In Vivo Reporters for Visualizing Alternative Splicing of Hormonal Genes
Abstract
1. Introduction
2. Results
2.1. Reporters for Visualizing Common Types of AS Events in Plant Hormonal Genes
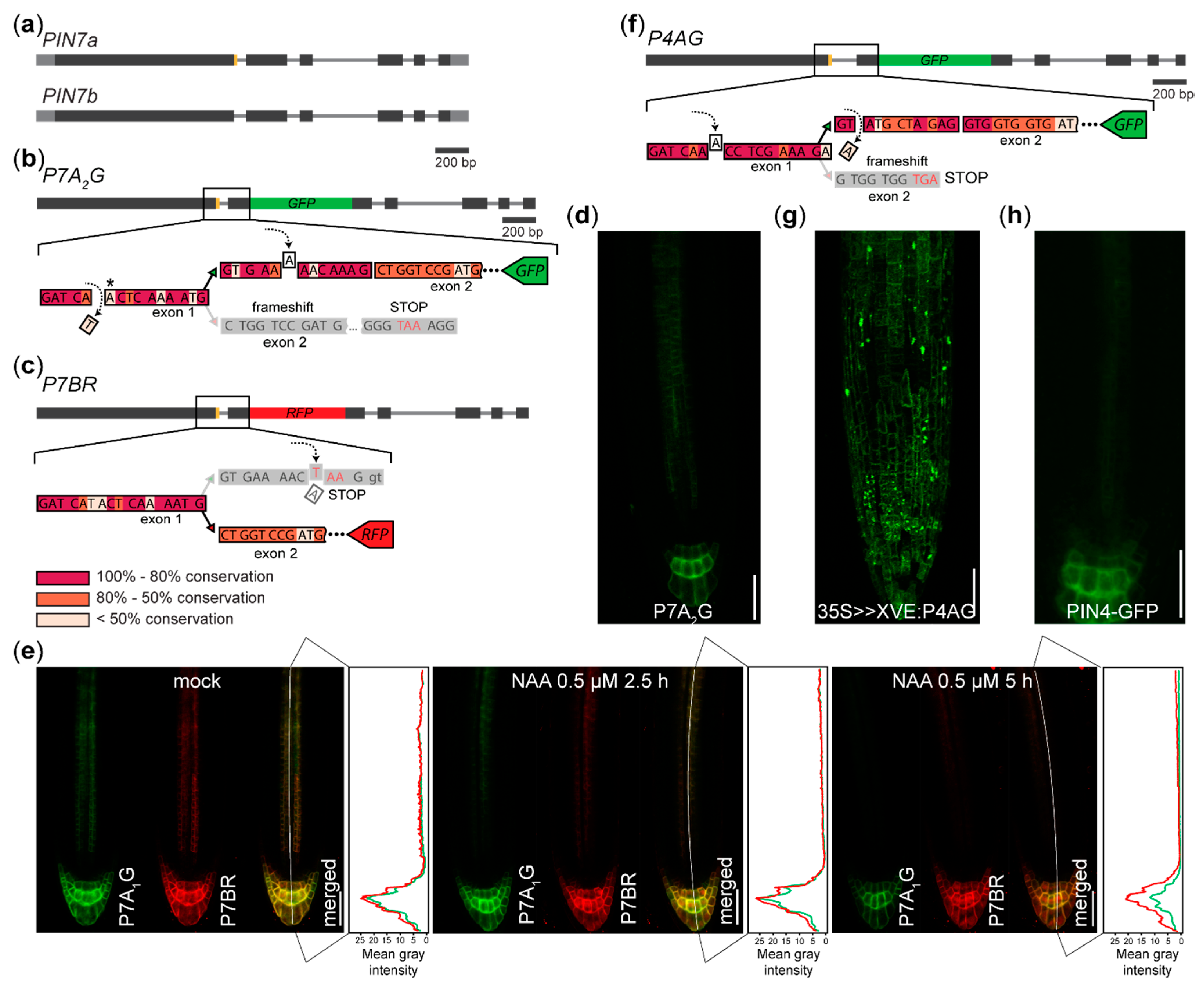
2.2. Reporters Monitoring AS of Auxin Transporters PIN4 and PIN7
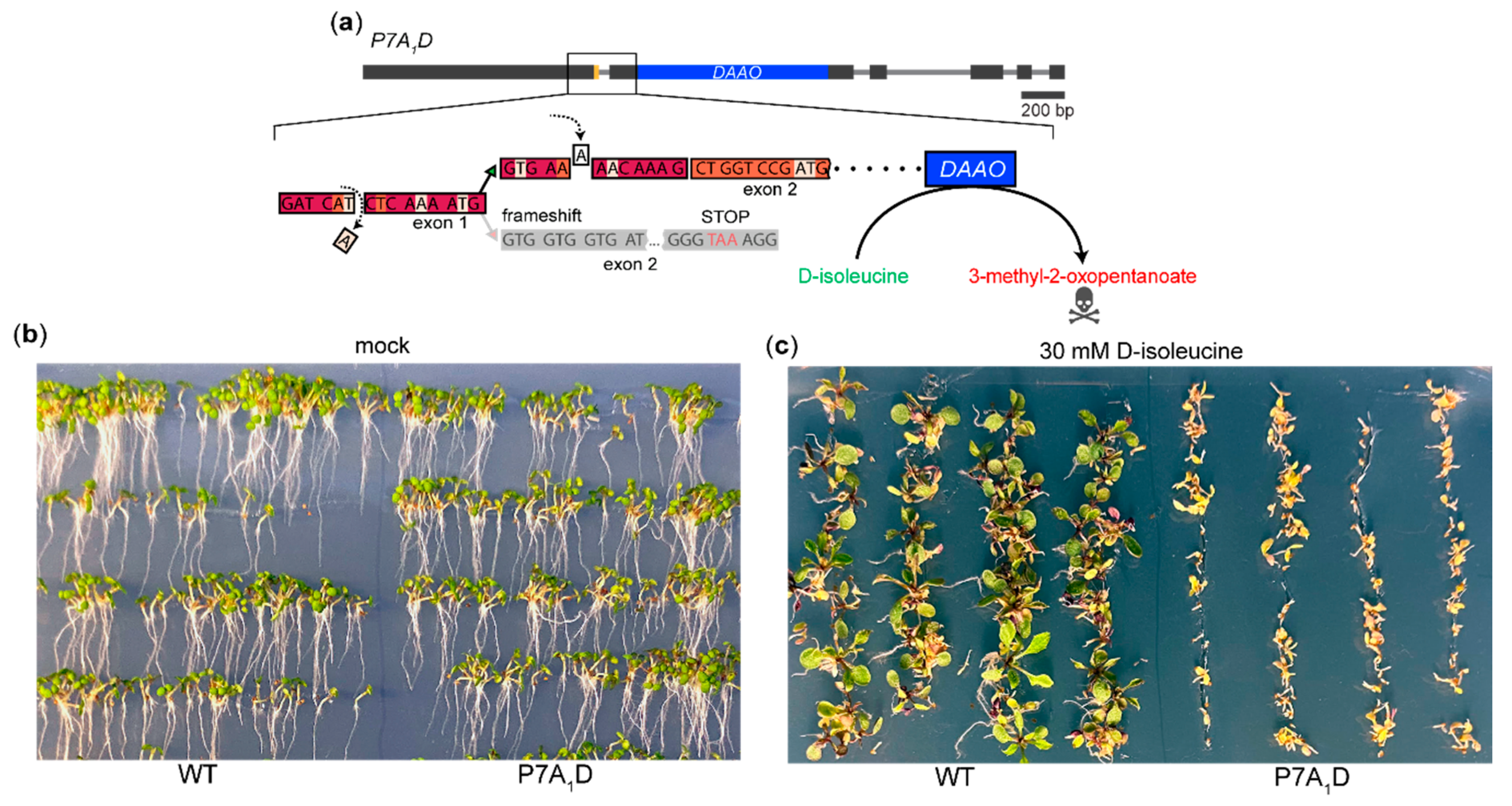
2.3. A System for Genetic Screening for the Factors Upstream of the PIN7 AS Event
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Microscopy
4.2. DNA Manipulations and Transgenic Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuroyanagi, H.; Kobayashi, T.; Mitani, S.; Hagiwara, M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods 2006, 3, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.D.; Gao, S.; Norris, M.L.; Ray, D.; Ramani, A.K.; Fraser, A.G.; Morris, Q.; Hughes, T.R.; Zhen, M.; Calarco, J.A. A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell 2014, 54, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Damoiseaux, R.; Chen, L.; Black, D.L. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res. 2013, 23, 998–1007. [Google Scholar] [CrossRef]
- Bonano, V.I.; Oltean, S.; Garcia-Blanco, M.A. A protocol for imaging alternative splicing regulation in vivo using fluorescence reporters in transgenic mice. Nat. Protoc. 2007, 2, 2166–2181. [Google Scholar] [CrossRef] [PubMed]
- Stoilov, P.; Lin, C.-H.; Damoiseaux, R.; Nikolic, J.; Black, D.L. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. PNAS 2008, 105, 11218–11223. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Wang, Q.; Kennedy, C.J.; Silver, P.A. An Alternative Splicing Network Links Cell-Cycle Control to Apoptosis. Cell 2010, 142, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.S.; Millard, S.S. Deterministic splicing of Dscam2 is regulated by Muscleblind. Sci. Adv. 2019, 5, eaav1678. [Google Scholar] [CrossRef]
- Göhring, J.; Jacak, J.; Barta, A. Imaging of Endogenous Messenger RNA Splice Variants in Living Cells Reveals Nuclear Retention of Transcripts Inaccessible to Nonsense-Mediated Decay in Arabidopsis. Plant Cell 2014, 26, 754–764. [Google Scholar] [CrossRef]
- Ushijima, T.; Hanada, K.; Gotoh, E.; Yamori, W.; Kodama, Y.; Tanaka, H.; Kusano, M.; Fukushima, A.; Tokizawa, M.; Yamamoto, Y.Y.; et al. Light Controls Protein Localization through Phytochrome-Mediated Alternative Promoter Selection. Cell 2017, 171, 1316–1325.e12. [Google Scholar] [CrossRef]
- Kashkan, I.; Hrtyan, M.; Filepova, R.; Vondrakova, Z.; Hejatko, J.; Simon, S.; Rombaut, D.; Jacobs, T.B.; Frilander, M.J.; Friml, J.; et al. Mutually opposing activity of PIN7 splicing isoforms is required for auxin-mediated tropic responses in Arabidopsis thaliana. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Ji, H.; Yuan, B.; Wang, S.; Su, C.; Yao, B.; Zhao, H.; Li, X. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 2015, 6, 8138. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Carvalho, S.D.; Duque, P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010, 154, 772–783. [Google Scholar] [CrossRef]
- Sugliani, M.; Brambilla, V.; Clerkx, E.J.M.; Koornneef, M.; Soppe, W.J.J. The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 2010, 22, 1936–1946. [Google Scholar] [CrossRef] [PubMed]
- Hrtyan, M.; Šliková, E.; Hejátko, J.; Růžička, K. RNA processing in auxin and cytokinin pathways. J. Exp. Bot. 2015, 66, 4897–4912. [Google Scholar] [CrossRef]
- Fujikura, U.; Jing, R.; Hanada, A.; Takebayashi, Y.; Sakakibara, H.; Yamaguchi, S.; Kappel, C.; Lenhard, M. Variation in Splicing Efficiency Underlies Morphological Evolution in Capsella. Dev. Cell 2018, 44, 192–203. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Wang, P.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef]
- Ghelli, R.; Brunetti, P.; Napoli, N.; Paolis, A.D.; Cecchetti, V.; Tsuge, T.; Serino, G.; Matsui, M.; Mele, G.; Rinaldi, G.; et al. A Newly Identified Flower-Specific Splice Variant of AUXIN RESPONSE FACTOR8 Regulates Stamen Elongation and Endothecium Lignification in Arabidopsis. Plant Cell 2018, 30, 620–637. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Friml, J.; Sá-Correia, I.; Duque, P. A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef]
- Mähönen, A.P.; Bishopp, A.; Higuchi, M.; Nieminen, K.M.; Kinoshita, K.; Törmäkangas, K.; Ikeda, Y.; Oka, A.; Kakimoto, T.; Helariutta, Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 2006, 311, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 Gene of Arabidopsis Encodes a Cytochrome P450 That Mediates Multiple 22α-Hydroxylation Steps in Brassinosteroid Biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Skerker, J.M.; Perchuk, B.S.; Siryaporn, A.; Lubin, E.A.; Ashenberg, O.; Goulian, M.; Laub, M.T. Rewiring the specificity of two-component signal transduction systems. Cell 2008, 133, 1043–1054. [Google Scholar] [CrossRef]
- Mähönen, A.P.; Higuchi, M.; Törmäkangas, K.; Miyawaki, K.; Pischke, M.S.; Sussman, M.R.; Helariutta, Y.; Kakimoto, T. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 2006, 16, 1116–1122. [Google Scholar] [CrossRef]
- Besnard, F.; Refahi, Y.; Morin, V.; Marteaux, B.; Brunoud, G.; Chambrier, P.; Rozier, F.; Mirabet, V.; Legrand, J.; Lainé, S.; et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 2014, 505, 417–421. [Google Scholar] [CrossRef]
- Doerner, P.; Jørgensen, J.-E.; You, R.; Steppuhn, J.; Lamb, C. Control of root growth and development by cyclin expression. Nature 1996, 380, 520–523. [Google Scholar] [CrossRef]
- Bancoş, S.; Nomura, T.; Sato, T.; Molnár, G.; Bishop, G.J.; Koncz, C.; Yokota, T.; Nagy, F.; Szekeres, M. Regulation of Transcript Levels of the Arabidopsis Cytochrome P450 Genes Involved in Brassinosteroid Biosynthesis. Plant Physiol. 2002, 130, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Kempe, K.; Gils, M. Pollination control technologies for hybrid breeding. Mol. Breed. 2011, 27, 417–437. [Google Scholar] [CrossRef]
- Debeaujon, I.; Nesi, N.; Perez, P.; Devic, M.; Grandjean, O.; Caboche, M.; Lepiniec, L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 2003, 15, 2514–2531. [Google Scholar] [CrossRef] [PubMed]
- Dotson, S.B.; Lanahan, M.B.; Smith, A.G.; Kishore, G.M. A phosphonate monoester hydrolase from Burkholderia caryophilli PG2982 is useful as a conditional lethal gene in plants. Plant J. 1996, 10, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Erikson, O.; Hertzberg, M.; Näsholm, T. A conditional marker gene allowing both positive and negative selection in plants. Nat. Biotechnol. 2004, 22, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Pollegioni, L.; Molla, G. New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol. 2011, 29, 276–283. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Zhang, R.; Calixto, C.P.G.; Marquez, Y.; Venhuizen, P.; Tzioutziou, N.A.; Guo, W.; Spensley, M.; Entizne, J.C.; Lewandowska, D.; ten Have, S.; et al. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Kuroyanagi, H.; Ohno, G.; Mitani, S.; Hagiwara, M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol. Cell. Biol. 2007, 27, 8612–8621. [Google Scholar] [CrossRef]
- Takeuchi, A.; Hosokawa, M.; Nojima, T.; Hagiwara, M. Splicing Reporter Mice Revealed the Evolutionally Conserved Switching Mechanism of Tissue-Specific Alternative Exon Selection. PLoS ONE 2010, 5, e10946. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Lin, W.-D.; Fu, J.L.; Chang, C.-L.; Matzke, A.J.M.; Matzke, M. A Genetic Screen for Pre-mRNA Splicing Mutants of Arabidopsis thaliana Identifies Putative U1 snRNP Components RBM25 and PRP39a. Genetics 2017. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Venhuizen, P.; Wen, T.-N.; Lin, W.-D.; Chiou, P.; Kalyna, M.; Matzke, A.J.M.; Matzke, M. PRP4KA, a Putative Spliceosomal Protein Kinase, Is Important for Alternative Splicing and Development in Arabidopsis thaliana. Genetics 2018, 210, 1267–1285. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Siligato, R.; Wang, X.; Yadav, S.R.; Lehesranta, S.; Ma, G.; Ursache, R.; Sevilem, I.; Zhang, J.; Gorte, M.; Prasad, K.; et al. MultiSite Gateway-Compatible Cell Type-Specific Gene-Inducible System for Plants. Plant Physiol. 2016, 170, 627–641. [Google Scholar] [CrossRef]
- Karimi, M.; Bleys, A.; Vanderhaeghen, R.; Hilson, P. Building Blocks for Plant Gene Assembly. Plant Physiol. 2007, 145, 1183–1191. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashkan, I.; Timofeyenko, K.; Kollárová, E.; Růžička, K. In Vivo Reporters for Visualizing Alternative Splicing of Hormonal Genes. Plants 2020, 9, 868. https://doi.org/10.3390/plants9070868
Kashkan I, Timofeyenko K, Kollárová E, Růžička K. In Vivo Reporters for Visualizing Alternative Splicing of Hormonal Genes. Plants. 2020; 9(7):868. https://doi.org/10.3390/plants9070868
Chicago/Turabian StyleKashkan, Ivan, Ksenia Timofeyenko, Eva Kollárová, and Kamil Růžička. 2020. "In Vivo Reporters for Visualizing Alternative Splicing of Hormonal Genes" Plants 9, no. 7: 868. https://doi.org/10.3390/plants9070868
APA StyleKashkan, I., Timofeyenko, K., Kollárová, E., & Růžička, K. (2020). In Vivo Reporters for Visualizing Alternative Splicing of Hormonal Genes. Plants, 9(7), 868. https://doi.org/10.3390/plants9070868




