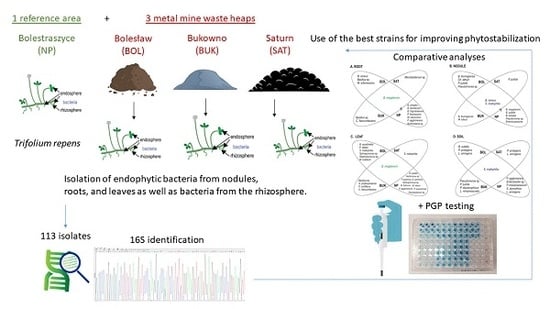
Trifolium repens-Associated Bacteria as a Potential Tool to Facilitate Phytostabilization of Zinc and Lead Polluted Waste Heaps
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Plant-Associated Bacteria
2.1.1. Isolation of Endophytes
2.1.2. Isolation of Rhizosphere Bacteria
2.2. Genetic Analysis
2.2.1. Determination of Taxonomic Position of Bacteria Based on 16S rRNA Gene Sequence
2.2.2. Phylogenetic and Genotypic Analysis
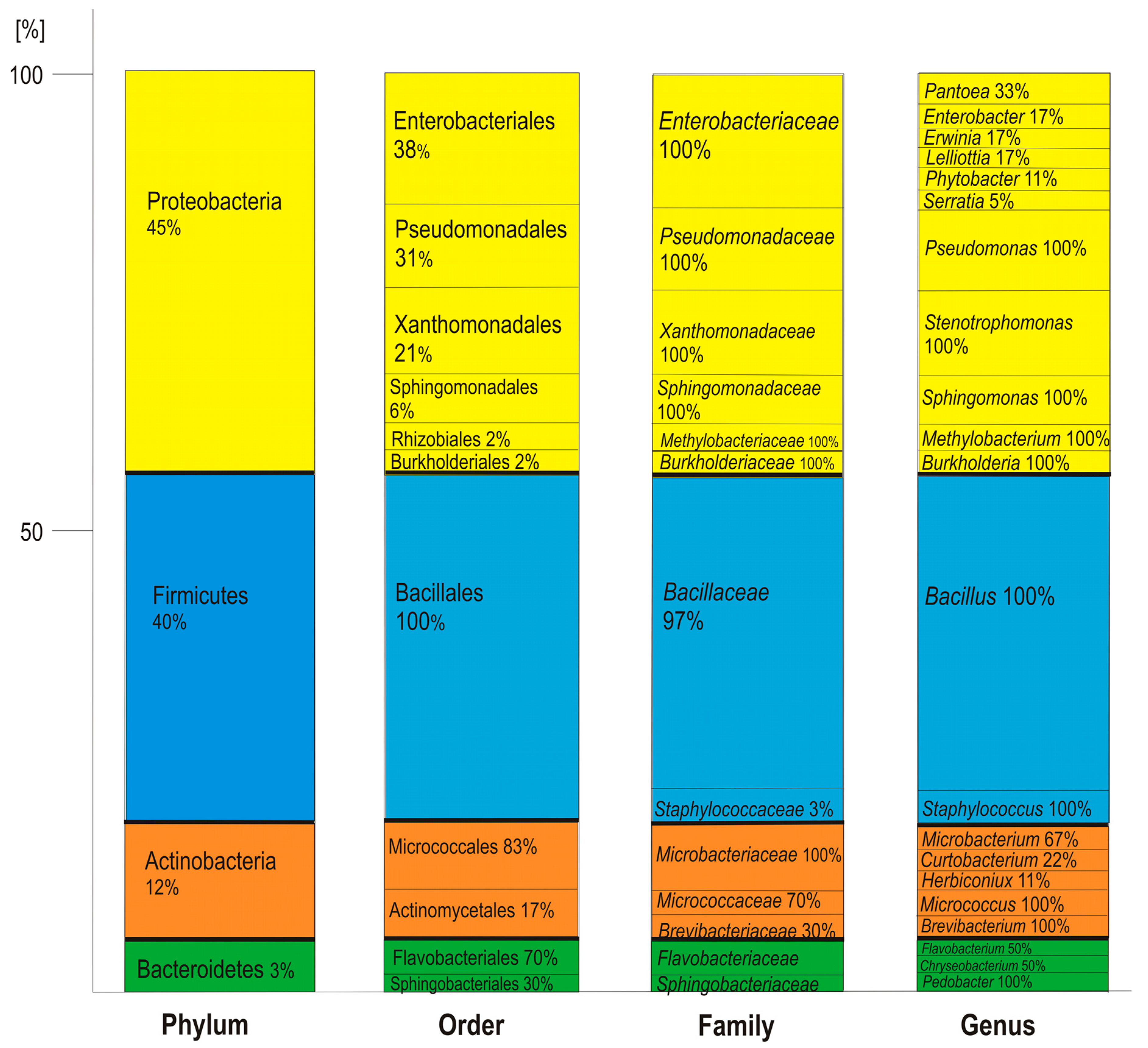
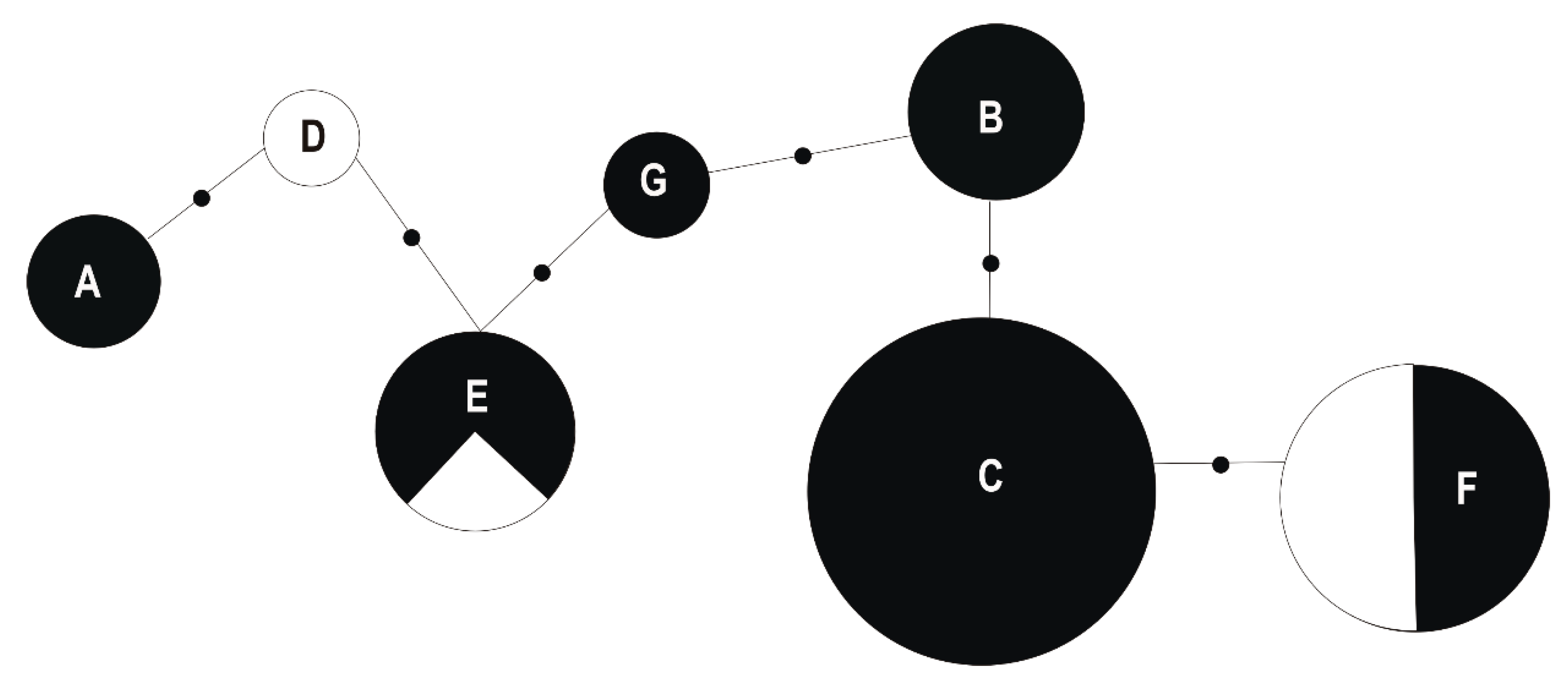
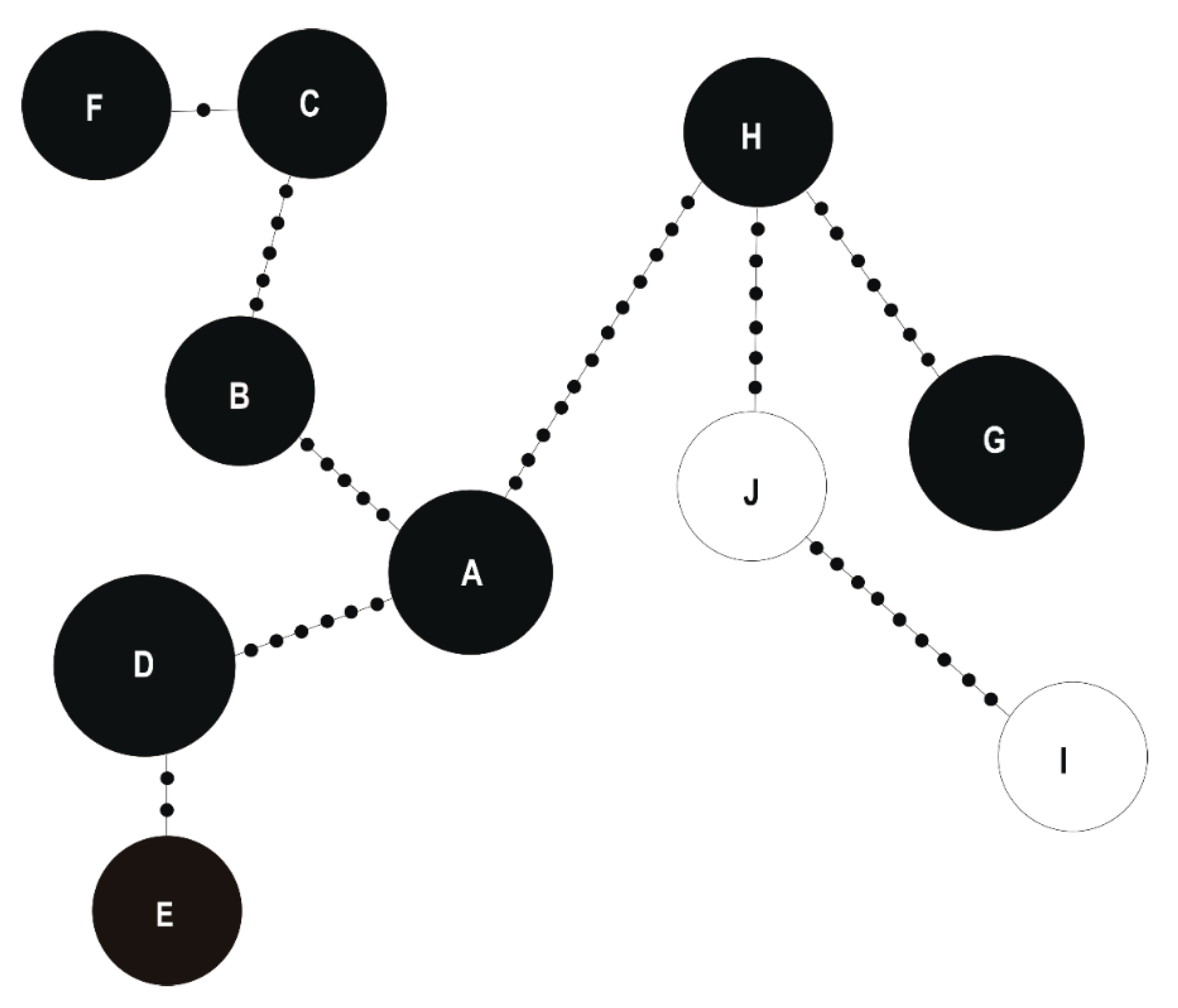
2.2.3. Determination of Relative Taxonomic Biodiversity in Communities
2.3. Phenotypic Traits of Isolated Bacterial Strains
2.3.1. In vitro Studies Testing the Plant Growth Promoting Properties
2.3.2. Metal Tolerance of Isolated Bacterial Strains
2.4. Analysis of Zinc, Lead, and Cadmium Concentration in Soils and T. repens Roots and Leaves
2.5. Data Availability
3. Results and Discussion
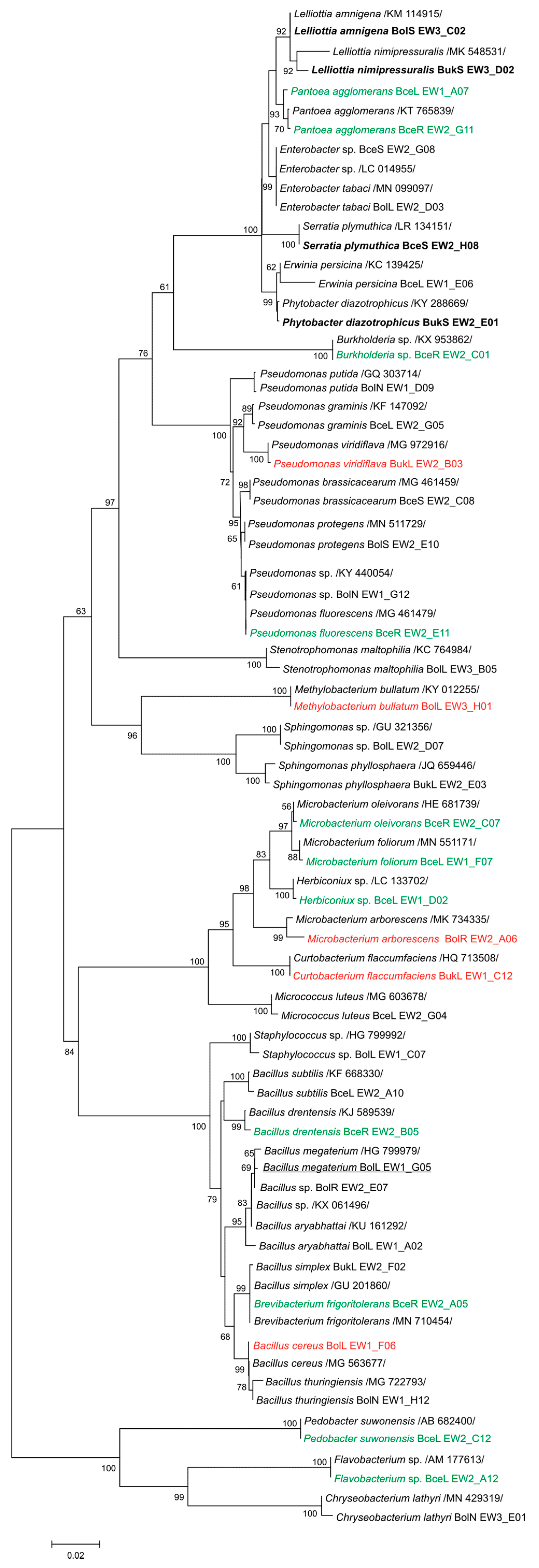
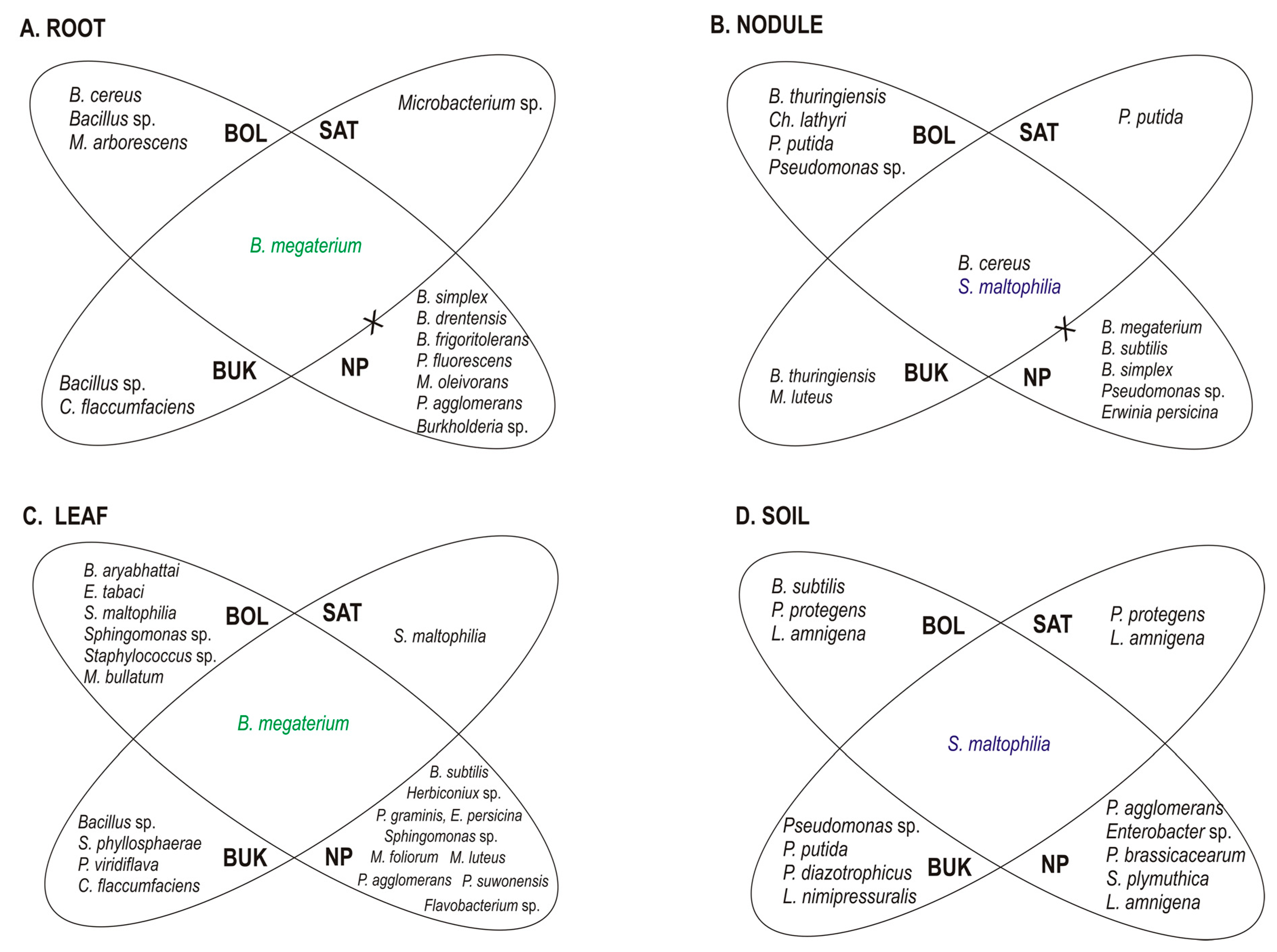
3.1. Taxonomic Identification and Genetic Analysis of Bacteria Associated with T. repens
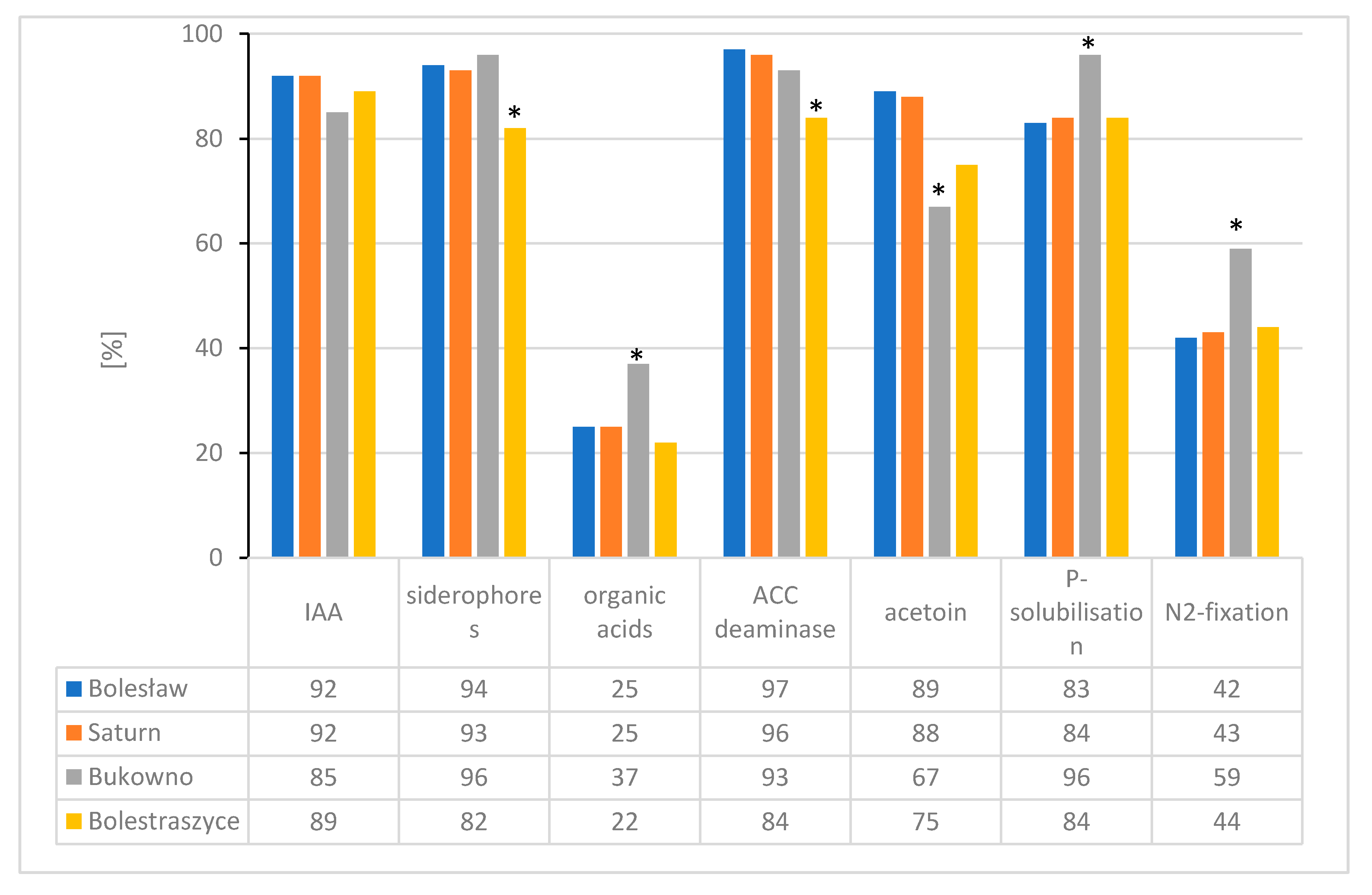
3.2. In Vitro Testing of Plant Growth Promotion Traits and Heavy Metal Tolerance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fuentes, A.; Almonacid, L.; Ocampo, J.A.; Arriagada, C. Synergistic interactions between a saprophytic fungal consortium and Rhizophagus irregularis alleviate oxidative stress in plants grown in heavy metal contaminated soil. Plant Soil 2016, 407, 355–366. [Google Scholar] [CrossRef]
- Etesami, H. Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: Mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 147, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.A.; Hussain, I.; Rasheed, R.; Iqbal, M.; Riaz, M.; Arif, M.S. Advances in microbe-assisted reclamation of heavy metal contaminated soils over the last decade: A review. J. Environ. Manag. 2017, 198, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Koul, B.; Taak, P. Chemical Methods of Soil Remediation. In Biotechnological Strategies for Effective Remediation of Polluted Soils; Koul, B., Taak, P., Eds.; Springer: Singapore, 2018; pp. 77–84. [Google Scholar]
- Vangronsveld, J.; Herzig, R.; Weyens, N.; Boulet, J.; Adriaensen, K.; Ruttens, A.; Thewys, T.; Vassilev, A.; Meers, E.; Nehnelajova, E.; et al. Phytoremediation of contaminated soils and groundwater: Lessons from the field. Environ. Sci. Poll. Res. 2009, 16, 765–794. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Vangronsveld, J. Phytoremediation: Plant-endophyte partnerships take the challenge. Curr. Opin. Biotechnol. 2009, 20, 248–254. [Google Scholar] [CrossRef]
- Rajkumar, M.; Sandhya, S.; Prasad, M.N.V.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef]
- Shah, V.; Daverey, A. Phytoremediation: A multidisciplinary approach to clean up heavy metal contaminated soil. Environ. Technol. Innov. 2020, 18, 100774. [Google Scholar] [CrossRef]
- Wójcik, M.; Gonnelli, C.; Selvi, F.; Dresler, S.; Rostański, A.; Vangronsveld, J. Metallophytes of serpentine and calamine soils—Their unique ecophysiology and potential for phytoremediation. Adv. Bot. Res. 2017, 83, 1–42. [Google Scholar]
- Ernst, W.H.O. Phytoextraction of mine waste—Options and impossibilities. Chem. Geochem. 2005, 65, 29–42. [Google Scholar] [CrossRef]
- Vangronsveld, J.; Sterckx, J.; Van Assche, F.; Clijsters, H. Rehabilitation studies on an old non-ferrous waste dumping ground: Effects of revegetation and metal immobilization by beringite. J. Geochem. Explor. 1995, 52, 221–229. [Google Scholar] [CrossRef]
- Vangronsveld, J.; Van Assche, F.; Clijsters, H. Reclamation of a bare industrial area contaminated by non ferrous metals—In situ metal immobilization and revegetation. Environ. Pollut. 1995, 87, 51–59. [Google Scholar] [CrossRef]
- Tam, N.F.Y.; Wong, Y.S. Effectiveness of bacterial inoculum and mangrove plants on remediation of sediment contaminated with polycyclic aromatic hydrocarbons. Mar. Poll. Bull. 2008, 57, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Ann. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef] [PubMed]
- Oleńska, E.; Małek, W. Sequence analysis of hypothetical lysine exporter genes of Rhizobium leguminosarum bv. trifolii from calamine old waste heaps and their evolutionary history. Curr. Microbiol. 2013, 66, 493–498. [Google Scholar] [PubMed]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant–microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Mishra, J.; Singh, R.; Arora, N.K. Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front. Microbiol. 2017, 8, 1706. [Google Scholar] [CrossRef]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Falmann, K.; Puschenreiter, M. The role of plant—Associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef]
- Sánchez-López, A.S.; Pintelon, I.; Stevens, V.; Imperato, V.; Timmermans, J.-P.; González-Chávez, C.; Carrillo-González, R.; Van Hamme, J.; Vangronsveld, J.; Thijs, S. Seed endophyte microbiome of Crotalaria pumila unpeeled: Identification of plant-beneficial Methylobacteria. Int. J. Mol. Sci. 2018, 19, 291. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W. Genetic differentiation of Trifolium repens microsymbionts deriving from Zn–Pb waste-heap and control area in Poland. J. Basic Microbiol. 2015, 55, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Oleńska, E.; Małek, W. Genomic polymorphism of Trifolium repens root nodule symbionts from heavy metal-abundant 100-year-old waste heap in southern Poland. Archiv. Microbiol. 2019, 201, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Bidar, G.; Garçon, G.; Pruvot, C.; Dewaele, D.; Cazier, F.; Douay, F.; Shirali, P. Behavior of Trifolium repens and Lolium perenne growing in a heavy metal contaminated field: Plant metal concentration and phytotoxicity. Environ. Pollut. 2007, 147, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hildago, P.; Hirsch, A.M. The nodule microbiome: N2—Fixing rhizobia do not live alone. Phytobiomes 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Kalita, M.; Małek, W.; Coutinho, T.A. Putative novel Bradyrhizobium and Phyllobacterium species isolated from root nodules of Chamaecytisus ruthenicus. Syst. Appl. Microbiol. 2020, 43, 126056. [Google Scholar] [CrossRef]
- Eevers, N.; Gielen, M.; Sánchez-López, A.; Jaspers, S.; White, J.C.; Vangronsveld, J.; Weyens, N. Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Microb. Biotechnol. 2015, 8, 707–715. [Google Scholar] [CrossRef]
- Imperato, V.; Portillo-Estrada, M.; McAmmond, B.M.; Douwen, Y.; Van Hamme, J.D.; Gawroński, S.W.; Vangronsveld, J.; Thijs, S. Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of Bóbrka (Poland). Genes 2019, 10, 443. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef]
- Weisburg, W.; Barns, S.M.; Pelletier, D.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Spada, E.; Sagliocca, L.; Sourdis, J.; Garbuglia, A.R.; Poggi, V.; De Fusco, C.; Mele, A. Use of the minimum spanning tree model for molecular epidemiological investigation of a nosocomial outbreak of hepatitis C virus infection. J. Clin. Microbiol. 2004, 42, 4230–4236. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischner, H.E.L. Arlequin suite ver. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Seguin, P.; Graham, P.H.; Sheaffer, C.C.; Ehlke, N.J.; Russelle, M.P. Genetic diversity of rhizobia nodulating Trifolium ambiguum in North America. Can. J. Microbiol. 2001, 47, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Farooq, F.T.; Vessey, J.K. Genetic diversity of Bradyrhizobium japonicum with soybean growing regions of the north-eastern Great Plains of North America as determined by REP-PCR and ERIC-PCR profiling. Symbiosis 2009, 48, 131–142. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Xie, G.H.; Cui, Z.; Yu, J.; Yan, J.; Hai, W.; Steinberger, Y. Identification of nif genes in N2-fixing bacterial strains isolated from rice fields along the Yangtze River Plain. J. Basic Microbiol. 2006, 46, 56–63. [Google Scholar] [CrossRef]
- Cunningham, J.E.; Kuiack, C. Production of citric and oxalic acids and solubilization of calcium phosphate by Penicillium bilaii. Appl. Environ. Microbiol. 1992, 58, 1451–1458. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Schlegel, H.; Gottschalk, G.; Von Bartha, R. Formation and utilization of poly-β-hydroxybutyric acid by knallgas bacteria (Hydrogenomonas). Nature 1961, 191, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indole-acetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Hontzeas, N.; Safronova, V.I.; Demchinskaya, S.V.; Piluzza, G.; Bullitta, S.; Glick, B.R. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Romick, T.L.; Fleming, H.P. Acetoin production as an indicator of growth and metabolic inhibition of Listeria monocytogenes. J. Appl. Microbiol. 1998, 84, 18–24. [Google Scholar] [CrossRef]
- Włostowski, T.; Kozłowski, P.; Łaszkiewicz-Tiszczenko, B.; Oleńska, E. Cadmium accumulation and pathological alterations in the midgut gland of terrestrial snail Helix pomatia L. from a zinc smelter area: Role of soil pH. Bull. Environ. Contam. Toxicol. 2016, 96, 484–489. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci. Rep. 2017, 7, 41766. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root associated microbiome of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 2013, 36, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tang, Y.; Wang, D.; Lin, N.; Zhou, J. Identification and characterization of a new Enterobacter onion bulb decay caused by Lelliottia amnigena in China. Appl. Micro. 2016, 2, 2. [Google Scholar] [CrossRef]
- Leys, N.M.E.J.; Ryngaert, A.; Bastiaens, L.; Verstraete, W.; Top, E.M.; Springael, D. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 2004, 70, 1944–1955. [Google Scholar] [CrossRef]
- Logan, N.A.; De Vos, P. Bacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Garrity, G.M., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; p. 41. [Google Scholar]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Muresu, R.; Polone, E.; Sulas, L.; Baldan, B.; Tondello, A.; Delogu, G.; Cappuccinelli, P.; Alberghini, S.; Benhizia, Y.; Benhizia, H.; et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 2008, 63, 383–400. [Google Scholar] [CrossRef]
- Schwartz, A.N.; Ortiz, I.; Maymon, M.; Herbold, C.W.; Fujishige, N.A.; Vijanderan, J.A.; Villella, W.; Hanamoto, K.; Diener, A.; Sanders, E.R.; et al. Bacillus simplex—A little known PGPB with anti-fungal activity—Alters Pea legume root architecture and nodule morphology when coinoculated with Rhizobium leguminosarum bv. viciae. Agronomy 2013, 3, 595–620. [Google Scholar] [CrossRef]
- Rajendran, G.; Patel, M.H.; Joshi, S.J. Isolation and characterization of nodule-associated Exiguobacterium sp. from the root nodules of fenugreek (Trigonella foenum-graecum) and their possible role in plant growth promotion. Int. J. Microbiol. 2012, 2012, 693982. [Google Scholar] [CrossRef]
- Sturz, A.V.; Christie, B.R.; Nowak, J. Bacterial endophytes: Potential role in developing sustainable system of crop production. Crit. Rev. Plant Sci. 2000, 19, 1–30. [Google Scholar] [CrossRef]
- Vyas, P.; Gulati, A. Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol. 2009, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Rekha, P.D.; Arun, A.B.; Shen, F.T.; Lai, W.A.; Young, C.C. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 2006, 34, 33–41. [Google Scholar] [CrossRef]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, W. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Lambrecht, M.; Okon, Y.; Vande Broek, A.; Vanderleyden, J. Indole-3-acetic acid: A reciprocal signaling molecule in bacteria-plant interactions. Trends Microbiol. 2000, 8, 298–300. [Google Scholar] [CrossRef]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef]
- Ferreira, M.; Silva, H.; Cunh, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Rudrappa, T.; Biedrzycki, M.L.; Kunjeti, S.G.; Donofrio, N.M.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Comm. Integr. Biol. 2010, 3, 130–138. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J.; Jiang, C.Y.; He, L.Y.; Qian, M. Characterization of heavy metal resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2009, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Xu, Y.; Chen, Y.; Zhan, Y.; Wei, X.; Li, L.; Wang, D.; He, P.; Li, S.; Chen, S. Metabolomics analysis reveals global acetoin stress response of Bacillus licheniformis. Metabolomics 2019, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Croes, S.; Weyens, N.; Janssen, J.; Vercampt, H.; Colpaert, J.V.; Carleer, R.; Vangronsveld, J. Bacterial communities associated with Brassica napus L. grown on trace element-contaminated and non-contaminated fields: A genotypic and phenotypic comparison. Microb. Biotechnol. 2013, 6, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Changes in the population of seed bacteria of transgenerationally Cd- exposed Arabidopsis thaliana. Plant Biol. 2013, 15, 971–981. [Google Scholar] [CrossRef]
- Carlos, M.-H.J.; Janette, A.-M.; Melani, M.-S. Assessing the effects of heavy metals in ACC deaminase and IAA production on plant growth-promoting bacteria. Microbiol. Res. 2016, 188, 53–61. [Google Scholar] [CrossRef]
- Akinrinlola, R.J.; Yuen, G.Y.; Drijber, R.A.; Adesemoye, A.O. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018, 2018, 5686874. [Google Scholar] [CrossRef]
- Ortíz-Castro, R.; Valencia-Cantero, E.; López-Bucio, J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signaling Behavior 2008, 3, 263–265. [Google Scholar] [CrossRef]
- Pueyo, M.T.; Bloch, C.J.; Carmona, R.A.M.; Masico, P. Lipopeptides produced by a soil Bacillus megaterium strain. Microb. Ecol. 2009, 57, 367–378. [Google Scholar] [CrossRef]
- Govindasamy, V.; Senthilkumar, M.; Magheshwaran, V.; Kumar, U.; Bose, P.; Sharma, V.; Annapurna, K. Bacillus and Paenibacillus spp.: Potential PGPR for Sustainable Agriculture. In Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Microbiology Monographs 18; Springer: Berlin/Heidelberg, Germany, 2010; pp. 333–364. [Google Scholar]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Elhalag, K.M.; Messiha, N.A.S.; Emara, H.M.; Abdallah, S.A. Evaluation of antibacterial activity of Stenotrophomonas maltophilia against Ralstonia solanacearum under different application conditions. J. Appl. Microbiol. 2016, 12, 1629–1645. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 2017, 8, 1945. [Google Scholar] [CrossRef] [PubMed]
| Soil | Root | Leaf | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Zn | Pb | Cd | Zn | Pb | Cd | Zn | Pb | Cd | |
| Bolesław | 52,795 ± 1197 a | 578 ± 5 a | 605 ± 73 a | 609 ± 41 a | 243 ± 80 a | 288 ± 161 a | 329 ± 22 a | 7.10 ± 1.12 a | 1.80 ± 0.16 a |
| Bukowno | 20,159 ± 1523 b | 35 ± 11 b | 18 ± 2 b | 405 ± 76 b | 2.85 ± 0.11 b | 1.25 ± 0.04 b | 190 ± 11 b | 0.59 ± 0.08 b | 0.36 ± 0.02 b |
| Saturn | 26,016 ± 2757 b | 48 ± 11 b | 22 ± 9 b | 499 ± 89 b | 3.26 ± 0.07 b | 5.89 ± 0.18 b | 221 ± 20 b | 0.62 ± 0.1 b | 1.20 ± 0.17 c |
| Bolestraszyce | 64 ± 25 c | 6.87 ± 1.65 c | 2.56 ± 0.74 c | 2.7 ± 0.39 c | 0.45 ± 0.25 c | 0.51 ± 0.05 c | 0.41 ± 0.09 c | 0.01 ± 0.005 c | 0.08 ± 0.02 d |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oleńska, E.; Imperato, V.; Małek, W.; Włostowski, T.; Wójcik, M.; Swiecicka, I.; Vangronsveld, J.; Thijs, S. Trifolium repens-Associated Bacteria as a Potential Tool to Facilitate Phytostabilization of Zinc and Lead Polluted Waste Heaps. Plants 2020, 9, 1002. https://doi.org/10.3390/plants9081002
Oleńska E, Imperato V, Małek W, Włostowski T, Wójcik M, Swiecicka I, Vangronsveld J, Thijs S. Trifolium repens-Associated Bacteria as a Potential Tool to Facilitate Phytostabilization of Zinc and Lead Polluted Waste Heaps. Plants. 2020; 9(8):1002. https://doi.org/10.3390/plants9081002
Chicago/Turabian StyleOleńska, Ewa, Valeria Imperato, Wanda Małek, Tadeusz Włostowski, Małgorzata Wójcik, Izabela Swiecicka, Jaco Vangronsveld, and Sofie Thijs. 2020. "Trifolium repens-Associated Bacteria as a Potential Tool to Facilitate Phytostabilization of Zinc and Lead Polluted Waste Heaps" Plants 9, no. 8: 1002. https://doi.org/10.3390/plants9081002
APA StyleOleńska, E., Imperato, V., Małek, W., Włostowski, T., Wójcik, M., Swiecicka, I., Vangronsveld, J., & Thijs, S. (2020). Trifolium repens-Associated Bacteria as a Potential Tool to Facilitate Phytostabilization of Zinc and Lead Polluted Waste Heaps. Plants, 9(8), 1002. https://doi.org/10.3390/plants9081002