Abstract
Doubled haploid technology is highly successful in maize breeding programs and is contingent on the ability of maize inducers to efficiently produce haploids. Knowledge of the genes involved in haploid induction is important for not only developing better maize inducers, but also to create inducers in other crops. The main quantitative trait loci involved in maize haploid induction are qhir1 and qhir8. The gene underlying qhir1 has been discovered and validated by independent research groups. Prior to initiation of this study, the gene associated with qhir8 had yet to be recognized. Therefore, this research focused on characterizing positional candidate genes underlying qhir8. Pursuing this goal, a strong candidate for qhir8, GRMZM2G435294 (MYO), was silenced by RNAi. Analysis of crosses with these heterozygous RNAi-transgenic lines for haploid induction rate revealed that the silencing of MYO significantly enhanced haploid induction rate by an average of 0.6% in the presence of qhir1. Recently, GRMZM2G465053 (ZmDMP) was identified by map-based gene isolation and shown to be responsible for qhir8. While our results suggest that MYO may contribute to haploid induction rate, results were inconsistent and only showing minor increases in haploid induction rate compared to ZmDMP. Instead, reciprocal crosses clearly revealed that the silencing of MYO causes male sterility.
1. Introduction
Doubled haploid (DH) lines are created when cells of haploid seedlings undergo genome doubling, resulting in completely homozygous diploid offspring after self-pollination. DH technology serves as a shortcut for inbred line development, as this procedure only takes two generations. Most haploids utilized in maize breeding programs are created by in vivo maternal haploid induction, which involves crossing a haploid inducer genotype as male to donor genotypes. Current inducers induce haploids at rates of >8% [1,2].
Genetic mechanisms controlling haploid induction rate (HIR) in maize are not well understood. Quantitative trait locus (QTL) mapping has located a major QTL on chromosome 1 [3,4]. The first comparative genome-wide QTL analysis involving inducers UH400 and CAUHOI confirmed that a major QTL, qhir1, is present on chromosome 1, and a minor QTL, qhir8, was found on chromosome 9 [1]. This minor QTL accounts for up to 20% of the genetic variation and acts to enhance HIR in the presence of qhir1. Unlike qhir8, qhir1 is required for haploid induction and accounts for up to 66% of the genotypic variance of this trait [1,5,6]. Dong et al. [7] conducted a fine mapping experiment of qhir1 using populations from crosses between UH400 and 1680, similar to those of Prigge et al. [1], and were able to narrow the QTL to a 243 kb region. Similarly, fine mapping experiments of qhir8 using crosses between UH400 and CAUHOI as a mapping population have narrowed qhir8 to a 789 kb region [5]. Additional regions on chromosome 1 have been detected by Hu et al. [6] with a novel genome wide association study approach detecting selective sweeps. This recently discovered region, qhir12, is 3.97 Mb long and is 985 kb downstream of the previously fine-mapped region for qhir1 [6,7].
In 2017, different research groups independently identified the gene MATRILINEAL (MTL), which is underlying qhir1, and explains the high HIR within inducer lines [8,9,10]. Fine-mapping identified MTL, a pollen-specific phospholipase, which was shown to contain a frame-shift mutation in inducer alleles. Furthermore, knockdown lines using RNA interference (RNAi) and complete knockout lines using transcription activator-like effector nuclease (TALEN) for MTL were created, resulting in HIRs of 2.7% and 6.7%, respectively.
Prior to the initiation of this study, the gene responsible for qhir8 had not been identified. However very recently, the gene has been discovered by Zhong et al. [11] who narrowed the fine-mapping region from 789 kb to 138 kb. Within this region, the gene GRMZM2G465053 (ZmDMP) was a strong candidate and expressed during late maturity of pollen grains. By knocking out this gene, HIR was increased 5–6-fold in the presence of qhir1, thereby confirming its likelihood to be responsible for qhir8. Additionally, the study shows a significant increase (10–40%) in aborted kernels when this gene is knocked out.
For the current study initiated prior to 2019, candidate genes for qhir8 were chosen from the study of Kelliher et al. [8], who also conducted RNA-seq experiments between inducer and non-inducer genotypes, resulting in a total of 60 differentially expressed genes, out of which 15 were co-expressed with MTL in inducer pollen. One of these genes is GRMZM2G435294 (MYO), located within the 789 kb qhir8 fine mapping region [5]. Therefore, MYO was considered a strong candidate that may be responsible for the enhancement of HIR, when both qhir1 and qhir8 are present.
MYO is predicted to be a myosin-11 like protein due to several conserved myosin domains, including the myosin N-terminal SH3-like domain (amino acids 10–48), motor head domain (76–720), gap junction protein N-terminal region, and the cargo binding tail domain (1110–1495) [12]. In plants, myosins act to mobilize various vesicular cargoes via attachment to the cargo binding tail domain [13]. MYO is likely involved with pollen tube growth, so the function of the cargo binding tail domain is to bind and carry the twin sperm cells. The pollen tube growth is driven by the head motor domain, which binds actin and hydrolyzes ATP, until it fuses with the plasma membrane and allows for pollen tube enlargement and cytoplasmic streaming [14]. Therefore, we predicted the knockdown of MYO would have an impact on male fertility. A complete knockout may thus result in male sterility. Instead, reducing MYO functions may allow both male fertility and haploid induction. For these reasons, RNA interference (RNAi) is a reasonable option for testing the influence of MYO on haploid induction rate.
Herein, we report results from transgenic experiments with altered MYO expression in defined genetic backgrounds with regard to presence or absence of qhir1. The specific objectives of this research were to (1) explore sequence differences within MYO between inducer and non-inducer genotypes, (2) develop transgenic events that successfully silence MYO by RNAi, and (3) evaluate these events for HIR and other reproductive phenotypes, such as male fertility.
2. Results
2.1. Sequence Alignment
MYO was confirmed as primary gene of interest because sequencing results revealed several polymorphic regions between inducer genotype RWS, and a non-inducer genotype, B73 RefGen_v3 [15]. There were five different single nucleotide polymorphisms (SNPs) scattered within exon 21. Four out of five SNPs were at the third degenerate position of the codon sequence, all leading to silent mutations. A small insertion of three nucleotides (AGG) was inserted into the RWS sequence at the beginning of exon 21, causing addition of a Glycine. Additionally, a large 60 bp deletion within exon 22 and 23 causes a deletion of 20 amino acids. These changes, however, are all within frame. A deletion of 5 bp within exon 23 causes a frameshift, resulting in 33 amino acid substitutions between B73 and RWS. Following these amino acid changes, there is an introduction of an early stop codon (TAA). The protein length is 1529 amino acids for B73, but with the introduction of the frameshift mutation and the subsequent early stop codon, the protein length is only 915 amino acids within RWS (Table S1), assuming the first exons are intact.
2.2. Silencing Efficiency
Two RNAi constructs (MYO16 and MYO18) were transformed into a non-inducing genetic background to knock down MYO. Five transgenic events, one from MYO16 and four from MYO18, were backcrossed as BC1F1 (MYOBC1F1) families with B73 and analyzed for silencing efficiency by quantitative PCR (qPCR). Anthers were dissected from immature tassels for RNA extraction. All transgenic events exhibited significantly (p = 0.05) decreased expression of MYO, and all were significantly different when compared to Viking, except for MYO18-4 (p = 0.16) (Table 1). Although only one transgenic family from MYO16 was included in this study, this construct was most efficient in silencing MYO. MYO16-1 showed values of −2.35 and 5.09 for calibrated expression levels (ΔΔCT) and relative expression levels (fold change), respectively. The average values for MYO18 events were −1.81 and 3.63 for calibrated expression levels (ΔΔCT) and relative expression levels (fold change), respectively (Figure 1). MYO18-4 was removed from the remaining analyses because it was the only transgenic event that did not successfully silence MYO.

Table 1.
qPCR results for different transgenic lines. Expression is normalized by house-keeping gene MEP and calibrated with Viking genotype.
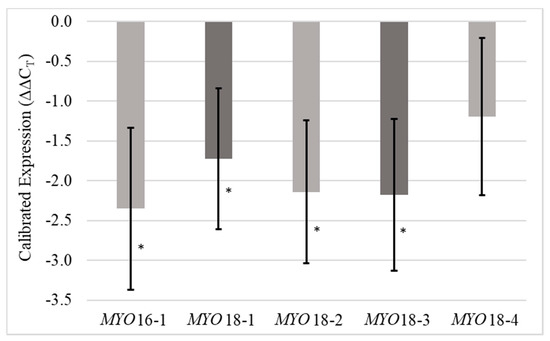
Figure 1.
Transgenic mRNA expression normalized to the expression of the house-keeping gene MEP and calibrated to the expression of these genes within the wild type (Viking). Error bars represent standard errors. Stars represent significance at the 0.05 level.
2.3. Haploid Induction Rate
Since qhir1 is required for haploid induction [1,5,6], it was necessary to evaluate the effect of the transgene (MYO) in the presence of qhir1. Isogenic lines, with or without MYO (RNAi transgene), were created within a qhir1-containing B73 genetic background and are denoted as qhir1,MYOBC2F1 and qhir1BC2F1, respectively. As controls, lines with similar B73 genetic background were created that contained: (1) MYO, denoted as MYOBC1F1, (2) qhir1 and qhir8, qhir1,qhir8B73, and (3) qhir8, qhir8B73.
Consistent with expectations [1,5,6], qhir1 was necessary to achieve higher values of HIR. Three out of the four MYOBC1F1 families were not significantly different from the spontaneous HIR of 0.1% (p-values ranged 0.03 to 0.19; Table 2). Pooled data from MYOBC1F1 families, however, did show significantly different HIR compared to spontaneous rates which was likely resulting from a larger data set and lower error rates (p = 0.0051). Pooled HIR data from qhir1BC2F1 and qhir,1MYOBC2F1 were both significantly higher than spontaneous rates as well, both with p-values of <0.0001. Both controls, qhir8B73 and qhir1,qhir8B73, also had significantly higher HIR than spontaneous rates with p-values of 0.0007 and 0.0016, respectively. Although significantly different compared to spontaneous rates, qhir8B73 HIR rates were still very low. qhir8B73, having an HIR of 0.24% (±0.07%), produced lower HIR than qhir1,qhir8B73, at 1.16% (±0.17%), and was most comparable to MYOBC1F1 events, which averaged 0.17% for those that were successfully silencing MYO. When comparing MYOBC1F1 events to qhir8B73, only one family showed significant differences with p-values of 0.03, 0.17, 0.10, and 0.19 for MYO16-1, MYO18-1, MYO18-2, and MYO18-3, respectively. qhir1,MYOBC2F1 families scored the highest HIR, averaging 1.53% for those that were successfully silencing MYO. In comparison, the isogenic qhir1BC2F1 families within these events typically scored lower HIR and averaged 0.96%. When contrasting HIR results from the pooled data of all isogenic families, qhir1,MYOBC2F1 was significantly higher than qhir1BC2F1 (p = 0.02). On average, the addition of MYO transgene in a qhir1 containing background increased HIR values by 0.6%. In addition, the pooled data of qhir1,MYOBC2F1 families were not significantly different than qhir1,qhir8B73 (p = 0.27). None of the families, however, displayed significant increases in HIR when comparing qhir1,MYOBC2F1 to qhir1BC2F1 when analyzed individually. Contrast statements for individual families resulted in p-values of 0.73, 0.14, 0.19, and 0.17 for MYO16-1, MYO18-1, MYO18-2, and MYO18-3, respectively.

Table 2.
Mean values and standard errors for HIR are reported along with the p-values, which indicate significance when compared to spontaneous HIR of 0.1% (* at the 0.05 level and ** at the 0.01 level).
2.4. Male Fertility
Significant differences were found between events, between differing male parents, and for event and male parent interactions with p-values of 0.0001, <0.0001, and 0.0001, respectively (Table 3). There were no significant differences found between events when B73 was the male parent (p = 0.4412). Therefore, survival rates were averaged between events and resulted in 47.45% (±1.16%) of offspring carrying the transgene and surviving the herbicide treatment, which was not significantly different than the expected 1:1 segregation ratio of the transgene (p-values ranging from 0.27 to 0.96). When MYOBC1F1 was the male parent, the portion of offspring carrying the transgene significantly varied between events (p < 0.0001) and ranged from 0% (±1.77%) to 20.11% (±2.97%) (Table 3, Figure 2. Interestingly, the transgenic event with the most efficient silencing effect (MYO16-1) also resulted in the fewest transgenic offspring when MYOBC1F1 was the male parent at 0% (±1.77%). With increasing silencing of MYO, there was a corresponding increase in HIR, but also an adverse effect of decreased male fertility (Figure S1).

Table 3.
Average offspring survival rate after herbicidal treatment, along with standard errors. p-values are also provided for transgenic events, female parent, and their interaction.
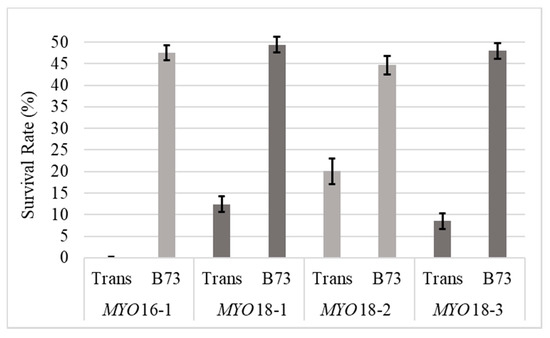
Figure 2.
Survival rate of offspring when either MYOF1 (Trans) or B73 were the male parent. All transgenic events were hemizygous for the transgene; therefore, the expected carry over would be 50%. Error bars represent standard errors.
3. Discussion
MYO is a strong candidate for the gene underlying qhir8, increasing HIR, because it is expressed specifically in anthers and was shown to be up-regulated in wild-type pollen with MATRILINEAL (MTL), a gene shown to substantially impact HIR and located in qhir1 [8]. Sequencing of the inducer allele of MYO revealed various polymorphic regions affecting the resulting protein. Other polymorphic regions may exist upstream of the gene region sequenced for this study. The frameshift mutation identified in the RWS allele would result in an early stop codon, if the upstream portion of this gene is intact. In addition, a large deletion of 20 amino acids within exons 22 and 23 was detected in RWS relative to B73. This mutation would likely be disruptive to the ability of MYO to bind and carry the twin sperm cells, because the truncated protein would be missing the entire cargo binding domain. However, because the motor domain is still present, the elongation of the pollen tube may still happen normally to deliver an “empty” cargo load.
Analysis of HIR among MYO-silencing RNAi transgenic families resulted in induction rates of at most 2.0%, which is a very low induction rate considering HIR for inducers exceed 8% [2]. These results were expected, however, because selection with each successive backcross event was focused on the presence of two QTL regions (qhir1, qhir8) and not for increased HIR, which involves additional QTL. Specifically, with each backcross, offspring were selected for qhir1 and/or qhir8 presence by molecular markers. Similar results were obtained by Gilles et al. [9], where MTL from inducer genotype PK6 was introgressed over four generations into a non-inducing line, and induction rates fell from 3.59% for BC0S1 to 0.5% for BC3S1. These inevitable consequences, due to selection on specific QTL regions, likely result from the absence of unknown QTL regions [1]. In addition, the QTL regions qhir1 and qhir8, as well as the transgene MYO were evaluated in the heterozygous or hemizygous state, which was necessary to ensure comparability of segregation patterns between groups. Pollen with induction ability was, therefore, diluted by one half. If two unlinked genetic components were involved, then pollen containing both was diluted even further to one quarter. Considering the comparison of most interest, qhir1,MYOBC2F1 vs. qhir1BC2F1, only half of the pollen produced from either possesses induction ability mediated by qhir1. In qhir1,MYOBC2F1, one quarter of the pollen will contain MYO only (theoretically having no effect), another quarter will contain qhir1 only (contributing partially to the HIR), and finally another quarter will have both MYO and qhir1 (MYO enhancing haploid induction of qhir1).
Results from HIR analysis show that there are two lines of evidence suggesting that MYO may be responsible for or contribute to qhir8: (1) the majority of MYOBC1F1 families were not significantly different from qhir8B73 while similarly qhir1,MYOBC2F1 were not significantly different than qhir1,qhir8B73, and (2) qhir1,MYOBC2F1 had significantly higher HIR than qhir1BC2F1. Although most MYOBC1F1 families had no significant differences in HIR when compared to spontaneous rates, it is well known that qhir1 is considered to be required by inducers [1,5,6]. Our results support these earlier findings that qhir8 depends on the presence of qhir1 to increase HIR. Thus, the response from qhir1,MYOBC2F1 compared to qhir1BC2F1 was of most interest. Although none of the transgenic families showed significant increases from comparing qhir1,MYOBC2F1 to qhir1BC2F1 when analyzed individually, pooled data resulted in significant differences. In addition, these families, including MYO16-1, MYO18-1, MYO18-2, and MYO18-3 exhibited similar or enhanced HIR with the addition of the RNAi construct in the qhir1 genetic background. This suggests that mutations of this gene in the inducer genotype may be responsible for enhancing HIR by qhir8 in the presence of qhir1.
However, recent findings by Zhong et al. [11] uncovered similar results for gene GRMZM2G465053 (ZmDMP). ZmDMP was initially located by narrowing the fine-mapping region of qhir8 from a 789 kb region [5] to a 138 kb region. ZmDMP falls within the narrowed region, however, MYO is located approximately 8.5 kb from the boundaries of this region. Zhong et al. [11] found that a knock-out of this gene resulted in a 5–6-fold increase in haploid induction in the presence of qhir1. Additionally, they found that when pollinated by the homozygous knock-out of zmdmp there was an increase in endosperm aborted kernels of 10–40%. However, there were no significant differences in pollen viability.
By silencing MYO, male fertility was affected, as shown by the significantly different percentages of transgenic offspring resulting from reciprocal crosses between transgenic MYOF1 and B73. All families exhibited a substantial and significant decrease in male fertility when MYOF1 was the male parent. The most affected family was MYO16-1, resulting in 0% transfer of the transgene when MYOF1 was the male parent, which complements the finding that MYO16-1 was also most effective in silencing MYO. When B73 is the male parent, the transgene was consistently transferred to approximately half of the offspring.
Considering the expected functions in pollen tube growth of the gene MYO, it is not surprising to see some impairment of fertility. It is therefore conceivable that the gene is not completely knocked out in the inducer genome. A complete knockout would likely arrest pollen tube growth, and therefore would result in a male sterile plant. It would be very unlikely that any seed would be produced when pollen tube growth is arrested.
To produce viable seed, including haploids, the pollen tube must reach the ovary to at least fertilize the central nucleate cell to create the endosperm. In that case, if MYO is involved with haploid induction, it seems plausible that the mutations of MYO within inducer genotype causes only impaired function of the protein. Together with the recent findings from Zhong et al. [11], it seems plausible that the silencing of MYO increased haploid induction by chance. For instance, since qhir1,MYOBC2F1 and qhir1BC2F1 were all maintained as heterozygotes, both qhir1 and the silencing of MYO would theoretically equally affect half of the pollen. However, for qhir1,MYOBC2F1, the MYO RNAi cassette would cause variable amounts of pollen containing qhir1 (assuming the RNAi cassette was inserted into a chromosome other than the qhir1-containg chromosome). Due to both crossing over and independent assortment, pollen containing qhir1 may range anywhere from 0% to 100% (Figure 3), which could explain our results. Alternatively, qhir8 may be comprised of two closely linked genes, both contributing to HIR. MYO and ZmDMP are closely linked, considering there is only 8.5 kb separating both genes. If both ZmDMP and MYO affect HIR, it would increase the chance for this genome region to be detected as QTL and could explain its consistent detection as major QTL. Similarly, an additional region (qhir12) within qhir1 on chromosome 1 is located 985 kb upstream of MTL and was found to affect HIR in two independent GWAS studies [6,16].
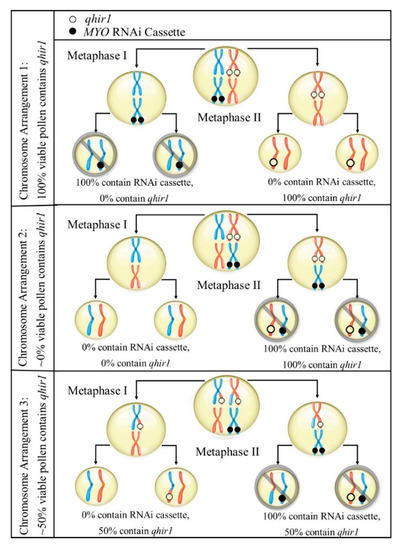
Figure 3.
Representation of some potential chromosome arrangements during microsporocyte meiosis within qhir1,MYOBC2F1 families. As heterozygotes, crossing over and independent assortment would affect arrangements of both qhir1 and the RNAi cassette (silencing MYO). The dispersal of MYO RNAi cassette may produce variable amounts of viable pollen containing qhir1 (assuming the RNAi cassette causes sterility).
Further studies may need to confirm or refute MYO’s correlation with HIR; however, results clearly establish the connection between MYO and male fertility/sterility, which has not been previously reported. Past studies have shown that there are many different myosin classes within plants, that all may have unique functions, especially considering many have specific localized expression [17]. Since MYO is expressed exclusively in the pollen, the function was expected to be related to male fertility; however, this is the first publication clearly establishing an association.
4. Materials and Methods
4.1. Identification of Gene of Interest
GRMZM2G435294 or Myosin-11 (MYO) was considered as a candidate gene for qhir8 because it is specifically expressed in anthers [18,19], co-expressed with MTL [8], and is located within the fine mapped region of qhir8 [5]. MYO was amplified from the haploid inducer genotype RWS/RWK-76 [20] and cloned into the pGEM-T vector system (Promega, Fitchburg, WI, USA) for sequencing and alignment with the B73 reference genome. A 1059 bp region downstream of the start of exon 19 was sequenced and aligned to B73 RefGen_v3 [15] and analyzed for polymorphic regions (Table S1).
4.2. Production of Transgenic Materials
Silencing of MYO was accomplished by delivering transgenic RNA interference (RNAi) constructs. The target sequences were designed using pssRNAit software [21]. The best siRNA options were chosen based on the most efficient silencing effect, the fewest off-target hits, as well as their location within the gene. Two target regions were chosen within exon 16 (126 bp) and exon 18 (96 bp), and designated as MYO16 and MYO18, respectively (Table S2). Both constructs were expected to accomplish the same goal of silencing MYO and are therefore considered equivalent, if qPCR results show similar silencing levels.
Corresponding RNAi vectors were constructed for creating transgenic events. Primers containing appropriate flanking restriction sites (Table S2) were used to amplify a fragment of MYO to be inserted in sense (SpeI and SmaI) and anti-sense orientations (BstZ17I and AvrII) within the pMCG1005 RNAi binary vector (obtained from the Plant Transformation Facility at ISU). This vector contains the first intron of maize alcohol dehydrogenase-1 (AdhI) upstream of the RNA hairpin-producing sequence, which is driven by the maize ubiquitin1 (ubi1) promotor. This vector also contains four copies of an enhanced CaMV 35S promoter that drives the bar gene. After these vectors were transformed via Agrobacterium into maize genotype HiII by the Plant Transformation Facility at ISU using the protocol described by Frame et al. [22], transgenic plants were identified based on glufosinate resistance. A total of 12 transgenic events were produced for each construct, but only 5 transgenic events (one for MYO16 and four for MYO18) are included in this study. These events were chosen for no other reason than early availability.
4.3. Development of Control Lines
Because qhir1 is required for haploid induction [1,5,6] and MYO is evaluated as a candidate for qhir8, it was necessary to evaluate the effect of the transgene in the presence of qhir1. qhir1-containing materials were created by crossing RWS/RWK-76 [20] with B73 and then backcrossing with B73 until reaching the BC2 generation. Simple sequence repeat (SSR) markers were designed to flank qhir1 (1.04_682414 and GSS_44), and public markers that flank qhir8 (umc1040 and bnlg1272) were used to select both QTL regions (Table S3). In each backcross generation, individuals were selected for presence of qhir1 and absence of qhir8 (qhir1B73) by marker analyses. Additional control lines were developed by selection for presence of qhir8 and absence of qhir1 (qhir8B73) or presence of both QTL, qhir1 and qhir8 (qhir,1qhir8B73). Finally, BC2F2–derived lines were created to fix qhir1 and/or qhir8, respectively.
4.4. Development of Transgenic Families
At the T0 stage, all events were crossed and then backcrossed to B73 (MSG 14786) [23] (MYOF1 and MYOBC1F1, respectively). Transgenic F1 offspring were selected by QualiPlate ELISA kit for the detection of PAT/bar (EnviroLogix, Portland, ME, USA), resulting in transgenic BC1 families segregating for hemizygous presence of the RNAi construct within the B73 genetic background.
Each of the five transgenic BC1 families were crossed with qhir1B73. BC1 materials were screened and selected for the presence of the transgene before crossing with qhir1B73. Finally, the resulting BC2 materials were screened and classified into those containing (qhir1,MYOBC2F1) or not containing (qhir1BC2F1) the MYO construct. Therefore, MYO and qhir1 are heterozygous in these lines. Both BC1 and BC2 offspring were screened for transgene presence with QualiPlate ELISA kit for the detection of PAT/bar (EnviroLogix, Portland, ME, USA) (Figure 4a). Controls included qhir8B73 (BC2F1), qhir1,qhir8B73 (BC2F1), and the MYOBC1F1 families. Prior to transplanting MYOBC1F1 families, the presence of the transgene was identified by spraying 0.5% glufosinate (190 L/ha).
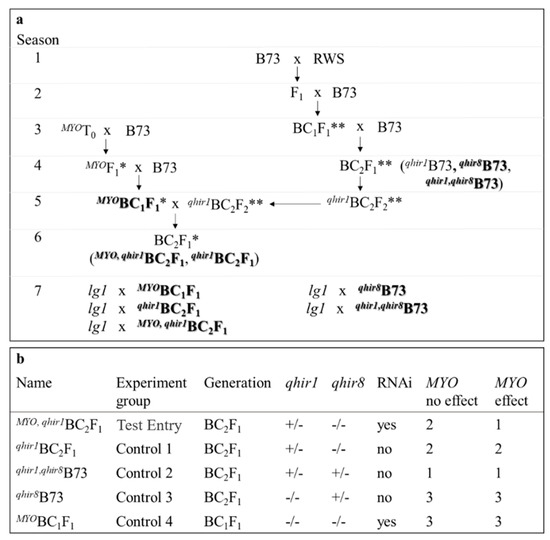
Figure 4.
Schematic development of transgenic families and control groups (a). Bolded families are those developed for the test-cross with lg1 (Season 7). Noted, are methods of selecting transgenic plants (ELISA PAT/bar; *) and those with desired QTL regions (marker selected for qhir1 and/or qhir8; **). A summary of details regarding each group tested in HIR is also provided (b). Expected results are specified if MYO is not responsible for qhir8 (MYO no effect) or if MYO is responsible for qhir8 (MYO effect). Expected outcomes are numbered, with “1” noted for the highest HIR levels and “3” for the lowest HIR levels. Equal numbers imply that HIR values are comparable between the two experimental groups.
4.5. Test Crossing and Evaluation of HIR
To analyze HIR, testcrosses were made between control inducers or potential inducers (male) and inbred line lg1 (female) (205B, MGS 14013) in the summer of 2017 in Ames, IA. The line lg1 contains B73 background and is homozygous recessive for ligule presence. Therefore, any haploids created by testcrosses can be identified by the lack of ligules.
4.6. Experimental Design and Statistical Analysis for HIR
This experiment was arranged in a modified incomplete block design including controls qhir8B73 and qhir1,qhir8B73, which were repeated in three biological replicates within each of three blocks to account for environmental variation between blocks. Transgenic families were included in two to four biological replicates within each block. Each replicate contained 10 plants, and when ≥5 plants were pollen shedding, pollen was bulked and used to randomly pollinate 3–6 lg1 plants. The female parent carrying lg1, was planted in a large area beside the crossing block. Subsequently, an average of 375 offspring were grown out for each biological replicate of each genotype in the greenhouse for analysis of ligule presence at the two to three leaf stage. All replicate data from the same control or transgenic family were combined within each incomplete block.
Haploid induction rates were compared between families or to the reference value for the spontaneous HIR rate (0.1% or 1 haploid in every 1000 seedlings) [24,25] by proc LOGISTICS in SAS 9.4 (SAS Institute Inc.). The following model was used for variance analysis:
where (nh/nt)ijk represents the observation of the ijth experimental unit, or haploid induction rate (number of haploid plants (nh) divided by the total number of plants (nt)). Main effects are represented by Bi or the ith block, and Fj or the ith family. The error term is defined as εij. The significance test was performed using Walds chi-square test.
(nh/nt)ij = µ + Bi + Fj + εij
The mean HIR for each family was computed with proc GLIMMIX in SAS 9.4 (SAS Institute Inc.). The following model was used for variance analysis:
where (nh/nt)ijk represents the observation of the ijkth experimental unit, or haploid induction rate (number of haploid plants (nh) divided by the total number of plants (nt)). The main effects are represented by Bi or the ith block, Fj or the ith family, and Ck or the kth cross type (stage of backcrossing). The error term is defined as εijk.
(nh/nt)ijk = µ + Bi + Fj + Ck + FCjk + εijk
To account for the potential contamination or accidental self-pollination of lg1 donor plants, pollination sources that were three times the standard deviation for any cross combination were considered outliers and removed [26]. Data from one pollination for each of the two genotypes MYO16-1 and MYO18-2 (both qhir1BC2F1) were removed from the data set.
4.7. Male Fertility Analysis
Reciprocal crosses were made between four hemizygous F1 plants (MYOF1) of each transgenic event and randomly selected B73 plants in the summer of 2016. Offspring from these reciprocal crosses were grown out in trays of 126 seeds per cross (4 crosses × 126 seeds = 504 seedlings analyzed in total for each event). Seedlings were sprayed with 0.5% glufosinate (190 L/ha) at the 2–3 leaf stage, and after 3–4 days the effects were visible enough to count survival rates. Data were analyzed by proc MIXED in SAS 9.4. Accuracy of the test was evaluated by a chi-square test with an expected segregation ratio of 1:1 when B73 was the male parent. This was performed using proc LOGISTICS in SAS 9.4 (SAS Institute Inc., Cary, NC, USA).
4.8. Silencing Efficiency by Quantitative PCR
Expression analysis by qPCR was performed using the ΔΔCT method [27]. For this method, the expression of transgenic events was compared to both a calibrator (wild type, i.e., Viking brand 60-01 by Albert Lea Seed) and a normalizer (housekeeping gene, i.e., MEP, Membrane protein PB1A10.07c) with the following equations:
where CT represents the designated sample’s cycle threshold, or the number of cycles required for the fluorescent signal to exceed the threshold level. Normalized expression is the expression after considering the house-keeping gene expression levels and is represented as ΔCT. Calibrated expression is the expression after considering expression levels for both the house-keeping gene and the reference genotype and is represented as ΔΔCT. Relative expression is the fold change in MYO expression between the genotype of interest and the reference genotype. Five transgenic (MYOBC1F1) families, segregating for the transgene, were grown in the greenhouse until the 2–3 leaf stage, sprayed with 0.5% glufosinate (190 L/ha) and surviving seedlings were transplanted to the field along with the Viking genotype. At approximately V10 stage, anthers were harvested for qPCR analysis. Three plants of appropriate size were harvested from Viking and from each transgenic family. Plant stalks were severed between nodes just above the ear shoots and stored in buckets of water, while being transported to the laboratory for anther dissection [28]. Since the upper anthers mature more quickly than the lower anthers, dissection was performed to harvest only the upper anthers for RNA extraction and qPCR analysis. Approximately 50–100 mg of dissected anthers, which ranged from 4–5 mm (binucleate microspore stage), were placed in a microfuge tube resting in liquid nitrogen. Samples were stored in −80 °C prior to grinding in liquid nitrogen for subsequent RNA extraction. RNA was purified using the RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) with on-column DNase digestion by following manufacturer’s instructions. RNA concentrations were measured by a NanoDrop 2000c Spectrophotometer prior to Reverse transcription-PCR (RT-PCR) reactions. RT-PCR was performed with SuperScriptTM III First-Strand Synthesis System (Invitrogen by life technologies, Carlsbad, CA, USA) using oligo(dT)20 primers. From the final 20 µl reaction containing cDNA, 1 µl was used for further qPCR reactions (for a target cDNA concentration of 10 ng). Reactions for qPCR were performed using Power SYBRTM Green PCR master mix (Applied Biosystems by life technologies, Foster City, CA, USA) and gene specific primers (Table S2) on a Mx3000P qPCR system. The primer pair chosen for MYO bridged an intronic region (81 bp) between exon 6 and 7 to further ensure the lack of DNA contamination during qPCR analysis. The primer pair utilized for MEP, the reference gene of choice, was published by Manoli et al. [29]. Gene-specific primers were optimized by conducting qPCR on a serial dilution (1 ng, 10 ng, 100 ng) with primer pairs for MYO and MEP. Primer optimization was performed by analyzing the linear trend of the difference between the expression of the MYO and MEP (ΔCT) as well as the analysis of standard curves for each individual primer pair. R2 values close to 1 indicate high amplification efficiency. The ΔCT curve produced an R2 value of 0.95, whereas both individual primer pairs for MYO and MEP scored R2 values of 0.99. A three-step PCR program of 95 °C (15 s), 55 °C (30 s), and 68 °C (30 s) for 40 cycles was used for both primer pairs with a 25 uL reaction size. There were three biological and three technical replicates for each genotype by primer pair combination. Statistical comparisons were performed in R using the ‘pcr’ package.
ΔCT = CT,MYO − CT,MEP
ΔΔCT = ΔCT,trans − ΔCT,Viking
relative expression = 2−ΔΔCT
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/6/773/s1, Figure S1: Correlations between relative expression (fold change) of MYO transgenic events and (a) survival rates after spraying a herbicidal treatment for transgene selection (data for B73 × MYOF1 crosses), or (b) haploid induction rates (% haploids found for qhir1,MYOBC2F1), Table S1: Protein alignment of B73 RefGen_v3 [15] with RWS. Sequenced region is shaded in grey, and remaining gaps were assumed to match reference sequence. Highlighted sequences indicate polymorphisms, Table S2: Sequence information for RNAi targets and primers, Table S3: Primers used for SSR marker analysis and the presence or absence of QTL regions. Relevant information provided here is based on B73 RefGen_v4 [30].
Author Contributions
K.V. carried out the majority of the experiment and wrote the manuscript with support from T.L. and U.K.F.; R.I. performed candidate gene sequencing. U.K.F. and T.L. conceived the original idea and supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by U.S. Department of Agriculture: IOW04714; and by U.S. Department of Agriculture: IOW01018.
Acknowledgments
Authors would like to thank USDA’s National Institute of Food and Agriculture (Project Numbers: IOW04714, IOW01018), as well as the RF Baker Center for Plant Breeding, the K.J. Frey Chair in Agronomy, and the Plant Sciences Institute Scholar program at Iowa State University for support this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prigge, V.; Xu, X.W.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 2012, 190, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Ren, J.; Mei, M.; Frei, U.K.; Trampe, B.; Lübberstedt, T.; Goldman, I.L.; Ortiz, R. Maize doubled haploids. In Plant Breeding Reviews; Janick, J., Ed.; Wiley Blackwell: Hoboken, NJ, USA, 2016; Volume 40, pp. 123–160. [Google Scholar]
- Deimling, S.; Röber, F.K.; Geiger, H.H. Methodology and genetics of in vivo haploid induction in maize. Vortr. Pflanzenzüchtg 1997, 38, 203–224. [Google Scholar]
- Barret, P.; Brinkmann, M.; Beckert, M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 2008, 117, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, W.; Zhong, Y.; Dong, X.; Hu, H.; Tian, X.; Wang, L.; Chen, B.; Chen, C.; Melchinger, A.E.; et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor. Appl. Genet. 2015, 128, 2507–2515. [Google Scholar] [CrossRef]
- Hu, H.; Schrag, T.A.; Peis, R.; Unterseer, S.; Schipprack, W.; Chen, S.; Lai, J.; Yan, J.; Prasanna, B.M.; Nair, S.; et al. The Genetic Basis of Haploid Induction in Maize Identified with a Novel Genome-Wide Association Method. Genetics 2016, 202, 1267–1276. [Google Scholar] [CrossRef]
- Dong, X.; Xu, X.; Miao, J.; Li, L.; Zhang, D.; Mi, X.; Liu, C.; Tian, X.; Melchinger, A.E.; Chen, S.; et al. Fine mapping of qhir1 influencing in vivo haploid induction in maize. Theor. Appl. Genet. 2013, 126, 1713–1720. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. Matrilineal, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Gilles, L.; Khaled, A.; Laffaire, J.B.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef]
- Liu, C.X.; Li, X.; Meng, D.X.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Peremyslov, V.V.; Morgun, E.A.; Kurth, E.G.; Makarova, K.S.; Koonin, E.V.; Dolja, V.V. Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 2013, 25, 3022–3038. [Google Scholar] [CrossRef] [PubMed]
- Shimmen, T. The sliding theory of cytoplasmic streaming: Fifty years of progress. J. Plant Res. 2007, 120, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar]
- Trentin, H.U. Study and Improvement of Maize Maternal Haploid Inducers (Doctoral Dissertation); Iowa State University: Ames, IA, USA, 2019. [Google Scholar]
- Kinkema, M.; Wang, H.; Schiefelbein, J. Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant Mol. Biol. 1994, 26, 1139–1153. [Google Scholar] [CrossRef][Green Version]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; De Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An expanded maize gene expression atlas based on RNA-sequencing and its use to. Plant Genome 2015, 1–16. [Google Scholar] [CrossRef]
- Röber, F.K.; Gordillo, G.A.; Geiger, H.H. In vivo haploid induction in maize-performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 2005, 50, 275–283. [Google Scholar]
- Dai, X.; Zhao, P.X. pssRNAMiner: A plant short small RNA regulatory cascade analysis server. Nucleic Acids Res. 2008, 36, W114–W118. [Google Scholar] [CrossRef]
- Frame, B.; Main, M.; Schick, R.; Wang, K. Genetic transformation using maize immature zygotic embryos. In Plant Embryo Culture: Methods and Protocols. Methods in Molecular Biology; Thorpe, A., Yeung, E.C., Eds.; Springer Science: New York, NY, USA, 2011; pp. 327–341. [Google Scholar]
- Russel, W.A. Crop Science. Madison, WI. Crop Sci. 1972, 12, 721. [Google Scholar]
- Chase, S.S. Monoploid frequencies in a commercial double cross hybrid maize, and in its component single cross hybrids and inbred lines. Genetics 1949, 34, 328–332. [Google Scholar] [PubMed]
- Chase, S.S. Monoploids in maize. In Heterosis; Gowen, J.W., Ed.; Iowa State College Press: Ames, IA, USA, 1952; pp. 389–399. [Google Scholar]
- Pukelsheim, F. The Three Sigma Rule. Am. Stat. 2012, 48, 88–91. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the Double Delta CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Duncan, D.; Morrow, D.J.; Fernandes, J.; Walbot, V. Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell types. Plant J. 2007, 50, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Manoli, A.; Sturaro, A.; Trevisan, S.; Quaggiotti, S.; Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Phys. 2012, 169, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.-S.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).