Google Scholar] [CrossRef] [PubMed]
- Davenport, R.J.; Reid, R.J.; Smith, F.A. Sodium-calcium interactions in two wheat species differing in salinity tolerance. Physiol. Plant. 1997, 99, 323–327. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Zhang, W.; Yan, S.; Wang, R.; Zhao, J.; Li, Y.; Qi, Z.; Sun, Z.; Zhu, Z. The putative auxin efflux carrier OsPIN3 tis involved in the drought stress response and drought tolerance. Plant J. 2012, 72, 805–816. [Google Scholar] [CrossRef]
- Fard, E.M.; Bakhshi, B.; Keshavarznia, R.; Nikpay, N.; Shahbazi, M.; Salekdeh, G.H. Drought responsive microRNAs in two barley cultivars differing in their level of sensitivity to drought stress. Plant Physiol. Bioch. 2017, 118, 121–129. [Google Scholar] [CrossRef]
- Chen, T.T.; Liu, F.F.; Xiao, D.W.; Jiang, X.Y.; Li, P.; Zhao, S.M.; Hou, B.K.; Li, Y.J. The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef]
- Dalal, M.; Sahu, S.; Tiwari, S.; Rao, A.R.; Gaikwad, K. Transcriptome analysis reveals interplay between hormones, ROS metabolism and cell wall biosynthesis for drought-induced root growth in wheat. Plant Physiol. Bioch. 2018, 130, 482–492. [Google Scholar] [CrossRef]
- Zhang, K.X.; Xu, H.H.; Yuan, T.T.; Zhang, L.; Lu, Y.T. Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J. 2013, 76, 308–321. [Google Scholar]
- Tognetti, V.B.; Aken, O.V.; Morreel, K.; Vandenbroucke, K.; Cotte, B.V.D.; Clercq, I.D.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Benny, J.; Marra, F.P.; Giovino, A.; Balan, B.; Caruso, T.; Martinelli, F.; Marchese, A. Transcriptome Analysis of Pistacia vera Inflorescence Buds in Bearing and Non-Bearing Shoots Reveals the Molecular Mechanism Causing Premature Flower Bud Abscission. Genes 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Ksouri, N.; Jiménez, S.; Wells, C.E.; Contreras-Moreira, B.; Gogorcena, Y. Transcriptional Responses in Root and Leaf of Prunus persica under Drought Stress Using RNA Sequencing. Front. Plant Sci. 2016, 7, 1715. [Google Scholar] [CrossRef] [PubMed]
- Khadka, V.S.; Vaughn, K.; Xie, J.; Swaminathan, P.; Ma, Q.; Cramer, G.R.; Fennell, A.Y. Transcriptomic response is more sensitive to water deficit in shoots than roots of Vitis riparia (Michx.). BMC Plant Biol. 2019, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Firouzabadi, F.N.; Shobbar, Z.-S. Transcriptome response of roots to salt stress in a salinity-tolerant bread wheat cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J.K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, J.M.; Yao, Q.H.; Xiong, F.; Sun, C.C.; Zho, X.R.; Zhang, J.; Xiong, A.S. Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Mol. Biol. Rep 2010, 38, 745–753. [Google Scholar] [CrossRef]
- Deokar, A.A.; Kondawar, V.; Jain, P.K.; Karuppayil, S.M.; Raju, N.L.; Vadez, V.; Varshney, R.K.; Srinivasan, R. Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and -susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biol. 2011, 11, 70. [Google Scholar] [CrossRef]
- Feng, Y.; Liang, C.; Li, B.; Wan, T.; Liu, T.; Cai, Y. Differential expression profiles and pathways of genes in drought resistant tree species Prunus mahaleb roots and leaves in response to drought stress. Sci. Hortic. 2017, 226, 75–84. [Google Scholar] [CrossRef]
- Wan, X.; Zou, L.H.; Zheng, B.Q.; Wang, Y. Circadian Regulation of Alternative Splicing of Drought-Associated CIPK Genes in Dendrobium catenatum (Orchidaceae). Int. J. Mol. Sci. 2019, 20, 688. [Google Scholar] [CrossRef]
- Xia, J.; Kong, D.; Xue, S.; Tian, W.; Li, N.; Bao, F.; Hu, Y.; Du, J.; Wang, Y.; Pan, X.; et al. Nitric oxide negatively regulates AKT1-mediated potassium uptake through modulating vitamin B6 homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 16196–16201. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, W.; Yan, J.; Zhang, J.; Liu, Z.L.X.; Yi, Y. Over-expression of Arabidopsis DnaJ (Hsp40) contributes to NaCl-stress tolerance. Afr. J. Biotechnol. 2010, 9, 972–978. [Google Scholar] [CrossRef]
- Bazakos, C.; Manioudaki, M.E.; Sarropoulou, E.; Spano, T.; Kalaitzis, P. 454 Pyrosequencing of Olive (Olea europaea L.) Transcriptome in Response to Salinity. PLoS ONE 2015, 10, e0143000. [Google Scholar] [CrossRef]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Anwar, S.; Yu, S.; Sun, M.; Yang, Z.; Gao, Z.Q. Development of Drought-Tolerant Transgenic Wheat: Achievements and Limitations. Int. J. Mol. Sci. 2019, 20, 3350. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.; Cho, D.; Park, J.; Kim, S. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Zuo, B.; Zheng, X.; He, P.; Wang, L.; Lei, Q.; Feng, C.; Zhou, J.; Li, Q.; Han, Z.; Kong, J. Overexpression of MzASMT improves melatonin production and enhances drought tolerance in transgenic Arabidopsis thaliana plants. J. Pineal Res. 2014, 57, 408–417. [Google Scholar] [CrossRef]
- Xiong, W.; Ye, T.; Yao, X.; Liu, X.; Ma, S.; Chen, X.; Chen, M.L.; Feng, Y.Q.; Wu, Y. The dioxygenase GIM2 functions in seed germination by altering gibberellin production in Arabidopsis. J. Integr. Plant Biol. 2018, 60, 276–291. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H.; Vafeados, D.; Chory, J. BIN2, a New Brassinosteroid-Insensitive Locus in Arabidopsis. Plant Physiol. 2001, 127, 14–22. [Google Scholar] [CrossRef]
- Sakamoto, H.; Matsuda, O.; Iba, K. ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant J. 2008, 56, 411–422. [Google Scholar] [CrossRef]
- Yang, X.; Sun, C.; Hu, Y.; Lin, Z. Molecular cloning and characterization of a gene encoding RING zinc finger ankyrin protein from drought-tolerant Artemisia desertorum. J. Biosci. 2008, 33, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and post germination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. WRKY Proteins: Signaling and Regulation of Expression during Abiotic Stress Responses. Sci. World J. 2015, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Şahin-Çevik, M. A WRKY transcription factor gene isolated from Poncirus trifoliata shows differential responses to cold and drought stresses. Plant Omics 2012, 5, 438–445. [Google Scholar]
- Kong, D.; Li, M.; Dong, Z.; Ji, H.; Li, X. Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 2014, 34, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Wi, J.; Jung, H.S.; Im, S.; Yang, S.; Park, E.J.; Hwang, M.S.; Jeong, W.J.; Choi, D.W. A nuclear fructosyltransferase-like protein, PtFUT, from marine red alga Pyropia tenera (Rhodophyta) confers osmotic stress tolerance. J. Appl. Phycol. 2017, 30, 717–727. [Google Scholar] [CrossRef]
- Hossain, M.A.; Bhattacharjee, S.; Armin, S.M.; Qian, P.; Xin, W.; Li, H.Y.; Burritt, D.J.; Fujita, M.; Tran, L.S.P. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front. Plant Sci. 2015, 6, 420. [Google Scholar] [CrossRef]
- Cimò, G.; Marchese, A.; Germanà, M.A. Microspore embryogenesis induced through in vitro anther culture of almond (Prunus dulcis Mill.). Plant Cell Tiss. Org. (PCTOC) 2017, 128, 85–95. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Gen. Biol. 2010, 1-1. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user–driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Steinhauser, D.; Gibon, Y.; Bläsing, O.E.; Redestig, H.; Sreenivasulu, N.; Krall, L.; Hannah, M.A.; Poree, F.; et al. PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinform. 2006, 7, 535. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.A.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Benner, M.J.; Hancock, R.E. NetworkAnalyst–integrative approaches for protein–protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef]


| Articles | Crops | Sample Information | ||
|---|---|---|---|---|
| Total | Down | Up | ||
| Drought | ||||
| Khadka et al. 2019 | Vitis riparia Michx | 5021 | 2950 | 2071 |
| Feng et al. 2017 | Prunus mahaleb L. | 6959 | 3056 | 3903 |
| Ksouri et al. 2016 | Prunus persica | 5856 | 2829 | 3027 |
| Salinity | ||||
| Bazakos et al. 2015 | Olea europaea L. cv. Kalamon | 6060 | 2982 | 3911 |
| Yaish et al. 2017 | Phoenix dactylifera L. cv. Khalas | 5504 | 3585 | 1919 |
| Radwan et al. 2015 | Phoenix dactylifera cv. Deglet Beida | 6676 | 3103 | 3573 |
| Commonly regulated in drought | 683 | 349 | 334 | |
| Commonly regulated in salinity | 750 | 390 | 360 | |
| Commonly regulated among both drought and salinity | 39 | 16 | 23 | |
| Common genes among drought and salinity | 82 | |||
| Gene ID | Description | Functional Term | Functional Category |
|---|---|---|---|
| Down-Regulated Genes | |||
| AT5G11700 | Ephrin type-B receptor ephrin type-B receptor | Vacuole | Cellular component |
| AT5G56270 | Probable WRKY transcription factor 2 | DNA-binding transcription factor activity | Molecular function |
| AT1G15060 | Alpha/beta hydrolase family protein | Hydrolase activity | Molecular function |
| AT1G04910 | O-Fructosyl transferase 1 | Carbohydrate metabolic process | Biological process |
| AT5G24090 | Acidic endo chitinase | Chitin catabolic process | Biological process |
| AT1G67180 | Zinc finger (C3HC4-type RING finger) protein | Cell cycle | Biological process |
| AT3G61790 | E3 ubiquitin protein ligase | Ubiquitin-dependent protein catabolic process | Biological process |
| AT4G02570 | Cullin 1 (CUL1) AT4G02570 protein | Auxin-activated signaling pathway | Biological process |
| AT2G42520 | DEAD-box adenosine triphosphate (ATP)-dependent RNA helicase 37 | Nucleic acid binding | Molecular function |
| AT5G47650 | Nudix hydrolase 2 | Metal ion binding | Molecular function |
| AT5G25930 | Leucine-rich repeat receptor-like protein kinase | Signaling receptor kinase | Biological process |
| AT4G32010 | B3 domain transcription repressor; Viviparous-1/Abscisic acid insensitive 3-like2 (VAL2) | Regulation of transcription, DNA-templated | Biological process |
| AT3G19840 | Pre-mRNA-processing protein 40C | Messenger RNA (mRNA) processing | Biological process |
| AT2G27900 | Ras-related protein (RABA)5d | Endocytic recycling | Biological process |
| AT5G14720 | Protein kinase superfamily protein | Phosphorylation | Biological process |
| AT4G32850 | Polynucleotide adenylyl transferase 4 | Nucleotidyltransferase activity | Molecular function |
| Up-Regulated Genes | |||
| AT4G03500 | Ankyrin repeat family protein | Membrane | Cellular component |
| AT1G15520 | Pleiotropic drug resistance 12 | Abscisic acid transport | Biological process |
| AT1G02520 | ATP-binding cassette B11 | Transmembrane transport | Biological process |
| AT3G06880 | Transducin/ tryptophan-aspartic (WD)40 repeat-like superfamily protein | Response to stress | Biological process |
| AT5G64813 | Light insensitive period 1 | Cytoplasm | Cellular component |
| AT4G31210 | DNA topoisomerase, type IA | Metal ion binding | Molecular function |
| AT5G07990 | Cytochrome P450 75B1 | Oxidation–reduction process | Biological process |
| AT5G52450 | Protein detoxification 16 | Response to nematode | Biological process |
| AT2G36690 | Germination insensitive to ABA mutant 2 (GIM2) | Oxidation–reduction process | Biological process |
| AT5G11040 | vascular network defective 4 (VAN4) | Cytokinesis by cell plate formation | Biological process |
| AT5G23150 | Enhancer of AG-4 2 (HUA2) | Regulation of transcription by RNA polymerase II | Biological process |
| AT3G14270 | forms aploid and binucleate cells 1B (FAB1B) | Phosphatidylinositol phosphorylation | Biological process |
| AT2G03810 | 18S pre-ribosomal assembly gar2-like protein | Regulation of asymmetric cell division | Biological process |
| AT4G26270 | phosphofructokinase 3 (PFK3) | Fructose 6-phosphate metabolic process | Biological process |
| AT5G58003 | C-terminal domain phosphatase-like 4 | Dephosphorylation of RNA polymerase II C-terminal domain | Biological process |
| AT4G35160 | N-Acetyl serotonin O-methyl transferase | Methylation | Biological process |
| AT2G45550 | Cytochrome P450 | Oxidation–reduction process | Biological process |
| AT2G19130 | S-locus lectin protein kinase family protein | Phosphorylation | Biological process |
| AT4G02590 | unfertilized embryo sac 12 (UNE12) | Regulation of defense response | Biological process |
| AT3G48190 | Serine/threonine protein kinase | DNA damage checkpoint | Biological process |
| AT5G54310 | ARF-GAP domain 5 (AGD5) | Activation of GTPase activity | Biological process |
| AT2G26330 | Quantitative resistance to plectosphaerella 1 | Receptor Serine/Threonine Kinase Binding | Molecular function |
| AT2G27920 | serine carboxypeptidase-like 51 (SCPL51) | Proteolysis | Biological process |
| Article | Stress | Species | No. of Samples | Control | Treated | Duration of Stress |
|---|---|---|---|---|---|---|
| Khadka et al. 2019 | Drought | Vitis riparia Michx | 6 | SRR6494883 (Control1) | SRR6494880 (Treated 1) | Roots were harvested at 14 days after stress (DAS) |
| SRR6494884 (Control2) | SRR6494881 (Treated 2) | |||||
| SRR6494885 (Control3) | SRR6494882 (Treated 3) | |||||
| Feng et al. 2017 | Drought | Prunus mahaleb L. | 6 | SRR5112808 (Control1) | SRR5112805 (Treated1) | Roots were harvested at 15 DAS |
| SRR5112809 (Control2) | SRR5112806 (Treated2) | |||||
| SRR5112810 (Control3) | SRR5112807 (Treated3) | |||||
| Ksouri et al. 2016 | Drought | Prunus persica | 6 | SAMEA3861653 (Control 1) | SAMEA3861656 (Treated1) | Roots were harvested at 16 DAS |
| SAMEA3861654 (Control2) | SAMEA3861657 (Treated2) | |||||
| SAMEA3861655 (Control3) | SAMEA3861658 (Treated3) | |||||
| SRR6770841 (Control 2) | SRR6770840 (Treated 2) | |||||
| Bazakos et al. 2015 | Salinity | Olea europaea L. cv. Kalamon | 2 | SRR891235 (Control1) | SRR886308 (Treated1) | Roots were harvested at 90 DAS |
| Yaish et al. 2017 | Salinity | Phoenix dactylifera L. cv. Khalas | 2 | SRR4034943 (Control1) | SRR4034944 (Treated1) | Roots were harvested at 45 DAS |
| Radwan et al. 2015 | Salinity | Phoenix dactylifera L. cv. Deglet Beida | 4 | SRR2027988 (Control 1) | SRR2029376 (Treated 1) | Roots were harvested at 60 DAS |
| SRR2029378 (Control 2) | SRR2029381 (Treated 2) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benny, J.; Marchese, A.; Giovino, A.; Marra, F.P.; Perrone, A.; Caruso, T.; Martinelli, F. Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops. Plants 2020, 9, 1059. https://doi.org/10.3390/plants9091059
Benny J, Marchese A, Giovino A, Marra FP, Perrone A, Caruso T, Martinelli F. Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops. Plants. 2020; 9(9):1059. https://doi.org/10.3390/plants9091059
Chicago/Turabian StyleBenny, Jubina, Annalisa Marchese, Antonio Giovino, Francesco Paolo Marra, Anna Perrone, Tiziano Caruso, and Federico Martinelli. 2020. "Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops" Plants 9, no. 9: 1059. https://doi.org/10.3390/plants9091059
APA StyleBenny, J., Marchese, A., Giovino, A., Marra, F. P., Perrone, A., Caruso, T., & Martinelli, F. (2020). Gaining Insight into Exclusive and Common Transcriptomic Features Linked to Drought and Salinity Responses across Fruit Tree Crops. Plants, 9(9), 1059. https://doi.org/10.3390/plants9091059