Two New Putative Plant Viruses from Wood Metagenomics Analysis of an Esca Diseased Vineyard
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA Extraction and Sequencing
2.3. Sequence Assembly and Analysis
2.4. Mechanical Transmission to Herbaceous Host
3. Results
3.1. Sequencing Results and Viral Identification
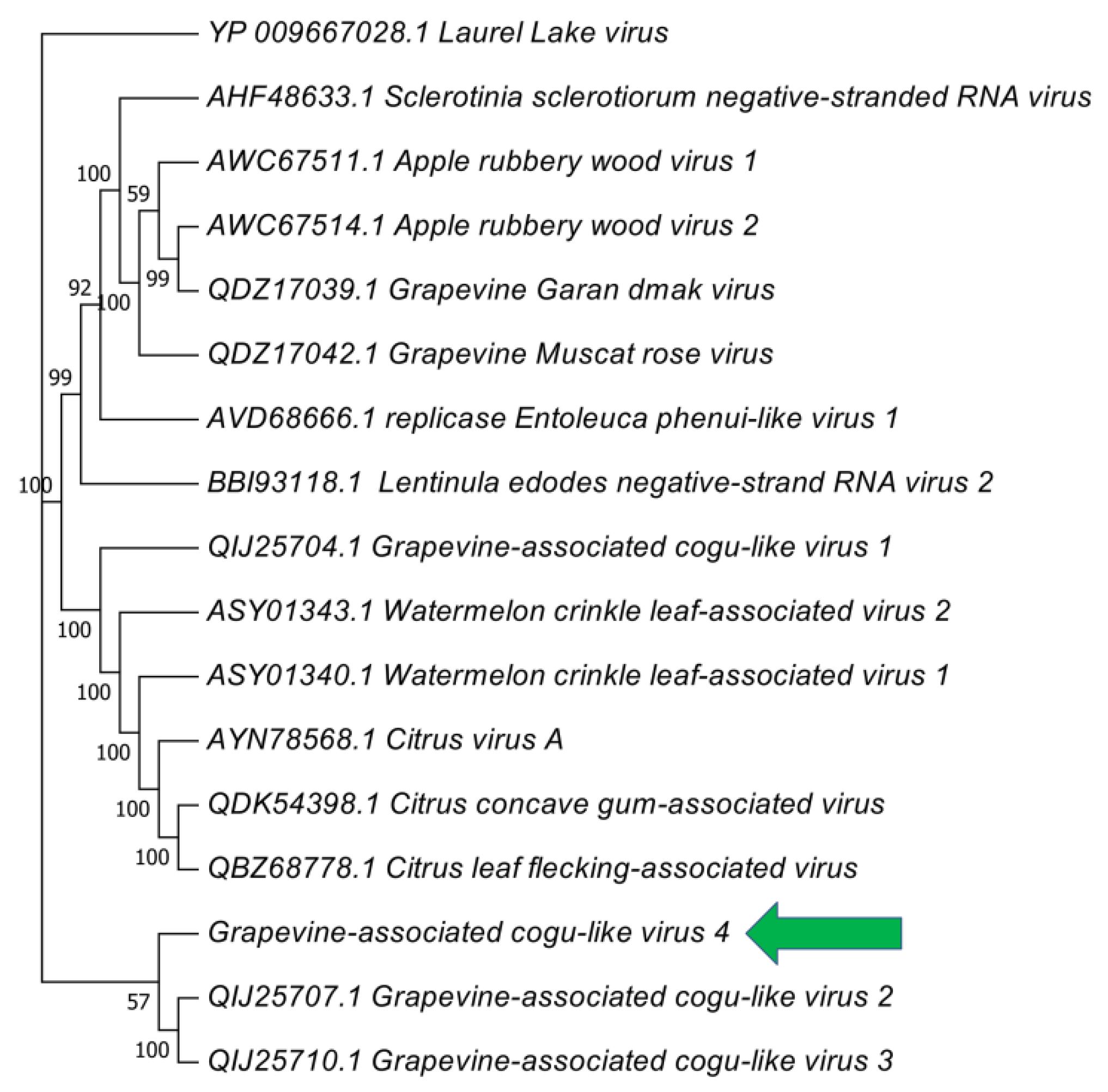
3.2. Phylogenetic Placement of Identified Viruses
3.3. Epidemiological Analysis and Mechanical Inoculation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Anderson, K. Changing varietal distinctiveness of the world’s wine regions: Evidence from a new global database. J. Wine Econ. 2014, 9, 249–272. [Google Scholar] [CrossRef]
- Guerrero, R.; Margulis, L.; Berlanga, M. Symbiogenesis: The holobiont as a unit of evolution. Int. Microbiol. 2013, 16, 133–143. [Google Scholar] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019, 17, 449–458. [Google Scholar] [CrossRef]
- Nerva, L.; Pagliarani, C.; Pugliese, M.; Monchiero, M.; Gonthier, S.; Gullino, M.L.; Gambino, G.; Chitarra, W. Grapevine phyllosphere community analysis in response to elicitor application against powdery mildew. Microorganisms 2019, 7, 662. [Google Scholar] [CrossRef]
- Nerva, L.; Turina, M.; Zanzotto, A.; Gardiman, M.; Gaiotti, F.; Gambino, G.; Chitarra, W. Isolation, molecular characterization and virome analysis of culturable wood fungal endophytes in esca symptomatic and asymptomatic grapevine plants. Environ. Microbiol. 2019, 21, 2886–2904. [Google Scholar] [CrossRef]
- Martelli, G. Where grapevine virology is heading to. In Proceedings of the 19th Congress of ICVG, Santiago, Chile, 9–12 April 2018; pp. 10–15. [Google Scholar]
- Mannini, F.; Digiaro, M. The effects of viruses and viral diseases on grapes and wine. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer: Berlin/Heidelberg, Germany, 2017; pp. 453–482. [Google Scholar]
- Perrone, I.; Chitarra, W.; Boccacci, P.; Gambino, G. Grapevine–virus–environment interactions: An intriguing puzzle to solve. New Phytol. 2017, 213, 983–987. [Google Scholar] [CrossRef]
- Chitarra, W.; Cuozzo, D.; Ferrandino, A.; Secchi, F.; Palmano, S.; Perrone, I.; Boccacci, P.; Pagliarani, C.; Gribaudo, I.; Mannini, F. Dissecting interplays between Vitis vinifera L. and grapevine virus B (GVB) under field conditions. Mol. Plant Pathol. 2018, 19, 2651–2666. [Google Scholar] [CrossRef]
- Fischer, M.; Peighami-Ashnaei, S. Grapevine, esca complex, and environment: The disease triangle. Phytopathol. Mediterr. 2019, 58, 17–37. [Google Scholar]
- Nerva, L.; Zanzotto, A.; Gardiman, M.; Gaiotti, F.; Chitarra, W. Soil microbiome analysis in an ESCA diseased vineyard. Soil Biol. Biochem. 2019, 135, 60–70. [Google Scholar] [CrossRef]
- Surico, G. Towards a redefinition of the diseases within the esca complex of grapevine. Phytopathol. Mediterr. 2009, 48, 5–10. [Google Scholar]
- Manici, L.; Saccà, M.; Caputo, F.; Zanzotto, A.; Gardiman, M.; Fila, G. Long-term grapevine cultivation and agro-environment affect rhizosphere microbiome rather than plant age. Appl. Soil Ecol. 2017, 119, 214–225. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef]
- Nerva, L.; Varese, G.C.; Turina, M. Different Approaches to Discover Mycovirus Associated to Marine Organisms. In Viral Metagenomics; Springer: Berlin/Heidelberg, Germany, 2018; pp. 97–114. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Picarelli, M.A.S.; Forgia, M.; Rivas, E.B.; Nerva, L.; Chiapello, M.; Turina, M.; Colariccio, A. Extreme diversity of mycoviruses present in isolates of Rhizoctonia solani AG2-2 LP from Zoysia japonica from Brazil. Front. Cell. Infect. Microbiol. 2019, 9, 244. [Google Scholar] [CrossRef]
- Bertazzon, N.; Filippin, L.; Forte, V.; Angelini, E. Grapevine Pinot gris virus seems to have recently been introduced to vineyards in Veneto, Italy. Arch. Virol. 2016, 161, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and evolution of the global RNA virome. MBio 2018, 9, e02329-18. [Google Scholar] [CrossRef]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Haydon, D.T.; Antia, R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol. Evol. 2005, 20, 238–244. [Google Scholar] [CrossRef]
- Vurro, M.; Bonciani, B.; Vannacci, G. Emerging infectious diseases of crop plants in developing countries: Impact on agriculture and socio-economic consequences. Food Secur. 2010, 2, 113–132. [Google Scholar] [CrossRef]
- Buoso, S.; Pagliari, L.; Musetti, R.; Fornasier, F.; Martini, M.; Loschi, A.; Fontanella, M.C.; Ermacora, P. With or without you: Altered plant response to boron-deficiency in hydroponically grown grapevines infected by grapevine pinot gris virus suggests a relation between grapevine Leaf mottling and deformation symptom occurrence and boron plant availability. Front. Plant Sci. 2020, 11, 226. [Google Scholar] [CrossRef]
- De, S.; Lõhmus, A.; Pollari, M.; Saha, S.; Mäkinen, K. Host–virus interactions from potyvirus replication to translation. In Plant Viruses Diversity, Interaction and Management; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Gibbs, A.; Ohshima, K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010, 48, 205–223. [Google Scholar] [CrossRef]
- Ala-Poikela, M.; Goytia, E.; Haikonen, T.; Rajamäki, M.-L.; Valkonen, J.P. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF (iso) 4E and eIF4E and contains a 4E binding motif. J. Virol. 2011, 85, 6784–6794. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HCP ro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Pruss, G.; Ge, X.; Shi, X.M.; Carrington, J.C.; Vance, V.B. Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 1997, 9, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Nerva, L.; Forgia, M.; Chitarra, W.; Ayllón, M.; Turina, M. Putative new plant viruses associated with Plasmopara viticola-infected grapevine samples. Ann. Appl. Biol. 2020, 176, 180–191. [Google Scholar] [CrossRef]
- Tanne, E.; Sela, I. Occurrence of a DNA sequence of a non-retro RNA virus in a host plant genome and its expression: Evidence for recombination between viral and host RNAs. Virology 2005, 332, 614–622. [Google Scholar] [CrossRef]
- Navarro, B.; Zicca, S.; Minutolo, M.; Saponari, M.; Alioto, D.; Di Serio, F. A negative-stranded RNA virus infecting citrus trees: The second member of a new genus within the order Bunyavirales. Front. Microbiol. 2018, 9, 2340. [Google Scholar] [CrossRef]
- Tokarz, R.; Sameroff, S.; Tagliafierro, T.; Jain, K.; Williams, S.H.; Cucura, D.M.; Rochlin, I.; Monzon, J.; Carpi, G.; Tufts, D. Identification of novel viruses in Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks. Msphere 2018, 3, e00614–e00617. [Google Scholar] [CrossRef]
- Xin, M.; Cao, M.; Liu, W.; Ren, Y.; Zhou, X.; Wang, X. Two negative-strand RNA viruses identified in watermelon represent a novel clade in the order Bunyavirales. Front. Microbiol. 2017, 8, 1514. [Google Scholar] [CrossRef]
- Fawcett, H.S. Citrus diseases and their control. Citrus Dis. Control 1936. [CrossRef]
- Navarro, B.; Minutolo, M.; de Stradis, A.; Palmisano, F.; Alioto, D.; di Serio, F. The first phlebo-like virus infecting plants: A case study on the adaptation of negative-stranded RNA viruses to new hosts. Mol. Plant Pathol. 2018, 19, 1075–1089. [Google Scholar] [CrossRef]
- Yugoslavia, I.; Vrabl, S. Rubbery wood—A new virus disease of apples. TT. 1966, 14, 65. [Google Scholar]
- Jakovljevic, V.; Otten, P.; Berwarth, C.; Jelkmann, W. Analysis of the apple rubbery wood disease by next generation sequencing of total RNA. Eur. J. Plant Pathol. 2017, 148, 637–646. [Google Scholar] [CrossRef]
- Diaz-Lara, A.; Navarro, B.; Di Serio, F.; Stevens, K.; Hwang, M.S.; Kohl, J.; Vu, S.T.; Falk, B.W.; Golino, D.; Al Rwahnih, M. Two Novel Negative-Sense RNA Viruses Infecting Grapevine Are Members of a Newly Proposed Genus within the Family Phenuiviridae. Viruses 2019, 11, 685. [Google Scholar] [CrossRef]
- Okada, R.; Ichinose, S.; Takeshita, K.; Urayama, S.; Fukuhara, T.; Komatsu, K.; Arie, T.; Ishihara, A.; Egusa, M.; Kodama, M. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 2018, 519, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Chitarra, W.; Siciliano, I.; Gaiotti, F.; Ciuffo, M.; Forgia, M.; Varese, G.; Turina, M. Mycoviruses mediate mycotoxin regulation in Aspergillus ochraceus. Environ. Microbiol. 2018. [CrossRef] [PubMed]
- Ahn, I.-P.; Lee, Y.-H. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant Microbe Interact. 2001, 14, 496–507. [Google Scholar] [CrossRef]
- Boulila, M.; Boscia, D.; Di Terlizzi, B.; Castellano, M.A.; Minafra, A.; Savino, V.; Martelli, G.P. Some properties of a phloem-limited non mechanically-transmissible grapevine virus. J. Phytopathol. 1990, 129, 151–158. [Google Scholar] [CrossRef]
- Castellano, M.A.; Martelli, G.P. Ultrastructure and nature of vesiculated bodies associated with isometric virus-like particles in diseased grapevines. J. Ultrastruct. Res. 1984, 89, 56–64. [Google Scholar] [CrossRef]
- Namba, S.; Yamashita, S.; Doi, Y.; Yora, K. A small spherical virus associated with ajinashika disease of Koshu grapevine. Annu. Phytopathol. Soc. Jpn. 1979, 45, 70–73. [Google Scholar] [CrossRef]
- Verderevskaya, T.D.; Marinesku, V.G.; Semtsghik, E.S. Aetiologie und Diagnose der Marmorierung der Weinrebe. Arch. Phytopathol. Pflanz. 1983, 1983 19, 221–226. [Google Scholar] [CrossRef]
- Granata, G.; Appiano, A. A grapevine disease in Italy resembling infectious necrosis. In Proceedings of the 9th Meeting ICVG, Kyriat Anavim, Israel, 6–11 September 1987. [Google Scholar]
- Belli, G.; Faoro, F.; Fortusini, A.; Tornaghi, R. Further data on grapevine leafroll etiology. Phytopathol. Mediterr. 1985, 24, 148–151. [Google Scholar]

| Country | Cultivar | GaPlV1-Infected/Total Samples |
|---|---|---|
| Italy | Bianco d’Alessano, Cesanese d’Affile, Chardonnay, Cornalin, Croatina, Glera, Gratena, Lambrusco Maestri, Lambrusco Salamino, Merlot, Nerello Mascalese, Pinot gris, Pinot noir, Primitivo, Traminer, Trebbiano toscano, Verdicchio, Vermentino | 21/28 |
| Portugal | Albariño, Cannonau, Chardonnay, Fernão Pires, Godello, Malvasia fine, Tinta barroca, Touriga Franca | 1/10 |
| Spain | Airen, Albariño, Fernão Pires, Macabeo, Parellada, Pedro Ximenez, Tinta barroca, Touriga Franca, Touriga National, Trepat | 8/10 |
| France | Chardonnay, Pinot gris, Pinot noir | 5/10 |
| Greece | Agiogirtiko, Assyrtiko, Kidonitsa, Kocifali, Roditis, Vilana | 7/10 |
| Bulgaria | Chardonnay, Dymiat, Brestovitsa, Pamid | 1/4 |
| Romania | Busuioaca de Bohotin, Galbena de Odobesti, Merlot, Rkatsiteli | 4/5 |
| Hungary | Franconia, Furmint, Harslevelu | 2/5 |
| Ukraine | Kokur Belji, Krasnostop zolotovsicij, Plecistik, Telebi koruk, Zimljansku cernj | 4/5 |
| Croatia | Malvasia, Plavac mali, Teran | 4/4 |
| Total | 64 | 57/91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertazzon, N.; Chitarra, W.; Angelini, E.; Nerva, L. Two New Putative Plant Viruses from Wood Metagenomics Analysis of an Esca Diseased Vineyard. Plants 2020, 9, 835. https://doi.org/10.3390/plants9070835
Bertazzon N, Chitarra W, Angelini E, Nerva L. Two New Putative Plant Viruses from Wood Metagenomics Analysis of an Esca Diseased Vineyard. Plants. 2020; 9(7):835. https://doi.org/10.3390/plants9070835
Chicago/Turabian StyleBertazzon, Nadia, Walter Chitarra, Elisa Angelini, and Luca Nerva. 2020. "Two New Putative Plant Viruses from Wood Metagenomics Analysis of an Esca Diseased Vineyard" Plants 9, no. 7: 835. https://doi.org/10.3390/plants9070835
APA StyleBertazzon, N., Chitarra, W., Angelini, E., & Nerva, L. (2020). Two New Putative Plant Viruses from Wood Metagenomics Analysis of an Esca Diseased Vineyard. Plants, 9(7), 835. https://doi.org/10.3390/plants9070835







