Heterologous Expression of a Soybean Gene RR34 Conferred Improved Drought Resistance of Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
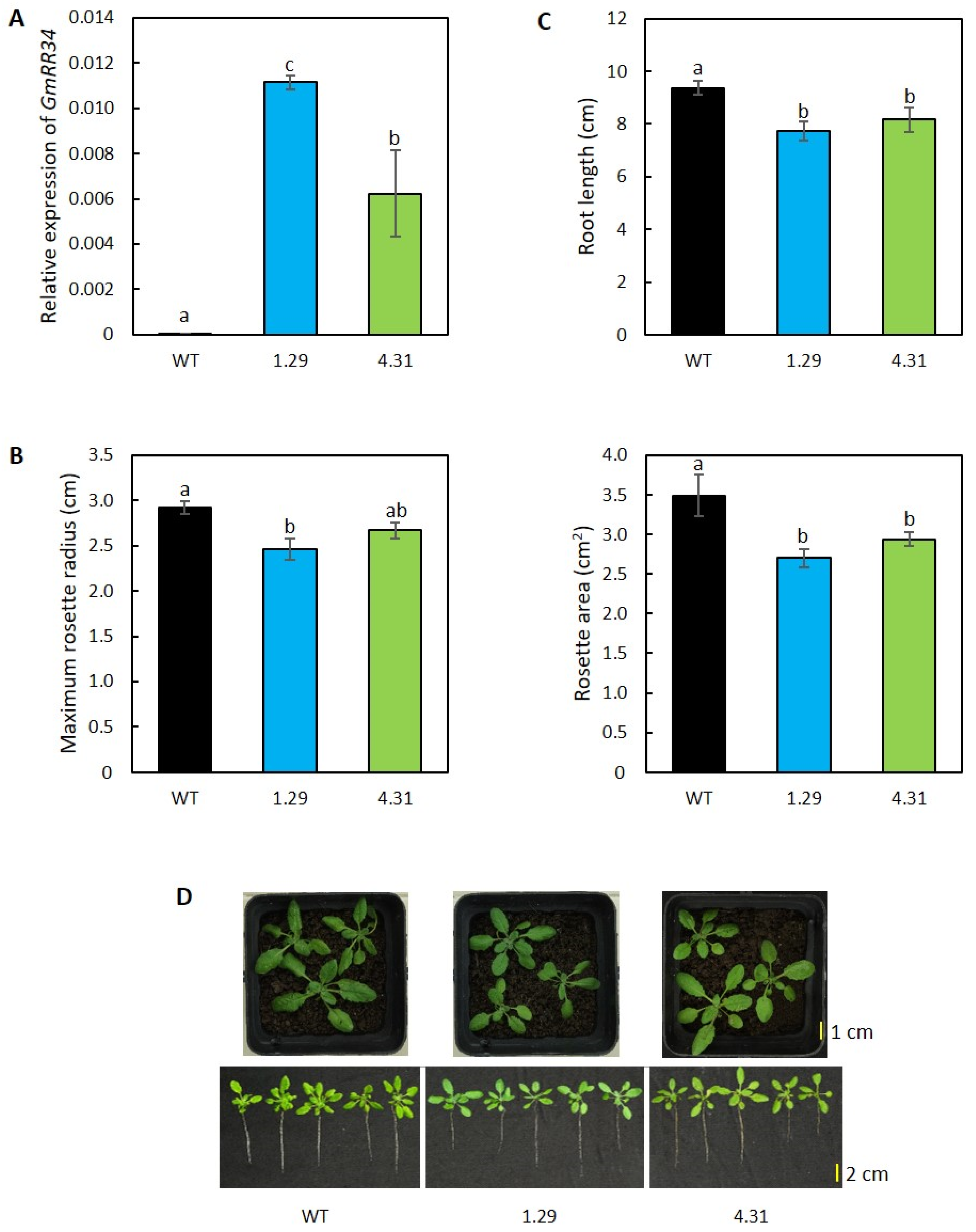
2.1. Ectopic Expression of GmRR34 Altered the Growth Characters in Transgenic Arabidopsis Plants
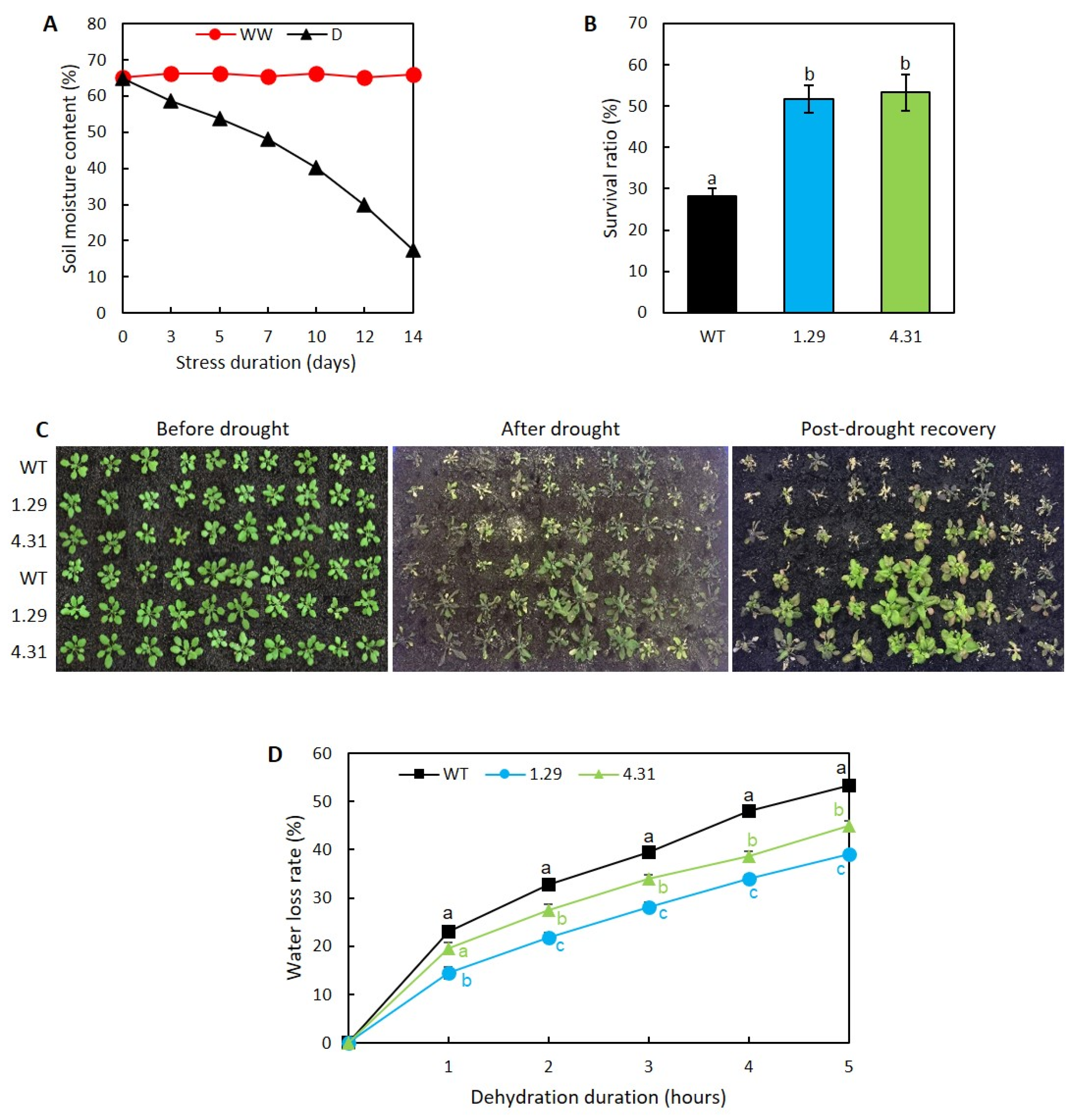
2.2. Ectopic Expression of GmRR34 Improved Post-Drought Survival Rates in Transgenic Arabidopsis
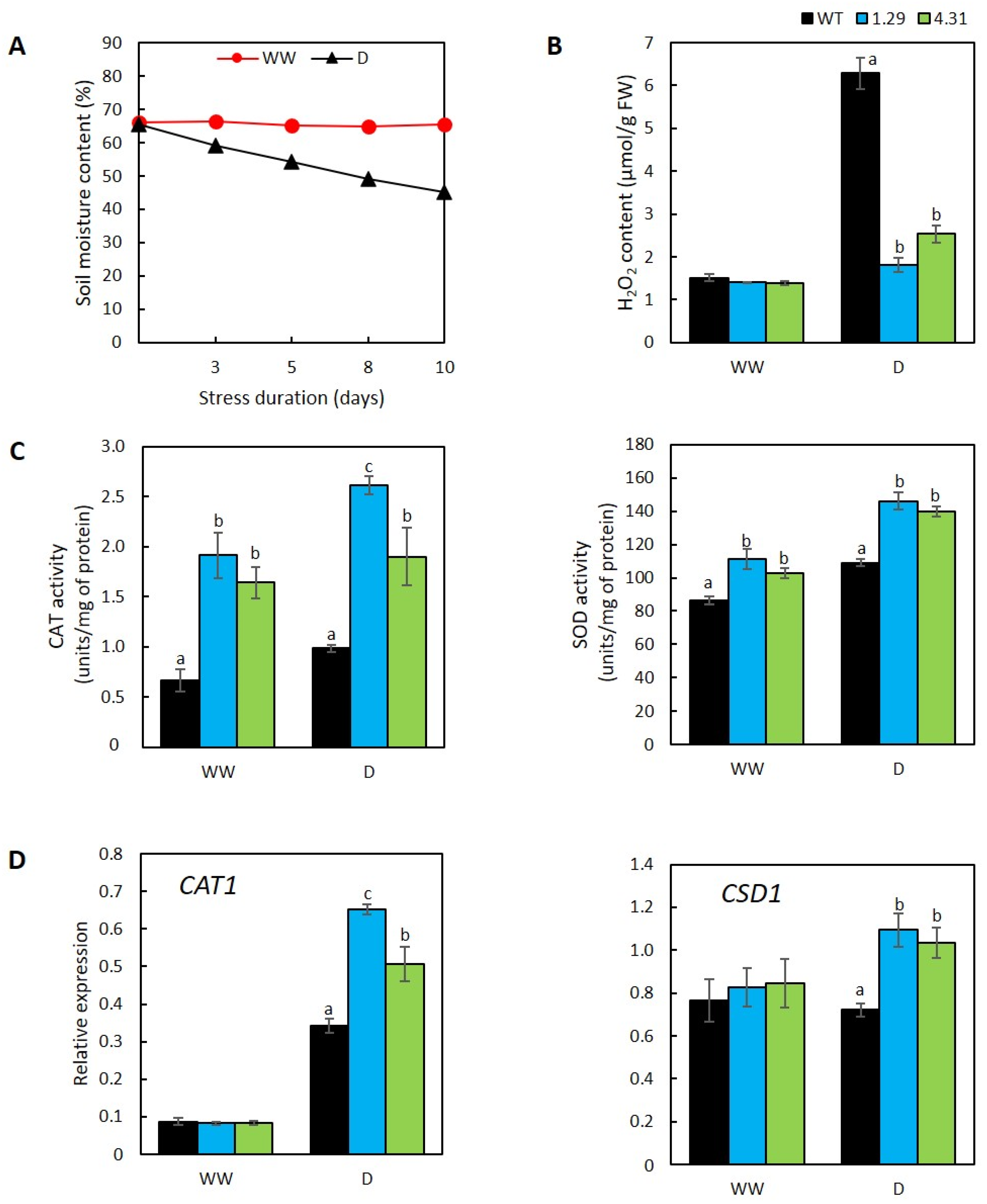
2.3. GmRR34-Ectopic Expression Lines Displayed More Active Enzymatic ROS-Scavenging Activities
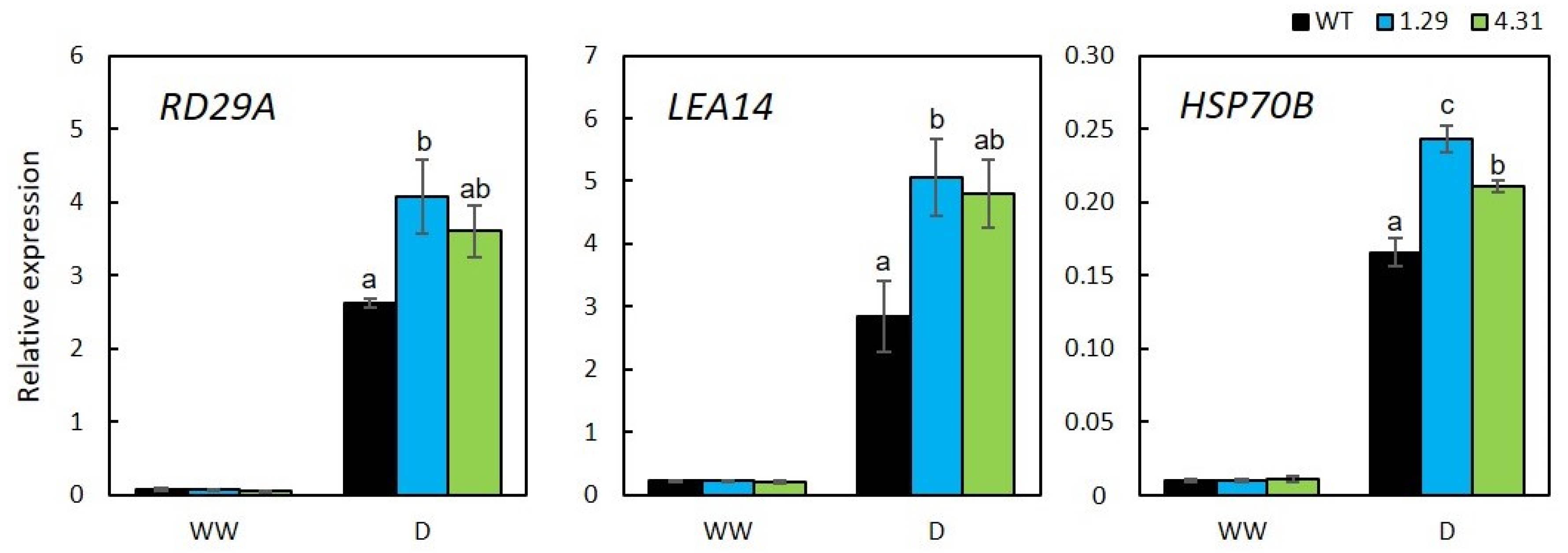
2.4. Stress-Related Genes with Enhanced Expression in GmRR34-Transgenic Arabidopsis
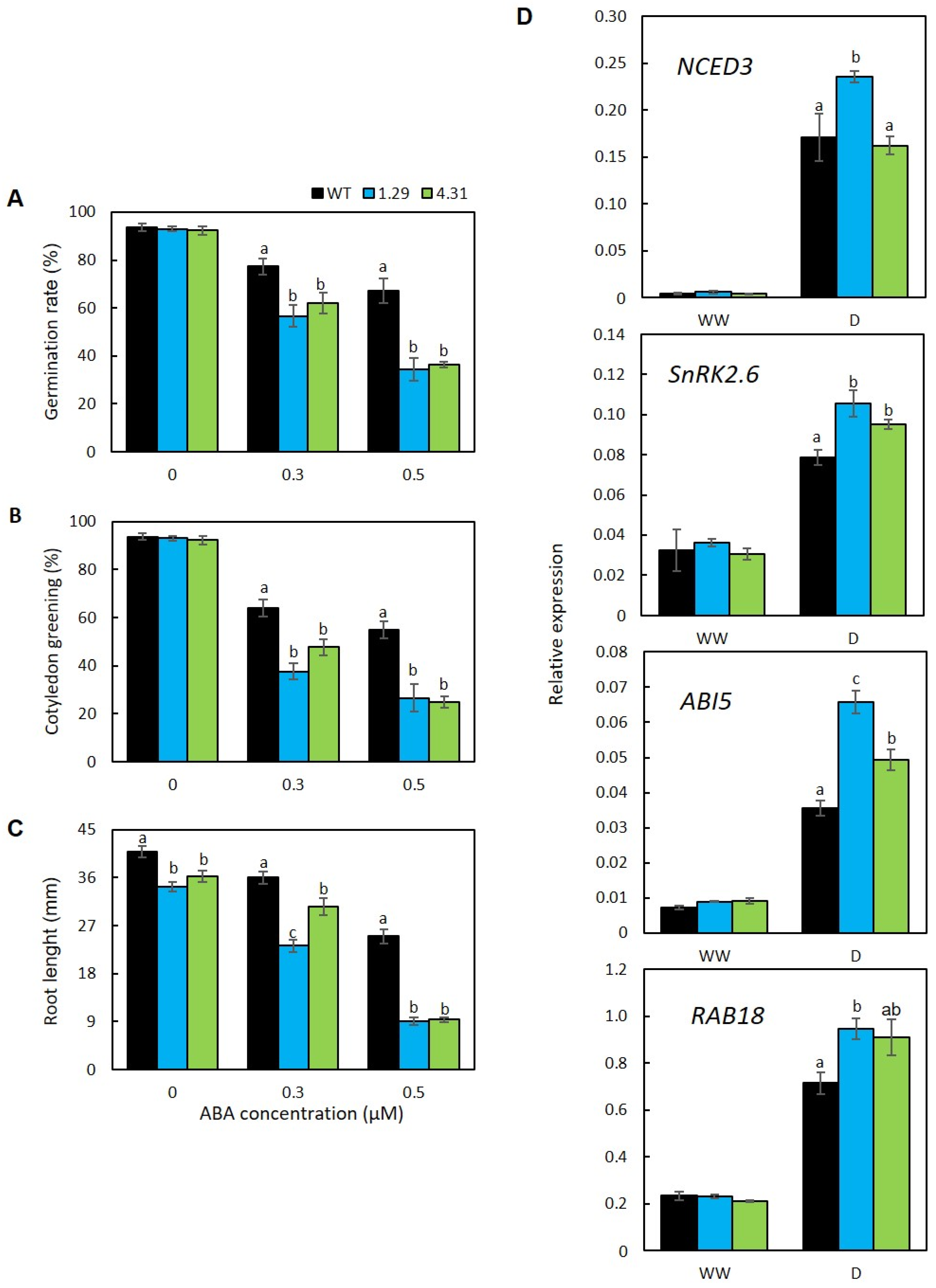
2.5. GmRR34 Ectopic Expression Lines Displayed Hypersensitivity to ABA
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Growth Conditions
4.3. Phenotypic Analyses of Ectopic Expression Lines under Normal Conditions
4.4. Survival Test
4.5. Determination of Excised-Leaf Water Loss Rate
4.6. Evaluation of H2O2 Content and ROS-Scavenging Enzymatic Activities
4.7. Expression Analysis of Transgene and Drought Response-Related Genes
4.8. ABA Sensitivity Assays
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Eliasson, J. The rising pressure of global water shortages. Nature 2015, 517, 6. [Google Scholar] [CrossRef] [PubMed]
- Vörösmarty, C.J.; McIntyre, P.B.; Gessner, M.O.; Dudgeon, D.; Prusevich, A.; Green, P.; Glidden, S.; Bunn, S.E.; Sullivan, C.A.; Liermann, C.R. Global threats to human water security and river biodiversity. Nature 2010, 467, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Hanjra, M.A.; Blackwell, J.; Carr, G.; Zhang, F.; Jackson, T.M. Wastewater irrigation and environmental health: Implications for water governance and public policy. Int. J. Hyg. Environ. Health 2012, 215, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Kummu, M.; Guillaume, J.H.A.; de Moel, H.; Eisner, S.; Flörke, M.; Porkka, M.; Siebert, S.; Veldkamp, T.I.E.; Ward, P.J. The world’s road to water scarcity: Shortage and stress in the 20th century and pathways towards sustainability. Sci. Rep. 2016, 6, 38495. [Google Scholar] [CrossRef]
- Rockström, J.; Falkenmark, M.; Karlberg, L.; Hoff, H.; Rost, S.; Gerten, D. Future water availability for global food production: The potential of green water for increasing resilience to global change. Water Resour. Res. 2009, 45. [Google Scholar] [CrossRef]
- Boutraa, T. Improvement of water use efficiency in irrigated agriculture: A review. J. Agron. 2010, 9, 1–8. [Google Scholar] [CrossRef]
- Blum, A. Drought resistance, water-use efficiency, and yield potential—Are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 56, 1159–1168. [Google Scholar] [CrossRef]
- Koyro, H.-W.; Khan, M.A.; Lieth, H. Halophytic crops: A resource for the future to reduce the water crisis? Emir. J. Food Agric. 2011, 23, 1–16. [Google Scholar] [CrossRef]
- Cominelli, E.; Tonelli, C. Transgenic crops coping with water scarcity. New Biotechnol. 2010, 27, 473–477. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Shao, H.-B.; Chu, L.-Y.; Jaleel, C.A.; Zhao, C.-X. Water-deficit stress-induced anatomical changes in higher plants. C. R. Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Thu, N.B.A.; Hoang, X.L.T.; Truc, M.T.; Sulieman, S.; Thao, N.P.; Tran, L.S.P. Cytokinin signaling in plant response to abiotic stresses. In Mechanism of Plant Hormone Signaling under Stress, 1st ed.; Pandey, G.K., Ed.; Wiley Online Library: Beverly, MA, USA, 2017; Volume 1, pp. 71–100. [Google Scholar]
- Miller, G.A.D.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R.O.N. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef]
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two-component systems in plant signal transduction. Trends Plant Sci. 2000, 5, 67–74. [Google Scholar] [CrossRef]
- Mizuno, T. Two-component phosphorelay signal transduction systems in plants: From hormone responses to circadian rhythms. Biosci. Biotechnol. Biochem. 2005, 69, 2263–2276. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Plant histidine kinases: An emerging picture of two-component signal transduction in hormone and environmental responses. Sci. STKE 2001, 2001, re18. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C. Light perception in plants: Cytokinins and red light join forces to keep phytochrome B active. Trends Plant Sci. 2002, 7, 143–145. [Google Scholar] [CrossRef]
- Grefen, C.; Harter, K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta 2004, 219, 733–742. [Google Scholar] [CrossRef]
- Ahmad, B.; Azeem, F.; Ali, M.A.; Nawaz, M.A.; Nadeem, H.; Abbas, A.; Batool, R.; Atif, R.M.; Ijaz, U.; Nieves-Cordones, M. Genome-wide identification and expression analysis of two component system genes in Cicer arietinum. Genomics 2020, 112, 1371–1383. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Ye, L.; Pan, C.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification and expression analysis of two-component system genes in tomato. Int. J. Mol. Sci. 2016, 17, 1204. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Zou, T.; Pan, C.; Qin, L.; Chen, L.; Lu, G. Genome-wide identification of two-component system genes in cucurbitaceae crops and expression profiling analyses in cucumber. Front. Plant Sci. 2016, 7, 899. [Google Scholar] [CrossRef]
- Dhar, Y.V.; Lakhwani, D.; Pandey, A.; Singh, S.; Trivedi, P.K.; Asif, M.H. Genome-wide identification and interactome analysis of members of two-component system in banana. BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Mochida, K.; Yoshida, T.; Sakurai, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010, 17, 303–324. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav. 2010, 5, 148–150. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 2008, 20, 1101–1117. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Van Ha, C.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Nishiyama, R.I.E.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011, 18, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Thu, N.B.A.; Hoang, X.L.T.; Nguyen, T.-D.H.; Thao, N.P.; Tran, L.-S.P. Differential expression of two-component system–related drought-responsive genes in two contrasting drought-tolerant soybean cultivars DT51 and MTD720 under well-watered and drought conditions. Plant Mol. Biol. Rep. 2015, 33, 1599–1610. [Google Scholar] [CrossRef]
- McClung, C.R. Regulation of catalases in Arabidopsis. Free Radic. Biol. Med. 1997, 23, 489–496. [Google Scholar] [CrossRef]
- Huang, C.-H.; Kuo, W.-Y.; Weiss, C.; Jinn, T.-L. Copper chaperone-dependent and-independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 2012, 158, 737–746. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Hoang, X.L.T.; Nguyen, N.C.; Nguyen, Y.-N.H.; Watanabe, Y.; Tran, L.-S.P.; Thao, N.P. The soybean GmNAC019 transcription factor mediates drought tolerance in Arabidopsis in an abscisic acid-dependent manner. Int. J. Mol. Sci. 2020, 21, 286. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Mostofa, M.G.; Li, W.; Van Ha, C.; Watanabe, Y.; Le, D.T.; Thao, N.P.; Tran, L.-S.P. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 2018, 151, 12–20. [Google Scholar] [CrossRef]
- Nguyen, N.C.; Hoang, X.L.T.; Nguyen, Q.T.; Binh, N.X.; Watanabe, Y.; Thao, N.P.; Tran, L.-S.P. Ectopic expression of Glycine max GmNAC109 enhances drought tolerance and ABA sensitivity in Arabidopsis. Biomolecules 2019, 9, 714. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought-and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004, 45, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhou, S.; Yin, S.; Zhao, Z.; Han, Y.; Wang, W. Stress-inducible expression of an F-box gene TaFBA1 from wheat enhanced the drought tolerance in transgenic tobacco plants without impacting growth and development. Front. Plant Sci. 2016, 7, 1295. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-H.; Lee, J.-t.; Charng, Y.-y.; Chan, M.-T. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002, 130, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-H.; Lee, J.-T.; Yang, P.-T.; Chiu, L.-H.; Charng, Y.-y.; Wang, Y.-C.; Chan, M.-T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding Factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002, 129, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mason, H.S.; Bensen, R.J.; Boyer, J.S.; Mullet, J.E. Water deficit and abscisic acid cause differential inhibition of shoot versus root growth in soybean seedlings: Analysis of growth, sugar accumulation, and gene expression. Plant Physiol. 1990, 92, 205–214. [Google Scholar] [CrossRef]
- Zhang, J.; Davies, W.J. Does ABA in the xylem control the rate of leaf growth in soil-dried maize and sunflower plants? J. Exp. Bot. 1990, 41, 1125–1132. [Google Scholar] [CrossRef]
- Skirycz, A.; Inzé, D. More from less: Plant growth under limited water. Curr. Opin. Plant Biol. 2010, 21, 197–203. [Google Scholar] [CrossRef]
- Murti, G.S.R.; Upreti, K.K. Plant growth regulators in water stress tolerance. J. Hortic. Sci. 2007, 2, 73–93. [Google Scholar]
- Farber, M.; Attia, Z.; Weiss, D. Cytokinin activity increases stomatal density and transpiration rate in tomato. J. Exp. Bot. 2016, erw398. [Google Scholar] [CrossRef]
- Mao, H.; Yu, L.; Han, R.; Li, Z.; Liu, H. ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 105, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.S.; Sethi, G.S. Inheritance of excised-leaf water loss and relative water content in bread wheat (Triticum aestivum). Euphytica 1998, 104, 39–47. [Google Scholar] [CrossRef]
- Clarke, J.M.; Romagosa, I.; Jana, S.; Srivastava, J.P.; McCaig, T.N. Relationship of excised-leaf water loss rate and yield of durum wheat in diverse environments. Can. J. Plant Sci. 1989, 69, 1075–1081. [Google Scholar] [CrossRef]
- Czyczyło-Mysza, I.M.; Marcińska, I.; Skrzypek, E.; Bocianowski, J.; Dziurka, K.; Rančić, D.; Radošević, R.; Pekić-Quarrie, S.; Dodig, D.; Quarrie, S.A. Genetic analysis of water loss of excised leaves associated with drought tolerance in wheat. PeerJ 2018, 6, e5063. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Wu, J.; Zhang, B.; Li, X.; Yan, Y.; Li, L. Overexpression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging. Plant Physiol. Biochem. 2013, 70, 354–359. [Google Scholar] [CrossRef]
- Cao, H.; Wang, L.; Nawaz, M.A.; Niu, M.; Sun, J.; Xie, J.; Kong, Q.; Huang, Y.; Cheng, F.; Bie, Z. Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 2052. [Google Scholar] [CrossRef]
- Liu, X.; Hong, L.; Li, X.-Y.; Yao, Y.; Hu, B.; Li, L. Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci. Biotechnol. Biochem. 2011, 75, 443–450. [Google Scholar] [CrossRef]
- Li, X.-l.; Yang, X.; Hu, Y.-x.; Yu, X.-d.; Li, Q.-l. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Clarke, J.M.; Richards, R.A. The effects of glaucousness, epicuticular wax, leaf age, plant height, and growth environment on water loss rates of excised wheat leaves. Can. J. Plant Sci. 1988, 68, 975–982. [Google Scholar] [CrossRef]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef] [PubMed]
- Blum, A.; Ebercon, A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 1981, 21, 43–47. [Google Scholar] [CrossRef]
- Møller, I.M.; Sweetlove, L.J. ROS signalling–specificity is required. Trends Plant Sci. 2010, 15, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Mustilli, A.-C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Hajiboland, R. Reactive oxygen species and photosynthesis. In Oxidative Damage to Plants; Ahmad, P., Ed.; Elsevier: Oxford, UK, 2014; pp. 1–63. [Google Scholar]
- Luna, C.M.; Pastori, G.M.; Driscoll, S.; Groten, K.; Bernard, S.; Foyer, C.H. Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J. Exp. Bot. 2005, 56, 417–423. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.-A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef]
- Du, Y.Y.; Wang, P.C.; Chen, J.; Song, C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993, 236, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Qi, S.; Li, H.; Liu, P.; Li, P.; Wu, C.; Zheng, C.; Huang, J. Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014, 454, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef]
- Kumar, D.; Chattopadhyay, S. Glutathione modulates the expression of heat shock proteins via BZIP10 and MYB21 transcription factors in Arabidopsis thaliana. J. Exp. Bot. 2018, 69, 3729–3743. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, R.; Fernandes, J.L.; Abdelrahman, M.; Burritt, D.J.; Tran, L.-S.P. Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Crit. Rev. Biotechnol. 2020, 320–340. [Google Scholar] [CrossRef]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Rock, C.D. Abscisic acid biosynthesis and response. Arabidopsis Book 2002, 1, e0058. [Google Scholar] [CrossRef] [PubMed]
- Grondin, A.; Rodrigues, O.; Verdoucq, L.; Merlot, S.; Leonhardt, N.; Maurel, C. Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 2015, 27, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Sato, Y.; Fukao, Y.; Fujiwara, M.; Umezawa, T.; Shinozaki, K.; Hibi, T.; Taniguchi, M.; Miyake, H.; Goto, D.B. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009, 424, 439–448. [Google Scholar] [CrossRef]
- Hallouin, M.; Ghelis, T.; Brault, M.; Bardat, F.; Cornel, D.; Miginiac, E.; Rona, J.-P.; Sotta, B.; Jeannette, E. Plasmalemma abscisic acid perception leads to RAB18 expression via phospholipase D activation in Arabidopsis suspension cells. Plant Physiol. 2002, 130, 265–272. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Mantyla, E.; Lang, V.; Palva, E.T. Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 1995, 107, 141–148. [Google Scholar] [CrossRef]
- Bi, C.; Ma, Y.; Wu, Z.; Yu, Y.-T.; Liang, S.; Lu, K.; Wang, X.-F. Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol. Biol. 2017, 94, 197–213. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Tizaoui, K.; Kchouk, M.E. Genetic approaches for studying transgene inheritance and genetic recombination in three successive generations of transformed tobacco. Genet. Mol. Biol. 2012, 35, 640–649. [Google Scholar] [CrossRef]
- Li, Y.; Varala, K.; Moose, S.P.; Hudson, M.E. The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS ONE 2012, 7, e47043. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cai, H.; Liu, P.; Wang, C.; Gao, H.; Wu, C.; Yan, K.; Zhang, S.; Huang, J.; Zheng, C. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 2017, 484, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Patterson, B.D.; MacRae, E.A.; Ferguson, I.B. Estimation of hydrogen peroxide in plant extracts using titanium (IV). Anal. Biochem. 1984, 139, 487–492. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Wang, C.-J.; Yang, W.; Wang, C.; Gu, C.; Niu, D.-D.; Liu, H.-X.; Wang, Y.-P.; Guo, J.-H. Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting rhizobacterium strains. PLoS ONE 2012, 7, e52565. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Q.; Liu, Z.; Yang, H.; Wang, J.; Li, X.; Yang, Y. Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J. Integr. Plant Biol. 2016, 58, 67–80. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, M.-M.; Ye, Q.; Wu, X.-Q.; Wu, W.-H.; Chen, Y.-F. Abscisic acid modulates seed germination via ABA INSENSITIVE5-mediated PHOSPHATE1. Plant Physiol. 2017, 175, 1661–1668. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Zhang, Q.; Jiang, Y.; Zhang, H.; Li, R. Overexpression of HbMBF1a, encoding multiprotein bridging factor 1 from the halophyte Hordeum brevisubulatum, confers salinity tolerance and ABA insensitivity to transgenic Arabidopsis thaliana. Plant Mol. Biol. 2020, 102, 1–17. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nghia, D.H.T.; Chuong, N.N.; Hoang, X.L.T.; Nguyen, N.C.; Tu, N.H.C.; Huy, N.V.G.; Ha, B.T.T.; Nam, T.N.H.; Thu, N.B.A.; Tran, L.-S.P.; et al. Heterologous Expression of a Soybean Gene RR34 Conferred Improved Drought Resistance of Transgenic Arabidopsis. Plants 2020, 9, 494. https://doi.org/10.3390/plants9040494
Nghia DHT, Chuong NN, Hoang XLT, Nguyen NC, Tu NHC, Huy NVG, Ha BTT, Nam TNH, Thu NBA, Tran L-SP, et al. Heterologous Expression of a Soybean Gene RR34 Conferred Improved Drought Resistance of Transgenic Arabidopsis. Plants. 2020; 9(4):494. https://doi.org/10.3390/plants9040494
Chicago/Turabian StyleNghia, Duong Hoang Trong, Nguyen Nguyen Chuong, Xuan Lan Thi Hoang, Nguyen Cao Nguyen, Nguyen Huu Cam Tu, Nguyen Van Gia Huy, Bui Thi Thanh Ha, Thai Nguyen Hoang Nam, Nguyen Binh Anh Thu, Lam-Son Phan Tran, and et al. 2020. "Heterologous Expression of a Soybean Gene RR34 Conferred Improved Drought Resistance of Transgenic Arabidopsis" Plants 9, no. 4: 494. https://doi.org/10.3390/plants9040494
APA StyleNghia, D. H. T., Chuong, N. N., Hoang, X. L. T., Nguyen, N. C., Tu, N. H. C., Huy, N. V. G., Ha, B. T. T., Nam, T. N. H., Thu, N. B. A., Tran, L.-S. P., & Thao, N. P. (2020). Heterologous Expression of a Soybean Gene RR34 Conferred Improved Drought Resistance of Transgenic Arabidopsis. Plants, 9(4), 494. https://doi.org/10.3390/plants9040494





