Arabidopsis Plastid-RNA Polymerase RPOTp Is Involved in Abiotic Stress Tolerance
Abstract
1. Introduction
2. Results
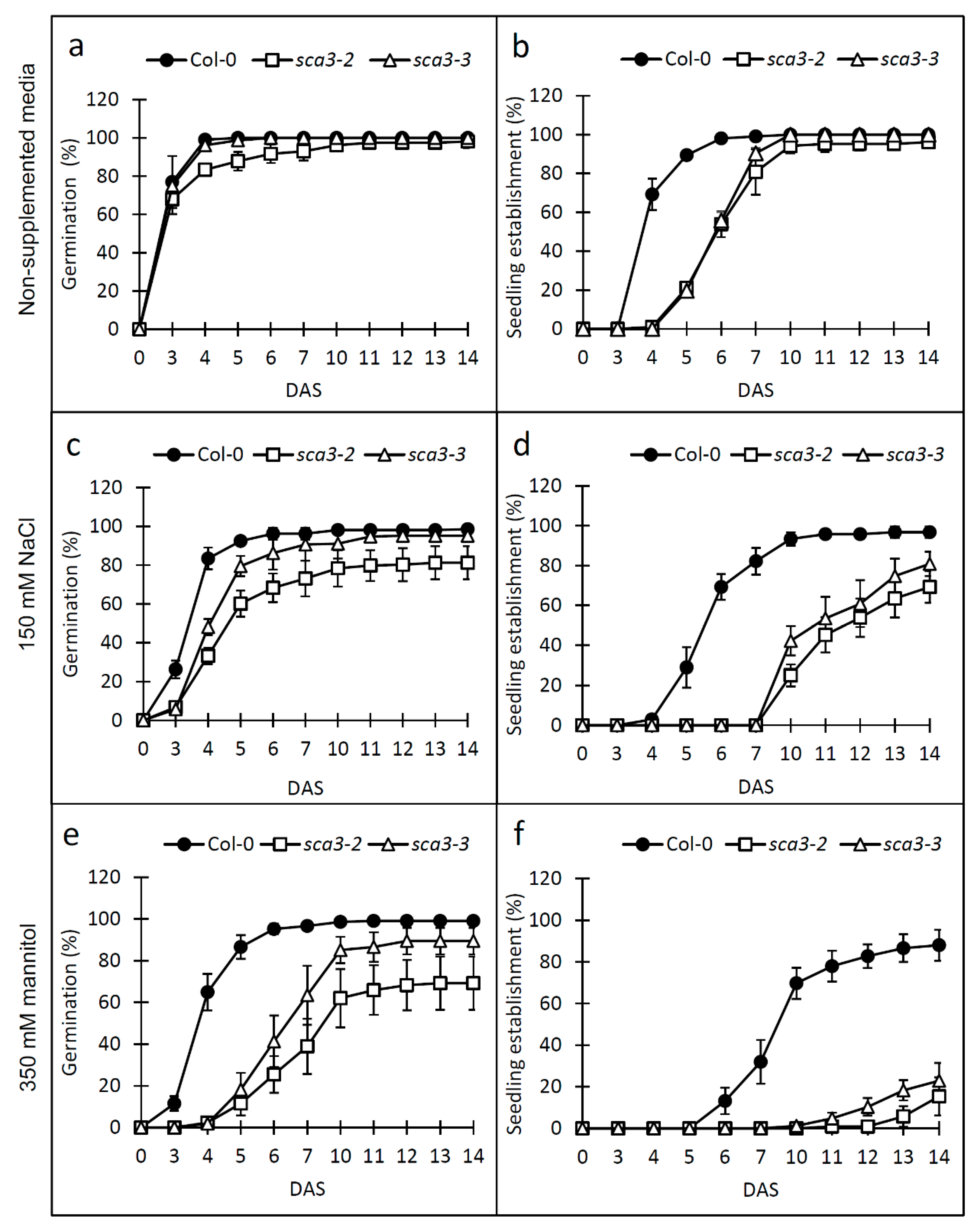
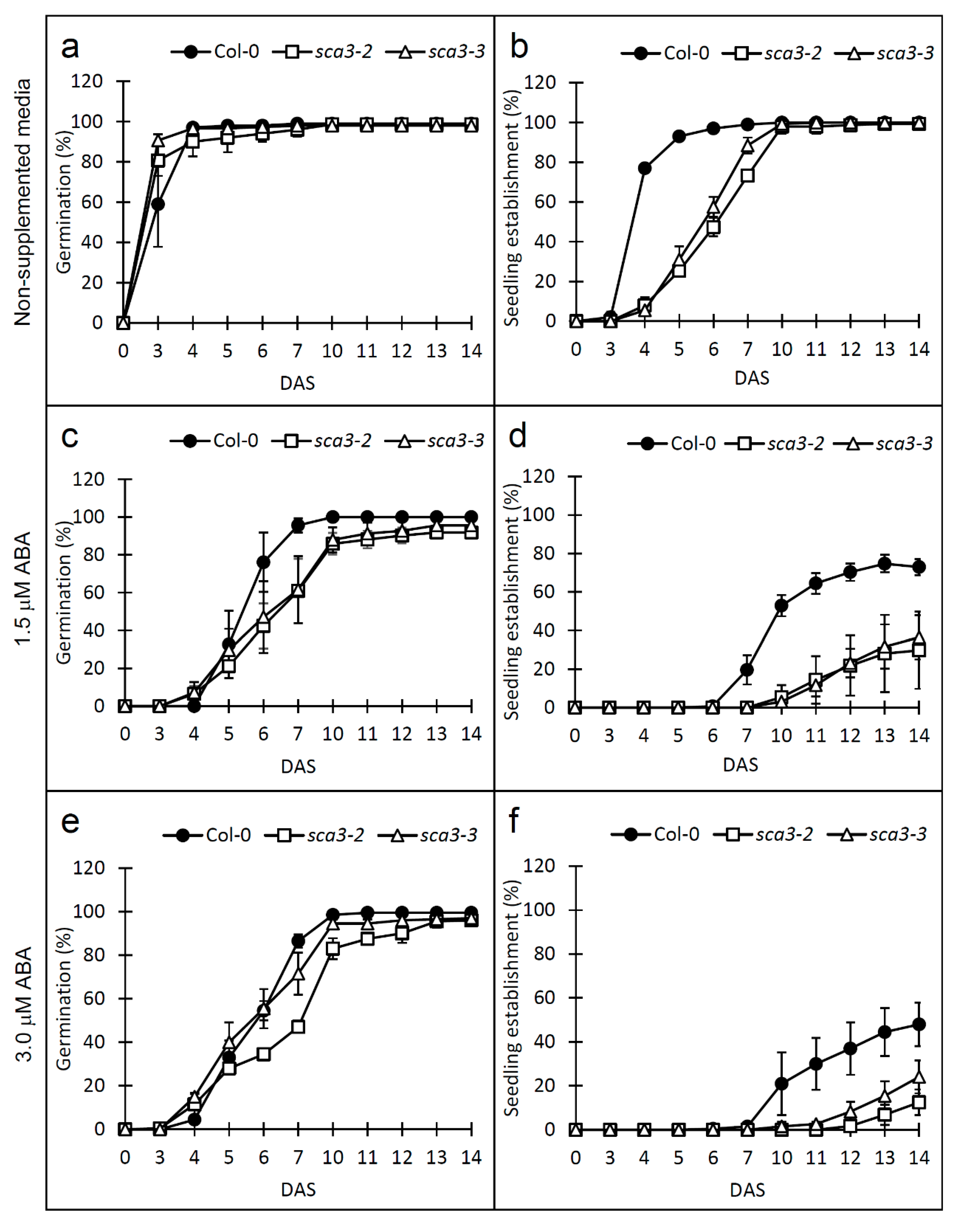
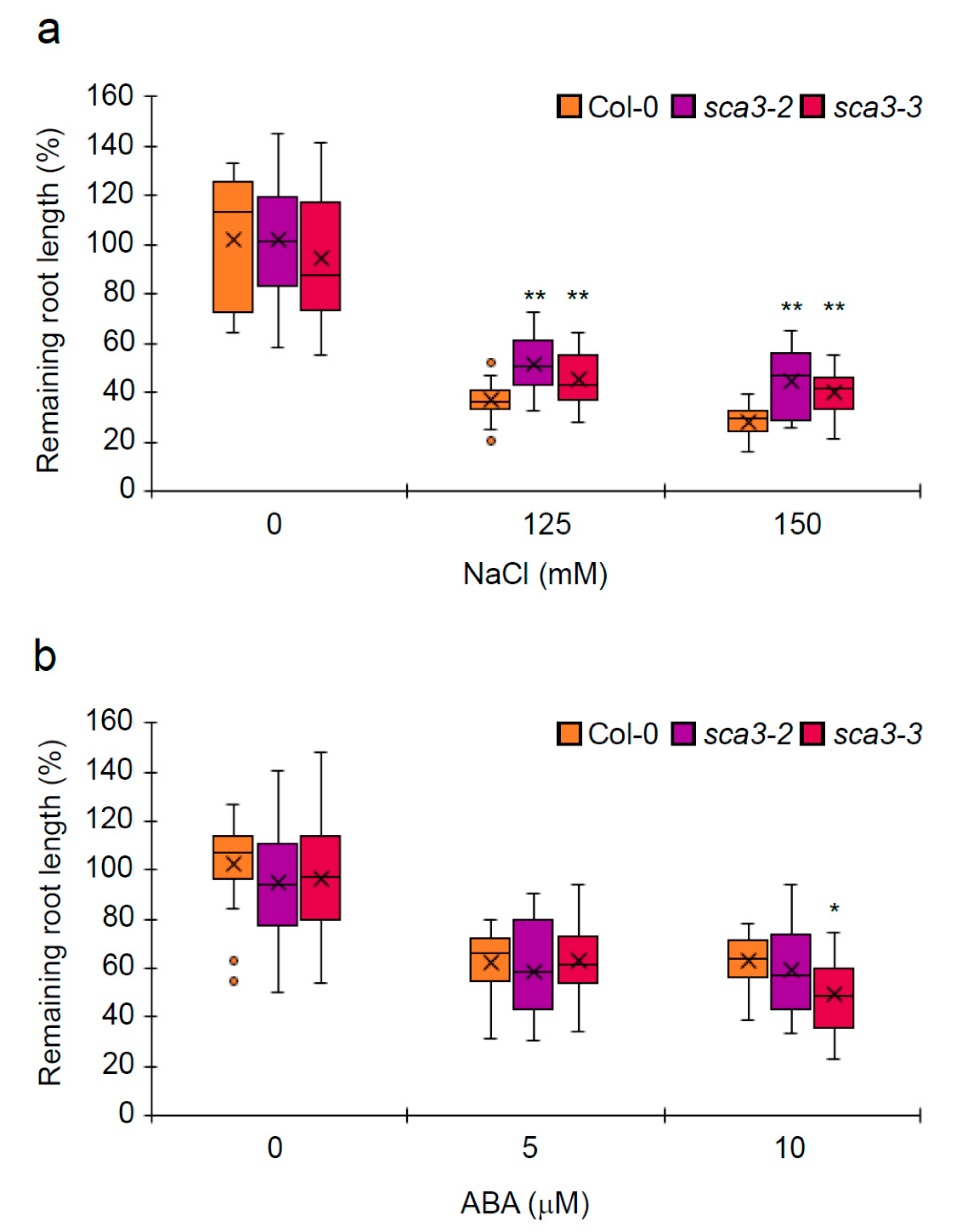
2.1. Knock-Down of RPOTp Alters NaCl, Mannitol and ABA Responses
2.2. Analysis of Gene Ontologies in the Transcriptome of the sca3-2 Mutant
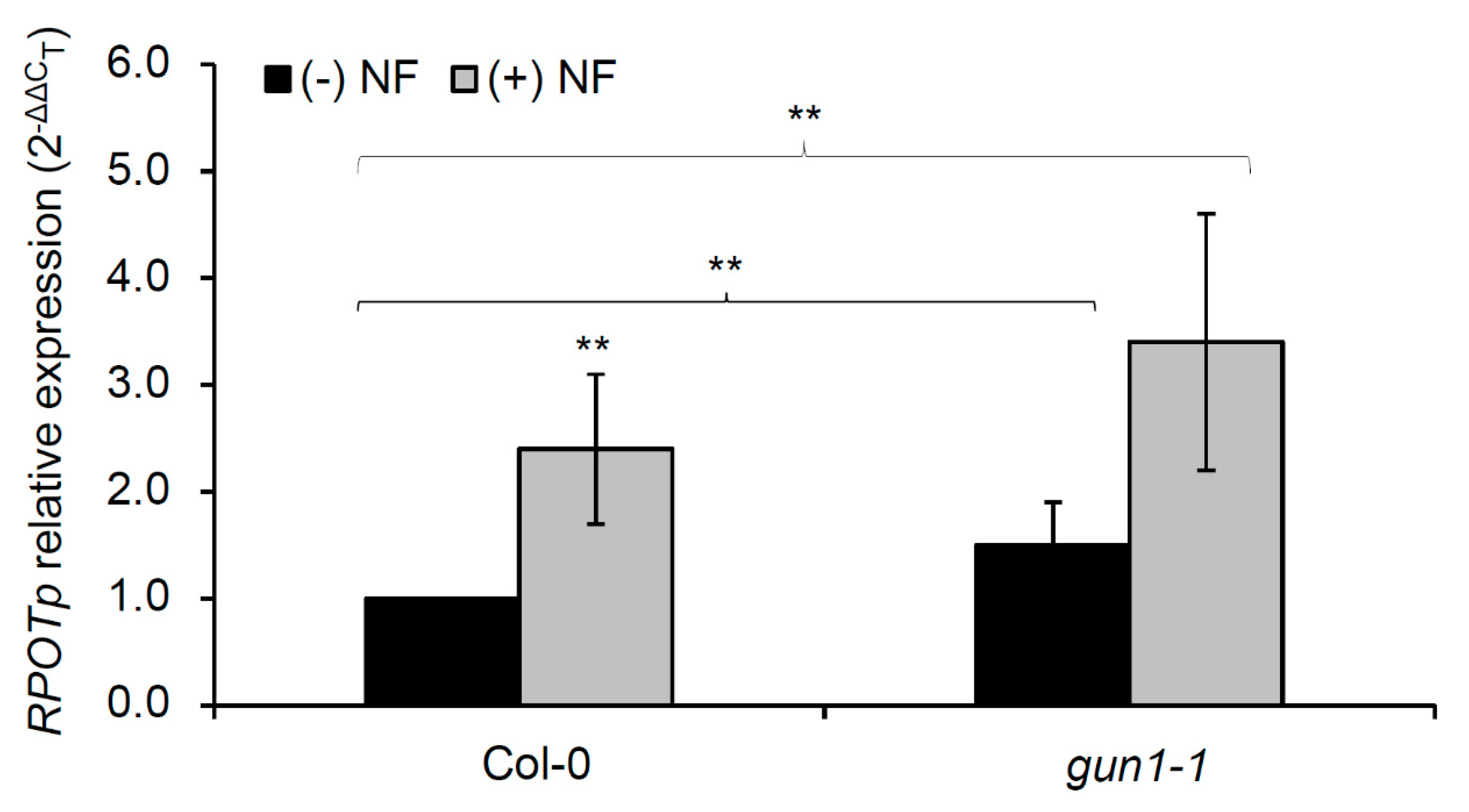
2.3. RPOTp Expression is Controlled by Plastid Retrograde Signaling in Response to Salt Stress
2.4. Salt Stress Differentially Affects the Transcript Abundance of Plastid-Encoded Genes
2.5. ABA Down-Regulates the Expression of NEP and Genes Encoding PEP Subunits
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Germination and Growth Sensitivity Assays
4.3. Quantitative RT-PCR (qRT-PCR)
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 1967, 14, 225-IN6. [Google Scholar] [CrossRef]
- Bock, R.; Timmis, J.N. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 2008, 30, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Sijben-Muller, G.; Hallick, R.B.; Alt, J.; Westhoff, P.; Herrmann, R.G. Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase alpha-subunit. Nucleic Acids Res. 1986, 14, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Pfannschmidt, T.; Link, G. The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol. Gen. Genet. 1997, 257, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Pfannschmidt, T.; Ogrzewalla, K.; Baginsky, S.; Sickmann, A.; Meyer, H.E.; Link, G. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur. J. Biochem. 2000, 267, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Ogrzewalla, K.; Piotrowski, M.; Reinbothe, S.; Link, G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur. J. Biochem. 2002, 269, 3329–3337. [Google Scholar] [CrossRef]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmuller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant. Cell 2006, 18, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Hedtke, B.; Börner, T.; Weihe, A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 1997, 277, 809–811. [Google Scholar] [CrossRef]
- Hedtke, B.; Wagner, I.; Börner, T.; Hess, W.R. Inter-organellar crosstalk in higher plants: Impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 1999, 19, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Hedtke, B.; Börner, T.; Weihe, A. One RNA polymerase serving two genomes. EMBO Rep. 2000, 1, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Dokiya, Y.; Sugita, M. Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem. Biophys. Res. Commun. 2001, 289, 1106–1113. [Google Scholar] [CrossRef]
- Liere, K.; Weihe, A.; Börner, T. The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 2011, 168, 1345–1360. [Google Scholar] [CrossRef]
- Steitz, T.A. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 2009, 19, 683–690. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.T.; Allison, L.A.; Maliga, P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997, 16, 4041–4048. [Google Scholar] [CrossRef]
- Emanuel, C.; Weihe, A.; Graner, A.; Hess, W.R.; Börner, T. Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 2004, 38, 460–472. [Google Scholar] [CrossRef]
- Allison, L.A.; Simon, L.D.; Maliga, P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996, 15, 2802–2809. [Google Scholar] [CrossRef]
- Legen, J.; Kemp, S.; Krause, K.; Profanter, B.; Herrmann, R.G.; Maier, R.M. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002, 31, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Baba, K.; Schmidt, J.; Espinosa-Ruiz, A.; Villarejo, A.; Shiina, T.; Gardeström, P.; Sane, A.P.; Bhalerao, R.P. Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J. 2004, 38, 38–48. [Google Scholar] [CrossRef]
- Hricová, A.; Quesada, V.; Micol, J.L. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 2006, 141, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Quesada, V. Transcriptional and Post-transcriptional Regulation of Organellar Gene Expression (OGE) and Its Roles in Plant Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 1056. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Wang, L.; Kleine, T. Organellar Gene Expression and Acclimation of Plants to Environmental Stress. Front. Plant Sci. 2017, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, G.; Kang, H. Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front. Plant Sci. 2017, 8, 871. [Google Scholar] [CrossRef]
- Suo, J.; Zhao, Q.; David, L.; Chen, S.; Dai, S. Salinity Response in Chloroplasts: Insights from Gene Characterization. Int. J. Mol. Sci. 2017, 18, 1011. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Heung, Y.; You, Q.; Yi, X.; Zhou, D.; Wenying, X.; Zhen, S. AgriGO v2.0: A GO analysis (toolkit for the agricultural community, 2017 update). Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive RD29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993, 236, 331–340. [Google Scholar] [CrossRef]
- Núñez-Delegido, E.; Robles, P.; Ferrández-Ayela, A.; Quesada, V. Functional analysis of mTERF5 and mTERF9 contribution to salt tolerance, plastid gene expression and retrograde signalling in Arabidopsis thaliana. Plant. Biol. 2020, 22, 459–471. [Google Scholar]
- Ding, S.; Zhang, Y.; Hu, Z.; Huang, X.; Zhang, B.; Lu, Q.; Wen, X.; Wang, Y.; Lu, C. mTERF5 Acts as a Transcriptional Pausing Factor to Positively Regulate Transcription of Chloroplast psbEFLJ. Mol. Plant 2019, 12, 1259–1277. [Google Scholar] [CrossRef]
- Sun, X.; Xu, D.; Liu, Z.; Kleine, T.; Leister, D. Functional relationship between mTERF4 and GUN1 in retrograde signalling. J. Exp. Bot. 2016, 67, 3909–3924. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Van Aken, O.; Ho, L.H.; Whelan, J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009, 150, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Padmasree, K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003, 8, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Susek, R.E.; Ausubel, F.M.; Chory, J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 1993, 74, 787–799. [Google Scholar] [CrossRef]
- Leister, D. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 2005, 354, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef]
- Kakizaki, T.; Matsumura, H.; Nakayama, K.; Che, F.S.; Terauchi, R.; Inaba, T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009, 151, 1339–1353. [Google Scholar] [CrossRef]
- Pesaresi, P.; Kim, C. Current understanding of GUN1: A key mediator involved in biogenic retrograde signaling. Plant Cell Rep. 2019, 38, 819–823. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Micol, J.L.; Quesada, V. Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant 2015, 154, 297–313. [Google Scholar] [CrossRef]
- Emanuel, C.; von Groll, U.; Müller, M.; Börner, T.; Weihe, A. Development- and tissue specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 2006, 223, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Navarro-Cartagena, S.; Ferrández-Ayela, A.; Núñez-Delegido, E.; Quesada, V. The Characterization of Arabidopsis mterf6 Mutants Reveals a New Role for mTERF6 in Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 2388. [Google Scholar] [CrossRef]
- Romani, I.; Manavski, N.; Morosetti, A.; Tadini, L.; Maier, S.; Kühn, K.; Ruwe, H.; Schmitz-Linneweber, C.; Wanner, G.; Leister, D.; et al. A Member of the Arabidopsis Mitochondrial Transcription Termination Factor Family Is Required for Maturation of Chloroplast Transfer RNAIle (GAU). Plant Physiol. 2015, 169, 627–646. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, Y.L.; Zhang, X.L.; Yu, Q.B.; Wang, X.; Yuan, X.B.; Qin, X.M.; He, X.F.; Huang, C.; Yang, Z.N. A nuclear-encoded protein, mTERF6, mediates transcription termination of rpoA polycistron for plastid-encoded RNA polymerase-dependent chloroplast gene expression and chloroplast development. Sci. Rep. 2018, 8, 11929. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, A.; Hanaoka, M.; Motohashi, R.; Seki, M.; Shinozaki, K.; Kanamaru, K.; Takahashi, H.; Tanaka, K. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci. Biotechnol. Biochem. 2004, 68, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Cui, R.; Xu, P.; Wu, J.; Mao, J.L.; Chen, Y.; Zhou, C.Z.; Yu, L.H.; Xiang, C.B. ATHB17 enhances stress tolerance by coordinating photosynthesis associated nuclear gene and ATSIG5 expression in response to abiotic stress. Sci. Rep. 2017, 7, 45492. [Google Scholar] [CrossRef]
- Robles, P.; Núñez-Delegido, E.; Ferrández-Ayela, A.; Sarmiento-Mañús, R.; Micol, J.L.; Quesada, V. Arabidopsis mTERF6 is required for leaf patterning. Plant Sci. 2018, 266, 117–129. [Google Scholar] [CrossRef]
- Danilova, M.N.; Kudryakova., N.V.; Andreeva, A.A.; Pojidaeva, E.S.; Kuznetsov, V.V. Differential impact of heat stress on the expression of chloroplast-encoded genes. Plant Physiol. Biochem. 2018, 129, 90–100. [Google Scholar] [CrossRef]
- Pfalz, J.; Pfannschmidt, T. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 2013, 18, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Yamburenko, M.V.; Zubo, Y.O.; Börner, T. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: Repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J. 2015, 82, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Yamburenko, M.V.; Zubo, Y.O.; Vanková, R.; Kusnetsov, V.V.; Kulaeva, O.N.; Börner, T. Abscisic acid represses the transcription of chloroplast genes. J. Exp. Bot. 2013, 64, 4491–4502. [Google Scholar] [CrossRef] [PubMed]
- Danilova, M.N.; Andreeva, A.A.; Doroshenko, A.S.; Kudryakova, N.V.; Kuznetsov, V.V.; Kusnetsov, V.V. Phytohormones Regulate the Expression of Nuclear Genes Encoding the Components of the Plastid Transcription Apparatus. Dokl. Biochem. Biophys. 2018, 478, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kasai, K.; Ochi, K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. USA 2004, 101, 4320–4324. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Takahashi, K.; Ochiai, Y.; Hosaka, T.; Ochi, K.; Nabeta, K. Bacterial alarmone, guanosine 5′-diphosphate 3′-diphosphate (ppGpp), predominantly binds the beta’ subunit of plastid-encoded plastid RNA polymerase in chloroplasts. ChemBioChem 2009, 10, 1227–1233. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Štefanic, P.P.; Koffler, T.; Adler, G.; Bar-Zvi, D. Chloroplasts of salt-grown Arabidopsis seedlings are impaired in structure, genome copy number and transcript levels. PLoS ONE 2013, 8, e82548. [Google Scholar]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Quesada, V.; Sarmiento-Mañús, R.; González-Bayón, R.; Hricová, A.; Pérez-Marcos, R.; Graciá-Martínez, E.; Medina-Ruiz, L.; Leyva-Díaz, E.; Ponce, M.R.; Micol, J.L. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 2011, 68, 738–753. [Google Scholar] [CrossRef]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef]
- Schweer, J.; Loschelder, H.; Link, G. A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 2006, 580, 6617–6622. [Google Scholar] [CrossRef] [PubMed]
- Tadini, L.; Peracchio, C.; Trotta, A.; Colombo, M.; Mancini, I.; Jeran, N.; Costa, A.; Faoro, F.; Marsoni, M.; Vannini, C.; et al. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 2020, 101, 1198–1220. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Feng, P.; Xu, X.; Guo, H.; Ma, J.; Chi, W.; Lin, R.; Lu, C.; Zhang, L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2011, 2, 477. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Moschopoulos, A.; Derbyshire, P.; Byrne, M.E. The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J. Exp. Bot. 2012, 63, 5233–5243. [Google Scholar] [CrossRef]
| Gene | Normalized Transcript Levels in Col-0 Plants Exposed to 100 mM NaCl Relative to Control Plants | ||
|---|---|---|---|
| Protein Product | Fold Change | p-Value | |
| Nuclear genes | |||
| RPOTp/SCA3 | NEP 1 RPOTp | 2.27 ± 0.84 | 4.113 × 10−5 ** |
| RPOTmp | NEP 1 RPOTmp | 3.50 ± 1.70 | 3.996 × 10−4 ** |
| RPOTm | NEP 1 RPOTm | 1.34 ± 0.33 | 0.036 * |
| AOX1A | Alternative oxidase 1A | 3.90 ± 1.00 | 4.113 × 10−5 ** |
| LHCB1 | Light-harvesting complex protein B1 | 0.70 ± 0.44 | 0.036 * |
| mTERF5 | mTERF5 | 0.77 ± 0.31 | 0.008 ** |
| mTERF9 | mTERF9 | 1.10 ± 0.30 | 0.015 * |
| COR15B | Cold-regulated 15B | 4.76 ± 2.11 | 2.20 × 10−10 ** |
| RD29A | Responsive to desiccation 29A | 2.74 ± 0.76 | 5.83 × 10−4 ** |
| Chloroplast genes | |||
| psaA | Photosystem I reaction center protein | 1.12 ± 0.49 | 0.709 |
| psaB | Photosystem I reaction center protein | 1.04 ± 0.28 | 0.709 |
| psbA | Chlorophyll binding protein D1 | 0.60 ± 0.22 | 4.113 × 10−5 ** |
| clpP | ATP-dependent protease | 1.18 ± 0.27 | 0.036 * |
| rps18 | Ribosomal protein S18 | 0.83 ± 0.23 | 0.709 |
| rpoA | PEP 2 α subunit | 0.63 ± 0.27 | 0.001 ** |
| rpoB | PEP 2 β subunit | 0.61 ± 0.09 | 0.002 ** |
| rpoC1 | PEP 2 β’ subunit | 0.86 ± 0.20 | 0.220 |
| accD | Carboxytransferase β subunit of the Acetyl-CoA carboxylase | 0.88 ± 0.35 | 0.709 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lidón-Soto, A.; Núñez-Delegido, E.; Pastor-Martínez, I.; Robles, P.; Quesada, V. Arabidopsis Plastid-RNA Polymerase RPOTp Is Involved in Abiotic Stress Tolerance. Plants 2020, 9, 834. https://doi.org/10.3390/plants9070834
Lidón-Soto A, Núñez-Delegido E, Pastor-Martínez I, Robles P, Quesada V. Arabidopsis Plastid-RNA Polymerase RPOTp Is Involved in Abiotic Stress Tolerance. Plants. 2020; 9(7):834. https://doi.org/10.3390/plants9070834
Chicago/Turabian StyleLidón-Soto, Abel, Eva Núñez-Delegido, Iván Pastor-Martínez, Pedro Robles, and Víctor Quesada. 2020. "Arabidopsis Plastid-RNA Polymerase RPOTp Is Involved in Abiotic Stress Tolerance" Plants 9, no. 7: 834. https://doi.org/10.3390/plants9070834
APA StyleLidón-Soto, A., Núñez-Delegido, E., Pastor-Martínez, I., Robles, P., & Quesada, V. (2020). Arabidopsis Plastid-RNA Polymerase RPOTp Is Involved in Abiotic Stress Tolerance. Plants, 9(7), 834. https://doi.org/10.3390/plants9070834







