Transcriptome Analysis of High-NUE (T29) and Low-NUE (T13) Genotypes Identified Different Responsive Patterns Involved in Nitrogen Stress in Ramie (Boehmeria nivea (L.) Gaudich)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and N Treatment
2.2. RNA Isolation and Library Preparation for DGE Sequencing
2.3. Quality Control and Quantification of Gene Expression Levels
2.4. Gene Annotation
2.5. Differential Expression Analysis
2.6. Pathway Enrichment Analysis of DEGs
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
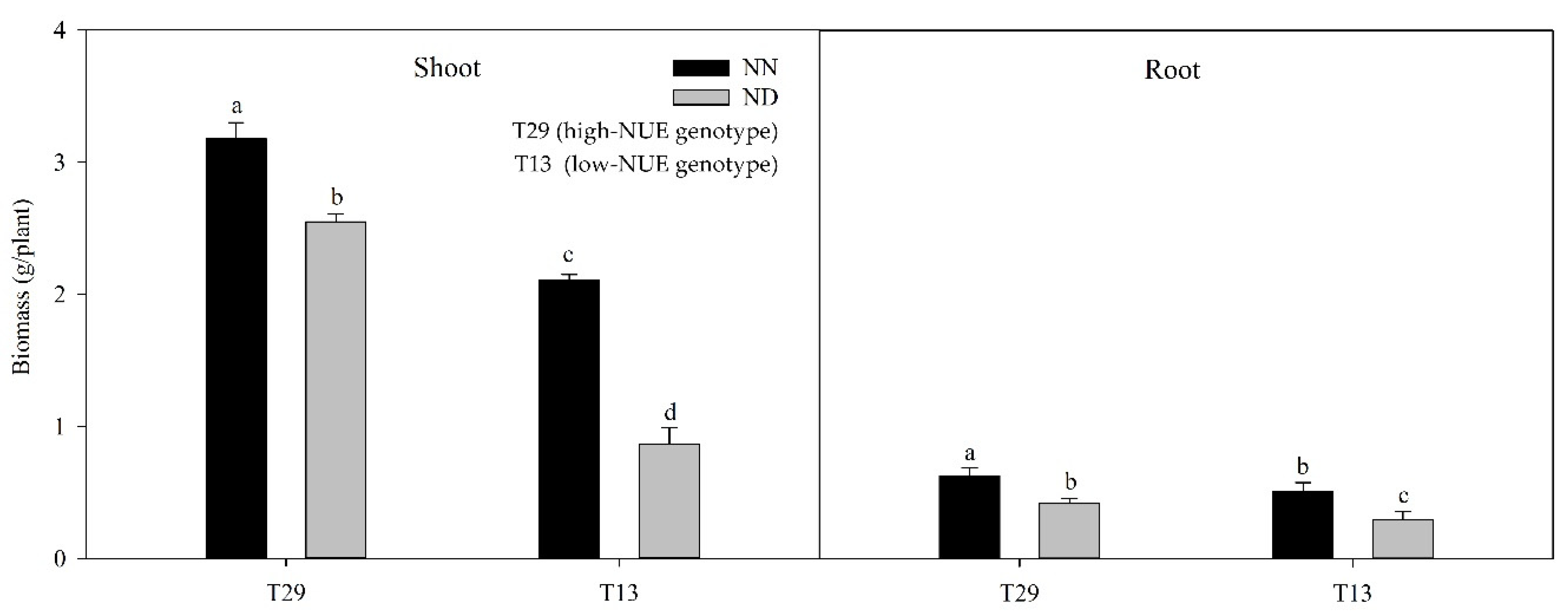
3.1. The Performance of Ramie Genotypes under Different Nitrogen Treatments
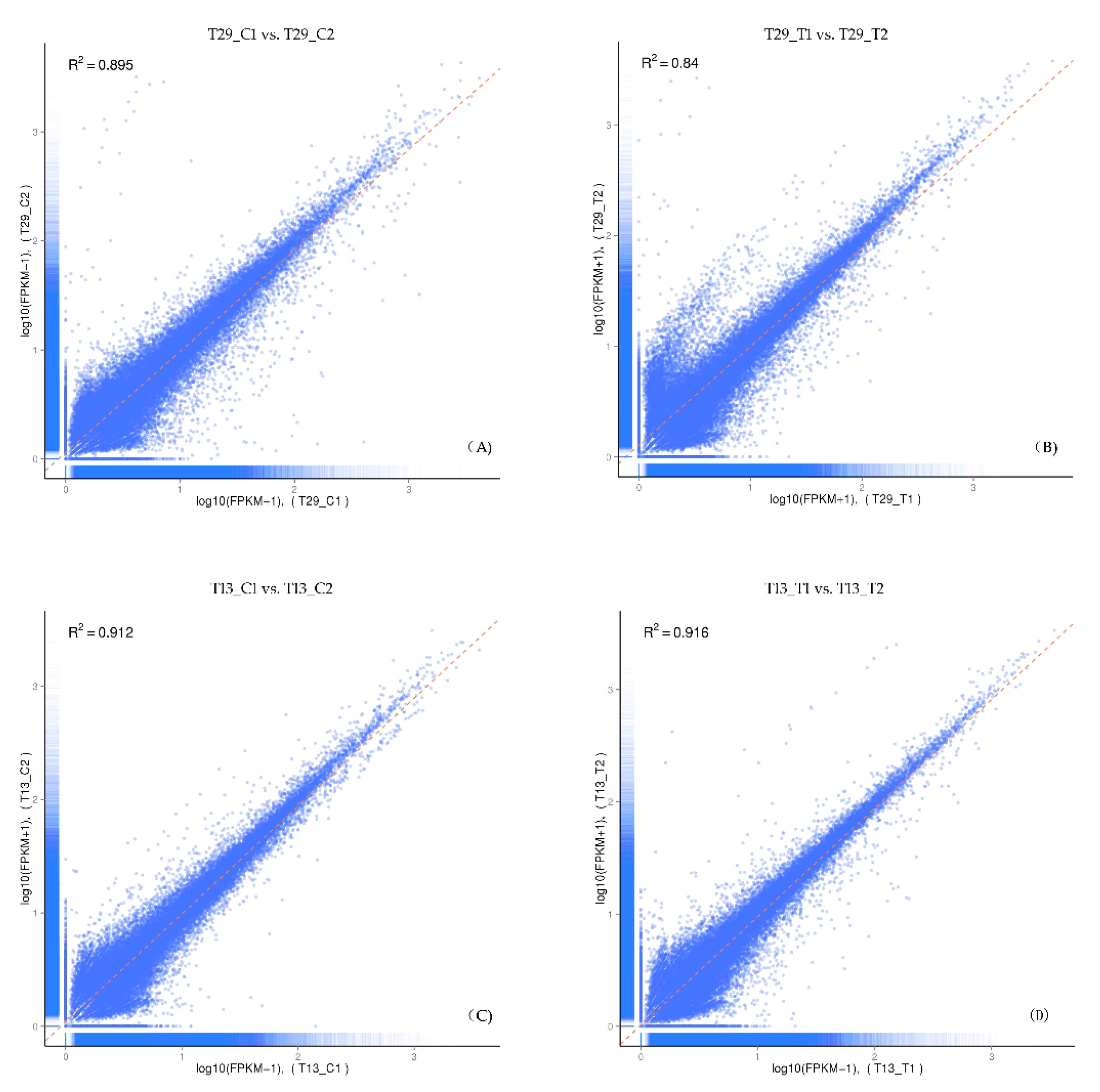
3.2. De Novo Transcriptome Sequencing in High-NUE and Low-NUE Ramie Genotypes
3.3. Global Analysis of Differential Gene Expression
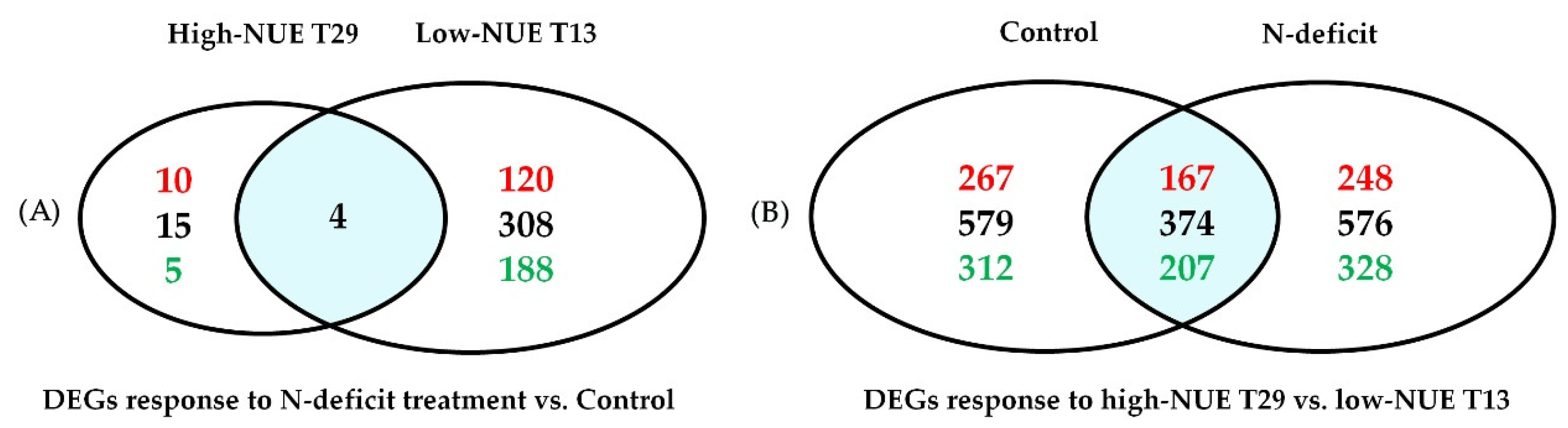
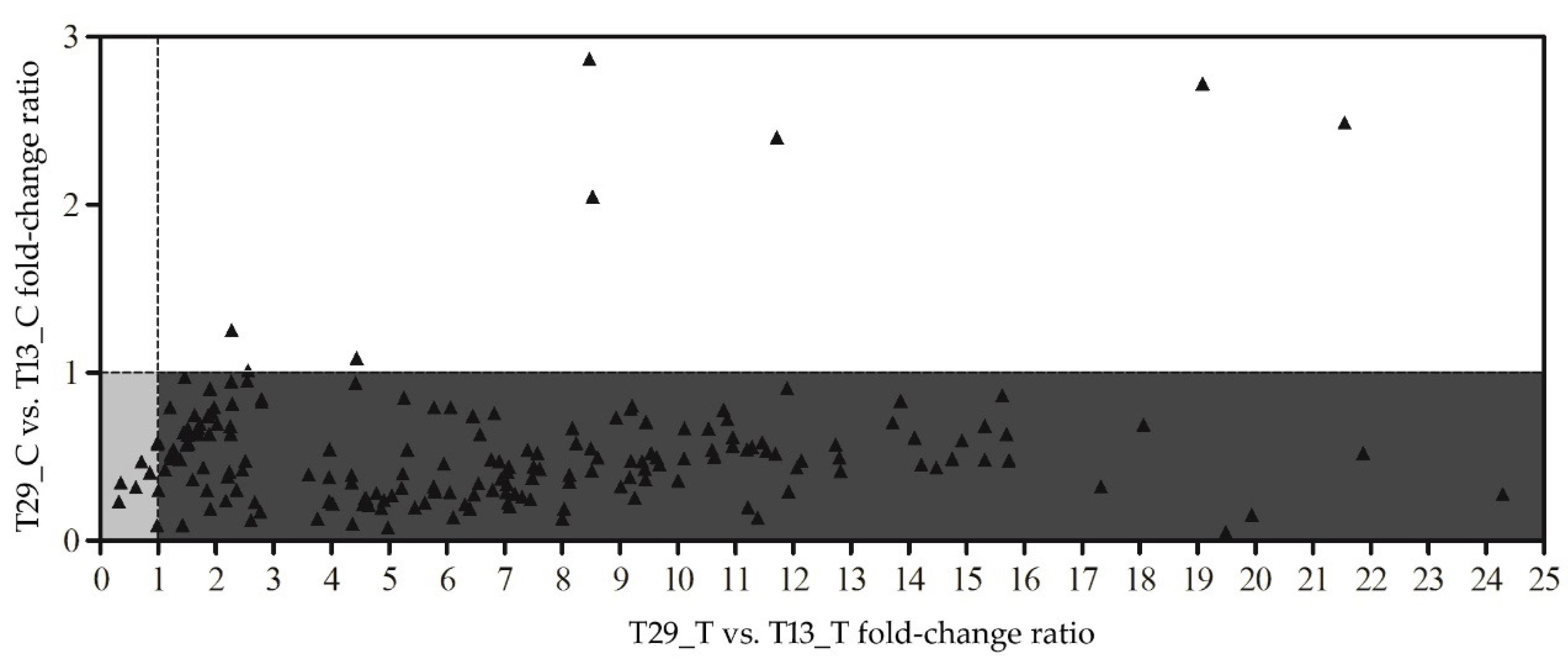
3.4. Genotype-Specific Gene Expression within Treatments
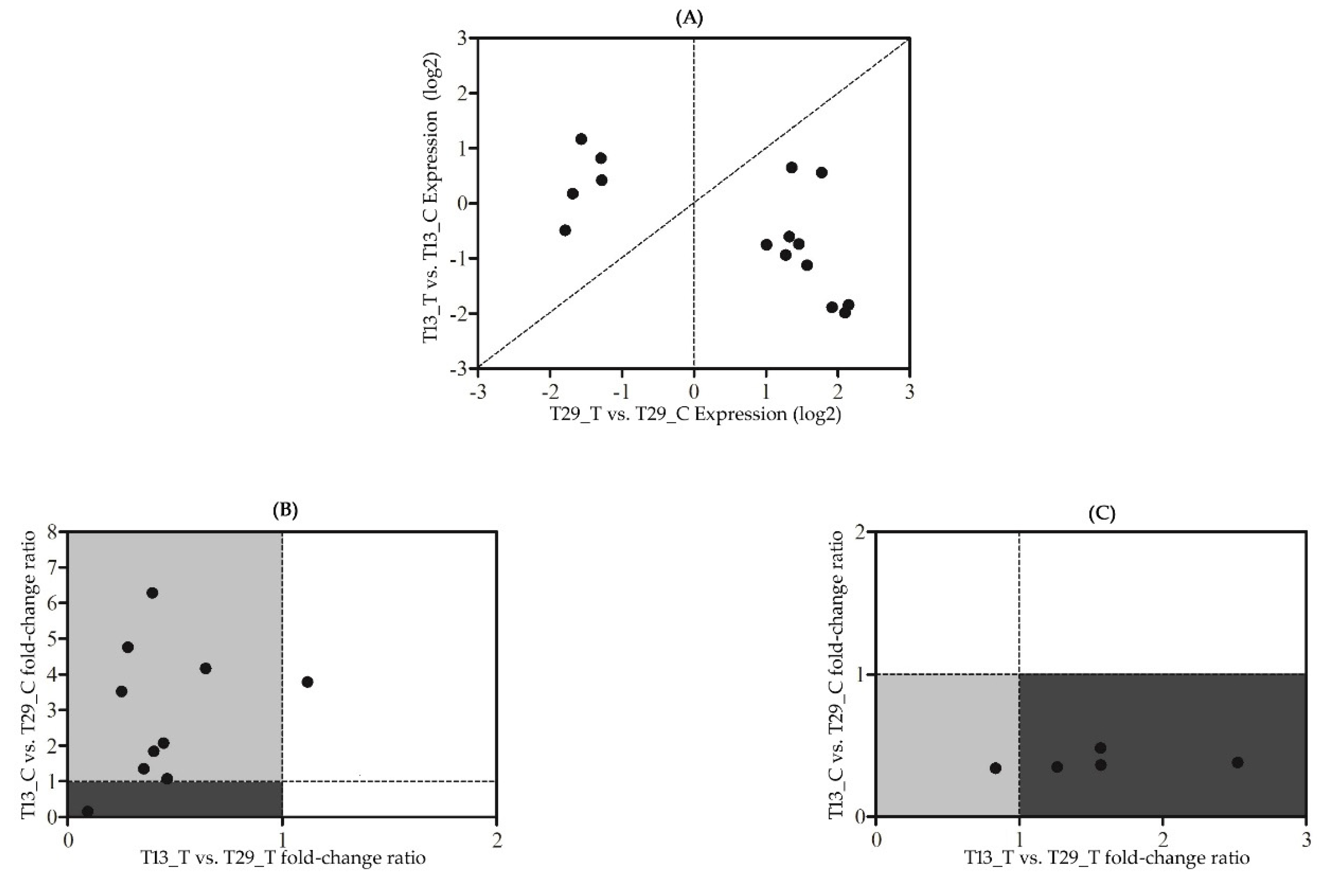
3.5. Variety-Specific Responses to N-Deficit Conditions
3.6. Quantitative Real-Time PCR
4. Discussion
4.1. Feasibility of Gene Expression Pattern in Ramie NUE
4.2. Functional Classification of N-Stress-Responsive Genes in Ramie
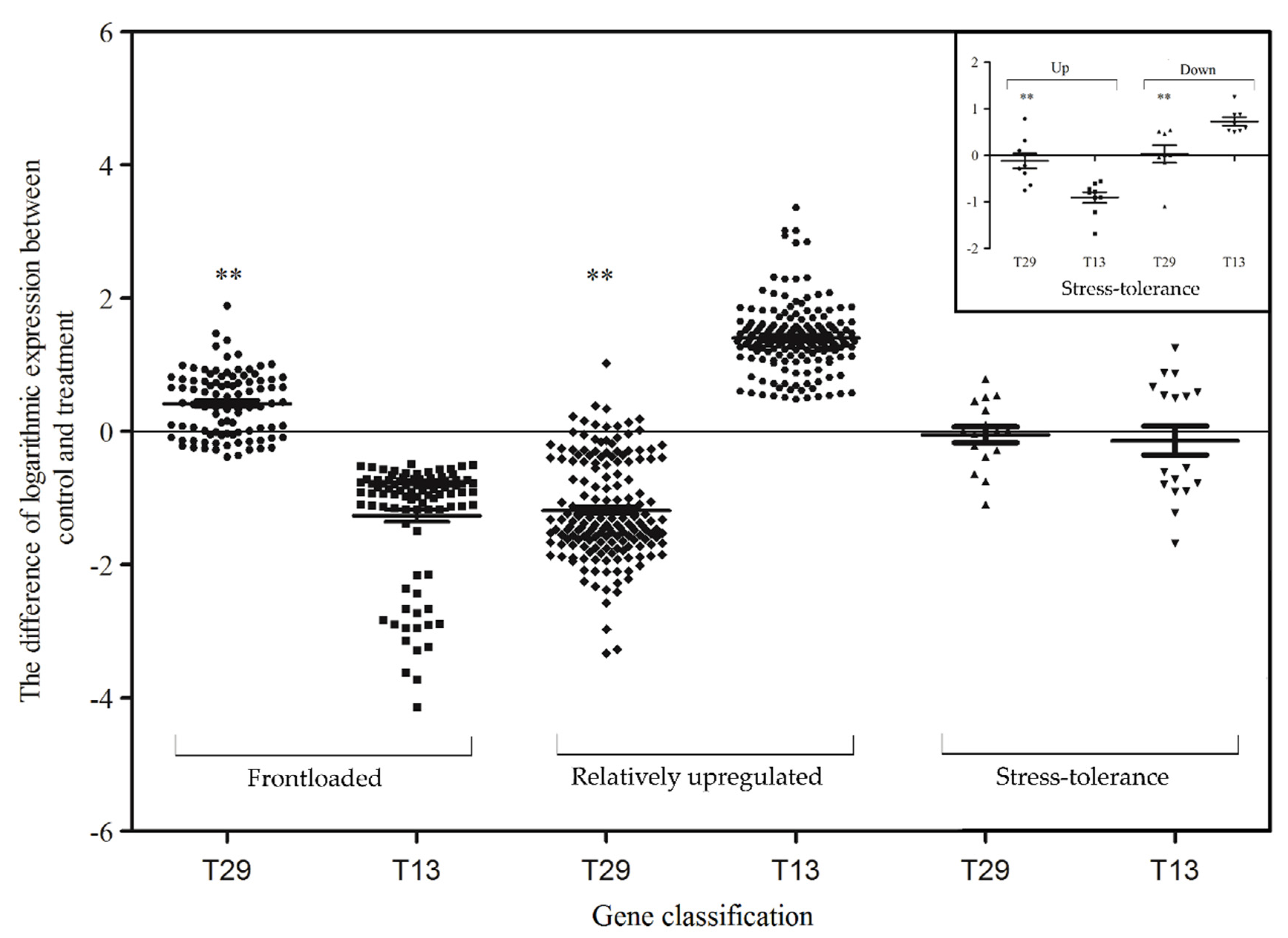
4.3. Frontloaded Genes
4.4. Relatively Upregulated Genes
4.5. Stress-Tolerance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O. Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Davidson, E.A.; Mauzerall, D.L.; Searchinger, T.D.; Dumas, P.; Shen, Y. Managing nitrogen for sustainable development. Nature 2015, 528, 51–59. [Google Scholar] [CrossRef]
- Adbelrahman, M.; Burritt, D.J.; Tran, L.P. The use of metabolomic quantitative trait locus mapping and osmotic adjustment traits for the improvement of crop yields under environmental stresses. Semin. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef]
- Mitra, S.; Saha, S.; Guha, B.; Chakrabarti, K.; Satya, P.; Sharma, A.K.; Gawande, S.P.; Kumar, M.; Saha, M. Ramie: The Strongest Bast Fibre of Nature. Tech. Bull. 2013, 8, 1–38. [Google Scholar] [CrossRef]
- Angelini, L.G.; Tavarini, S. Ramie [Boehmeria nivea (L.) Gaud.] as a potential new fibre crop for the Mediterranean region: Growth, crop yield and fibre quality in a long-term field experiment in Central Italy. Ind. Crop. Prod. 2013, 51, 138–144. [Google Scholar] [CrossRef]
- Kipriotis, E.; Heping, X.; Vafeiadakis, T.; Kiprioti, M.; Alexopoulou, E. Ramie and kenaf as feed crops. Ind. Crop. Prod. 2015, 68, 126–130. [Google Scholar] [CrossRef]
- Luan, M.; Jian, J.; Chen, P.; Chen, J.; Chen, J.; Gao, Q.; Gao, G.; Zhou, J.; Chen, K.; Guang, X.; et al. Draft genome sequence of ramie, Boehmeria nivea (L.) Gaudich. Mol. Ecol. Resour. 2018. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Gong, W.; Yan, L.; Zhu, Z.; Hu, Z.; Peng, Y. Biodegradation of ramie stalk by Flammulina velutipes: Mushroom production and substrate utilization. AMB Express 2017, 7, 171. [Google Scholar] [CrossRef]
- Jose, S.; Tajna, S.; Ghosh, P. Ramie Fibre Processing and Value Addition. Asian J. Text. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Angelini, L.G.; Lazzeri, A.; Levita, G.; Fontanelli, D.; Bozzi, C. Ramie (Boehmeria nivea (L.) Gaud.) and Spanish Broom (Spartium junceum L.) fibres for composite materials: Agronomical aspects, morphology and mechanical properties. Ind. Crop. Prod. 2000, 11, 145–161. [Google Scholar] [CrossRef]
- Özgür, T.; Emre, I.; Aykut, T.F.; Hamdi, A.; Önder, C. Impact of Different Nitrogen and Potassium Application on Yield and Fiber Quality of Ramie (Boehmeria nivea). Int. J. Agri. Biol. 2010, 12, 369–372. [Google Scholar]
- Subandi, M. The Effect of Fertilizers on the Growth and the Yield of Ramie (Boehmeria nivea L. Gaud). Asian Econ. Soc. Soc. 2012, 2, 126. [Google Scholar]
- Raun, W.R.; Johnson, G.V. Improving nitrogen use efficiency for cereal production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef]
- Stanton, M.L.; Roy, B.A.; Thiede, D.A. Evolution in stressful environments. I. Phenotypic variability, phenotypic selection, and response to selection in five distinct environmental stresses. Evolution 2000, 54, 93–111. [Google Scholar] [CrossRef]
- Huang, C.; Wei, G.; Luo, Z.; Xu, J.; Zhao, S.; Wang, L.; Jie, Y. Effects of nitrogen on ramie (Boehmeria nivea) hybrid and its parents grown under field conditions. J. Agr. Sci. (Tor.) 2014, 6, 230–243. [Google Scholar] [CrossRef]
- Cabangbang, R.P. Fiber yield and agronomic characters of ramie as affected by plant density and fertilizer level. Philipp. J. Crop. Sci. 1978, 3, 78–89. [Google Scholar]
- Wang, R.; Guegler, K.; LaBrie, S.T.; Crawford, N.M. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 2000, 12, 1491–1509. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Wang, S.; Zhang, J.; Feng, Q.; Zhang, L.; Fan, D.; Li, X.; Yuan, D.; Han, B.; Zhang, Q. Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol. Biol. 2006, 60, 617–631. [Google Scholar] [CrossRef]
- Bi, Y.M.; Kant, S.; Clark, J.; Gidda, S.; Ming, F.; XU, J.Y.; Rochon, A.; Shelp, B.J.; Hao, L.X.; Zhao, R.; et al. Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ. 2009, 32, 1749–1760. [Google Scholar] [CrossRef]
- Hao, Q.N.; Zhou, X.A.; Sha, A.H.; Wang, C.; Zhou, R.; Chen, S.L. Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genom. 2011, 12, 525. [Google Scholar] [CrossRef]
- Chen, R.; Tian, M.; Wu, X.; Huang, Y. Differential global gene expression changes in response to low nitrogen stress in two maize inbred lines with contrasting low nitrogen tolerance. Genes Genom. 2011, 33, 491–497. [Google Scholar] [CrossRef]
- Deng, G.; Liu, L.J.; Zhong, X.Y.; Lao, C.Y.; Wang, H.Y.; Wang, B.; Zhu, C.; Shah, F.; Peng, D.X. Comparative proteome analysis of the response of ramie under N, P and K deficiency. Planta 2014, 239, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genom. 2014, 15, 179. [Google Scholar] [CrossRef]
- Chandna, R.; Ahmad, A. Nitrogen stress-induced alterations in the leaf proteome of two wheat varieties grown at different nitrogen levels. Physiol. Mol. Biol. Plants 2015, 21, 19–33. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Liu, L.; Anwar, S.; Tuo, X.; Khan, S.; Wang, B.; Peng, D. Effects of Fertilization on Ramie (Boehmeria nivea L.) Growth, Yield and Fiber Quality. Sustainability 2016, 8, 887. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, H.; Chen, X.; Zhang, C.; Ma, W.; Huang, C.; Zhang, W.; Mi, G.; Miao, Y.; Li, X.; et al. Pursuing sustainable productivity with millions of smallholder farmers. Nature 2018, 555, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Xiong, H.; Chen, K.; Chen, J.; Chen, P.; Yu, C.; Zhu, A. Gene expression profiling of ramie roots during hydroponic induction and adaption to aquatic environment. Genom. Data 2017, 14, 32–35. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, J.; Jie, Y.; Xing, H.; Zhong, Y.; She, W.; Wei, G.; Yu, W.; Ma, Y. A ramie (Boehmeria nivea) bZIP transcription factor BnbZIP3 positively regulates drought, salinity and heavy metal tolerance. Mol. Breed. 2016, 36. [Google Scholar] [CrossRef]
- Zhu, S.; Zheng, X.; Dai, Q.; Tang, S.; Liu, T. Identification of quantitative trait loci for flowering time traits in ramie (Boehmeria nivea L. Gaud). Euphytica 2016, 210, 367–374. [Google Scholar] [CrossRef]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Seneca, F.O.; Traylor-Knowles, N.; Palumbi, S.R. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 1387–1392. [Google Scholar] [CrossRef]
- Naranjo, M.A.; Forment, J.; Roldan, M.; Serrano, R.; Vicente, O. Overexpression of Arabidopsis thaliana LTL1, a salt-induced gene encoding a GDSL-motif lipase, increases salt tolerance in yeast and transgenic plants. Plant Cell Environ. 2006, 29, 1890–1900. [Google Scholar] [CrossRef]
- Hong, J.K.; Choi, H.W.; Hwang, I.S.; Kim, D.S.; Kim, N.H.; Choi, D.S.; Kim, Y.J.; Hwang, B.K. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 2008, 227, 539–558. [Google Scholar] [CrossRef]
- Zhu, S.; Tang, S.; Tang, Q.; Liu, T. Genome-wide transcriptional changes of ramie (Boehmeria nivea L. Gaud) in response to root-lesion nematode infection. Gene 2014, 552, 67–74. [Google Scholar] [CrossRef]
- Huang, S.; Raman, A.S.; Ream, J.E.; Fujiwara, H.; Cerny, R.E.; Brown, S.M. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998, 118, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Coles, J.P.; Phillips, A.L.; Croker, S.J.; Garcia-Lepe, R.; Lewis, M.J.; Hedden, P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999, 17, 547–556. [Google Scholar]
- Sakamoto, T. An Overview of Gibberellin Metabolism Enzyme Genes and Their Related Mutants in Rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef]
- Fagoaga, C.; Tadeo, F.R.; Iglesias, D.J.; Huerta, L.; Lliso, I.; Vidal, A.M.; Talon, M.; Navarro, L.; Garcia-Martinez, J.L.; Pena, L. Engineering of gibberellin levels in citrus by sense and antisense overexpression of a GA 20-oxidase gene modifies plant architecture. J. Exp. Bot. 2007, 58, 1407–1420. [Google Scholar] [CrossRef]
- Langenbach, C.; Campe, R.; Schaffrath, U.; Goellner, K.; Conrath, U. UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi. New Phytol. 2013, 198, 536–545. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic Acquired Resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Chen, S.; Zimei, L.; Cui, J.; Jiangang, D.; Xia, X.; Liu, D.; Yu, J. Alleviation of chilling-induced oxidative damage by salicylic acid pretreatment and related gene expression in eggplant seedlings. Plant Growth Regul. 2011, 65, 101–108. [Google Scholar] [CrossRef]
- de Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; Rolim Medeiros, J.; Gomes-Filho, E. Hydrogen peroxide pre-treatment induces salt-stress acclimation in maize plants. J. Plant Physiol. 2005, 162, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Liu, Z.; Xu, P.; Fang, Y.; Bai, J. Paraquat pre-treatment increases activities of antioxidant enzymes and reduces lipid peroxidation in salt-stressed cucumber leaves. Acta Physiol. Plant 2011, 33, 295–304. [Google Scholar] [CrossRef]
- Wagner, T.A.; Kohorn, B.D. Wall-Associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef]
- Lally, D.; Ingmire, P.; Tong, H.Y.; He, Z.H. Antisense expression of a cell wall-Associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 2001, 13, 1317–1331. [Google Scholar]
- Hou, X. Involvement of a Cell Wall-Associated Kinase, WAKL4, in Arabidopsis Mineral Responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Zhang, H.; Yang, Y.; Ge, X.; Song, F. A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene 2008, 420, 57–65. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Starrett, D.A.; Laties, G.G. Involvement of Wound and Climacteric Ethylene in Ripening Avocado Discs. Plant Physiol. 1991, 97, 720–729. [Google Scholar] [CrossRef][Green Version]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Phys. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Kaur, J. A comprehensive review on metabolic syndrome. Cardiol. Res. Pract. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
| Salts | Concentration (mg·L−1) | ||
|---|---|---|---|
| N0 | N10 | ||
| Macro-elements | Ca(NO3)2 4(H2O) | - | 944.6 |
| KNO3 | - | 202.2 | |
| KCl | - | 223.65 | |
| K2SO4 | 435.65 | - | |
| KH2PO4 | 136.07 | 136.07 | |
| MgSO4 7H2O | 492.96 | 492.96 | |
| CaCl2 | 554.9 | 110.98 | |
| Micro-elements | H3BO3 | 2.86 | 2.86 |
| MnSO4·H2O | 1.55 | 1.55 | |
| ZnSO4·7H2O | 0.22 | 0.22 | |
| CuSO4·5H2O | 0.08 | 0.08 | |
| H2MoO4·4H2O | 0.09 | 0.09 | |
| FeNa·EDTA | 13.00 | 13.00 | |
| Database | Number of Unigenes | Matching Proportion |
|---|---|---|
| NCBI Non-redundant Protein Sequences (NR) | 30,919 | 50.3% |
| NCBI Nucleotide Sequences (NT) | 14,730 | 24.0% |
| KEGG Ortholog (KO) | 11,856 | 19.3% |
| Swiss-Prot | 23,502 | 38.3% |
| Protein family (Pfam) | 22,973 | 37.4% |
| Gene Ontology (GO) | 23,968 | 39.0% |
| Clusters of Orthologous Groups of proteins (COGs) | 14,230 | 23.2% |
| Total | 34,251 | 55.8% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, L.; Gao, G.; Yu, C.; Zhu, A.; Chen, P.; Chen, K.; Chen, J.; Xiong, H. Transcriptome Analysis of High-NUE (T29) and Low-NUE (T13) Genotypes Identified Different Responsive Patterns Involved in Nitrogen Stress in Ramie (Boehmeria nivea (L.) Gaudich). Plants 2020, 9, 767. https://doi.org/10.3390/plants9060767
Tan L, Gao G, Yu C, Zhu A, Chen P, Chen K, Chen J, Xiong H. Transcriptome Analysis of High-NUE (T29) and Low-NUE (T13) Genotypes Identified Different Responsive Patterns Involved in Nitrogen Stress in Ramie (Boehmeria nivea (L.) Gaudich). Plants. 2020; 9(6):767. https://doi.org/10.3390/plants9060767
Chicago/Turabian StyleTan, Longtao, Gang Gao, Chunming Yu, Aiguo Zhu, Ping Chen, Kunmei Chen, Jikang Chen, and Heping Xiong. 2020. "Transcriptome Analysis of High-NUE (T29) and Low-NUE (T13) Genotypes Identified Different Responsive Patterns Involved in Nitrogen Stress in Ramie (Boehmeria nivea (L.) Gaudich)" Plants 9, no. 6: 767. https://doi.org/10.3390/plants9060767
APA StyleTan, L., Gao, G., Yu, C., Zhu, A., Chen, P., Chen, K., Chen, J., & Xiong, H. (2020). Transcriptome Analysis of High-NUE (T29) and Low-NUE (T13) Genotypes Identified Different Responsive Patterns Involved in Nitrogen Stress in Ramie (Boehmeria nivea (L.) Gaudich). Plants, 9(6), 767. https://doi.org/10.3390/plants9060767







