Diversity, Function and Regulation of Cell Surface and Intracellular Immune Receptors in Solanaceae
Abstract
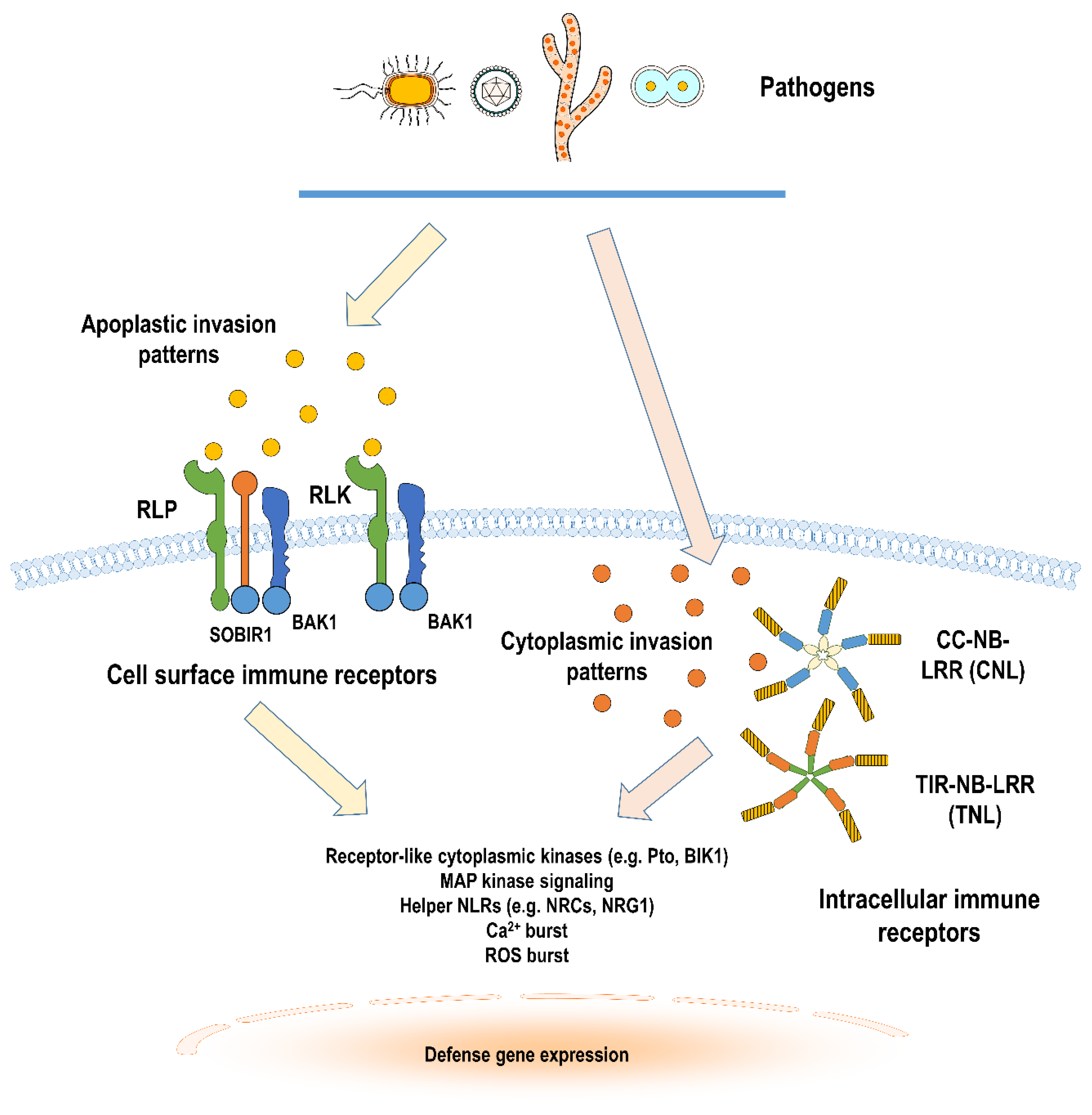
1. Introduction
2. Cell Surface Immune Receptors in Solanaceous Plants
2.1. Structure
2.2. Ligand Recognition and Signaling
2.3. Expression and Regulation
3. Intracellular Immune Receptors in Solanaceous Plants
3.1. Structure
3.2. Ligand Recognition, Signaling, and Regulation
4. Immune Receptor Crosstalk
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.F.; Mackey, D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [PubMed]
- Albert, I.; Hua, C.; Nurnberger, T.; Pruitt, R.; Zhang, L. Surface sensor systems in plant immunity. Plant Physiol. 2019, in press. [Google Scholar] [CrossRef]
- Hind, S.R.; Strickler, S.R.; Boyle, P.C.; Dunham, D.M.; Bao, Z.; O’Doherty, I.M.; Baccile, J.A.; Hoki, J.S.; Viox, E.G.; Clarke, C.R.; et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2016, 2, 16128. [Google Scholar] [CrossRef]
- Robatzek, S.; Bittel, P.; Chinchilla, D.; Köchner, P.; Felix, G.; Shiu, S.H.; Boller, T. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 2007, 64, 539–547. [Google Scholar] [CrossRef]
- Wang, L.; Einig, E.; Almeida-Trapp, M.; Albert, M.; Fliegmann, J.; Mithöfer, A.; Kalbacher, H.; Felix, G. The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants 2018, 4, 152–156. [Google Scholar] [CrossRef]
- Liao, D.; Sun, X.; Wang, N.; Song, F.; Liang, Y. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2018, 9, 1004. [Google Scholar] [CrossRef]
- Sanabria, N.M.; van Heerden, H.; Dubery, I.A. Molecular characterisation and regulation of a Nicotiana tabacum S-domain receptor-like kinase gene induced during an early rapid response to lipopolysaccharides. Gene 2012, 501, 39–48. [Google Scholar] [CrossRef]
- Chen, D.; Cao, Y.; Li, H.; Kim, D.; Ahsan, N.; Thelen, J.; Stacey, G. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Park, Y.S.; Lee, S.; Song, G.C.; Ryu, C.M. Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 2016, 209, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Zeng, L.; Velásquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012, 69, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Shan, L.; He, P.; de Vries, S.; Kemmerling, B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009, 14, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Albert, M.; Einig, E.; Fürst, U.; Krust, D.; Felix, G. The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Berg, G.C.; Zhang, Z.; Smit, P.; Cordewener, J.H.; America, A.H.; Sklenar, J.; Jones, A.M.; Tameling, W.I.; Robatzek, S.; et al. Receptor-Like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef]
- Du, J.; Verzaux, E.; Chaparro-Garcia, A.; Bijsterbosch, G.; Keizer, L.C.; Zhou, J.; Liebrand, T.W.; Xie, C.; Govers, F.; Robatzek, S.; et al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 2015, 1, 15034. [Google Scholar] [CrossRef]
- Liebrand, T.W.H.; van den Burg, H.A.; Joosten, M.H.A.J. Two for all: Receptor-Associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014, 19, 123–132. [Google Scholar] [CrossRef]
- Hegenauer, V.; Fürst, U.; Kaiser, B.; Smoker, M.; Zipfel, C.; Felix, G.; Stahl, M.; Albert, M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 2016, 353, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.-M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.-C.; Kaloshian, I. The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS ONE 2014, 9, e93302. [Google Scholar] [CrossRef] [PubMed]
- Nietzschmann, L.; Gorzolka, K.; Smolka, U.; Matern, A.; Eschen-Lippold, L.; Scheel, D.; Rosahl, S. Early Pep-13-induced immune responses are SERK3A/B-dependent in potato. Sci. Rep. 2019, 9, 18380. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef]
- Abuqamar, S.; Chai, M.F.; Luo, H.; Song, F.; Mengiste, T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef]
- Schwizer, S.; Kraus, C.M.; Dunham, D.M.; Zheng, Y.; Fernandez-Pozo, N.; Pombo, M.A.; Fei, Z.; Chakravarthy, S.; Martin, G.B. The tomato kinase Pti1 contributes to production of reactive oxygen species in response to two flagellin-derived peptides and promotes resistance to Pseudomonas syringae infection. Mol. Plant Microbe Interact. 2017, 30, 725–738. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J. 2011, 66, 642–655. [Google Scholar] [CrossRef]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-Wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Pan, C.; Guan, X.; Wang, Y.; Liu, S.; He, Y.; Chen, J.; Chen, L.; Lu, G. Genome-Wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE 2014, 9, e103032. [Google Scholar] [CrossRef]
- Hu, Z.; Lv, X.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y. Genome-Wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 2016, 7, 469. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Lindo, L.; Cardoza, R.E.; Lorenzana, A.; Casquero, P.A.; Gutiérrez, S. Identification of plant genes putatively involved in the perception of fungal ergosterol-squalene. J. Integr. Plant Biol. 2019. [Google Scholar] [CrossRef]
- Katou, S.; Asakura, N.; Kojima, T.; Mitsuhara, I.; Seo, S. Transcriptome analysis of WIPK/SIPK-suppressed plants reveals induction by wounding of disease resistance-related genes prior to the accumulation of salicylic acid. Plant Cell Physiol. 2013, 54, 1005–1015. [Google Scholar] [CrossRef]
- Nazar, R.N.; Castroverde, C.D.M.; Xu, X.; Kurosky, A.; Robb, J. Wounding induces tomato Ve1 R-gene expression. Planta 2019, 249, 1779–1797. [Google Scholar] [CrossRef]
- Castroverde, C.D.; Xu, X.; Nazar, R.N.; Robb, J. Biotic factors that induce the tomato Ve1 R-gene. Plant Sci. 2017, 265, 61–69. [Google Scholar] [CrossRef]
- Andolfo, G.; Ferriello, F.; Tardella, L.; Ferrarini, A.; Sigillo, L.; Frusciante, L.; Ercolano, M.R. Tomato genome-wide transcriptional responses to Fusarium wilt and tomato mosaic virus. PLoS ONE 2014, 9, e94963. [Google Scholar] [CrossRef]
- Li, K.; Wu, G.; Li, M.; Ma, M.; Du, J.; Sun, M.; Sun, X.; Qing, L. Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus. Virol. J. 2018, 15, 138. [Google Scholar] [CrossRef]
- Zuluaga, A.P.; Vega-Arreguín, J.C.; Fei, Z.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K. Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 2016, 17, 42–54. [Google Scholar] [CrossRef]
- Thara, V.K.; Seilaniantz, A.R.; Deng, Y.; Dong, Y.; Yang, Y.; Tang, X.; Zhou, J.M. Tobacco genes induced by the bacterial effector protein AvrPto. Mol. Plant Microbe Interact. 2004, 17, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, A.; Bergelson, J. Modulation of R-gene expression across environments. J. Exp. Bot. 2016, 67, 2093–2105. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Z.; Zhang, Z.; Lv, Y.; Yang, N.; Zhang, G.; Wu, M.; Lv, S.; Pan, L.; Joosten, M.H.; et al. Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 3339–3351. [Google Scholar] [CrossRef] [PubMed]
- Cambiagno, D.A.; Nota, F.; Zavallo, D.; Rius, S.; Casati, P.; Asurmendi, S.; Alvarez, M.E. Immune receptor genes and pericentromeric transposons as targets of common epigenetic regulatory elements. Plant J. 2018, 96, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, N.M.; Dubery, I.A. Alternative splicing of the receptor-like kinase Nt-Sd-RLK in tobacco cells responding to lipopolysaccharides: Suggestive of a role in pathogen surveillance and perception? FEBS Lett. 2016, 590, 3628–3638. [Google Scholar] [CrossRef]
- Giska, F.; Martin, G.B. PP2C phosphatase Pic1 negatively regulates the phosphorylation status of Pti1b kinase, a regulator of flagellin-triggered immunity in tomato. Biochem. J. 2019, 476, 1621–1635. [Google Scholar] [CrossRef]
- Orosa, B.; Yates, G.; Verma, V.; Srivastava, A.K.; Srivastava, M.; Campanaro, A.; De Vega, D.; Fernandes, A.; Zhang, C.; Lee, J.; et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 2018, 9, 5185. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. The tomato U-box type E3 ligase PUB13 acts with group III Ubiquitin E2 enzymes to modulate FLS2-mediated immune signaling. Front. Plant Sci. 2018, 9, 615. [Google Scholar] [CrossRef]
- Taylor, K.W.; Kim, J.G.; Su, X.B.; Aakre, C.D.; Roden, J.A.; Adams, C.M.; Mudgett, M.B. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 2012, 8, e1002768. [Google Scholar] [CrossRef]
- Chen, L.; Hamada, S.; Fujiwara, M.; Zhu, T.; Thao, N.P.; Wong, H.L.; Krishna, P.; Ueda, T.; Kaku, H.; Shibuya, N.; et al. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 2010, 7, 185–196. [Google Scholar] [CrossRef]
- Huang, S.; Nie, S.; Wang, S.; Liu, J.; Zhang, Y.; Wang, X. SlBIR3 negatively regulates PAMP responses and cell death in tomato. Int. J. Mol. Sci. 2017, 18, 1966. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Leibman-Markus, M.; Schuster, S.; Bar, M.; Meltz, T.; Avni, A. Tomato prenylated RAB acceptor protein 1 modulates trafficking and degradation of the pattern recognition receptor LeEIX2, affecting the innate immune response. Front. Plant Sci. 2018, 9, 257. [Google Scholar] [CrossRef] [PubMed]
- Bar, M.; Avni, A. EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. Plant J. 2009, 59, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; McMichael, C.; Walker, K.A.; Anderson, J.C.; Bednarek, S.Y.; Heese, A. Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J. Biol. Chem. 2010, 285, 23342–23350. [Google Scholar] [CrossRef] [PubMed]
- Spallek, T.; Beck, M.; Ben Khaled, S.; Salomon, S.; Bourdais, G.; Schellmann, S.; Robatzek, S. ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 2013, 9, e1004035. [Google Scholar] [CrossRef]
- Saur, I.M.; Kadota, Y.; Sklenar, J.; Holton, N.J.; Smakowska, E.; Belkhadir, Y.; Zipfel, C.; Rathjen, J.P. NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2016, 113, 3389–3394. [Google Scholar] [CrossRef]
- Catanzariti, A.M.; Lim, G.T.; Jones, D.A. The tomato I-3 gene: A novel gene for resistance to Fusarium wilt disease. New Phytol. 2015, 207, 106–118. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Wu, X.; Long, Z.; Sun, C.; Wang, S.; Birch, P.R.J.; Tian, Z. A potato STRUBBELIG-RECEPTOR FAMILY member, StLRPK1, associates with StSERK3A/BAK1 and activates immunity. J. Exp. Bot. 2018, 69, 5573–5586. [Google Scholar] [CrossRef]
- Lori, M.; van Verk, M.C.; Hander, T.; Schatowitz, H.; Klauser, D.; Flury, P.; Gehring, C.A.; Boller, T.; Bartels, S. Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: Interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 2015, 66, 5315–5325. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Z.; Lei, C.; Zheng, C.; Wang, J.; Shao, S.; Li, X.; Xia, X.; Cai, X.; Zhou, J.; et al. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca. Plant Cell 2018, 30, 652–667. [Google Scholar] [CrossRef]
- Sun, M.; Voorrips, R.E.; Steenhuis-Broers, G.; Van’t Westende, W.; Vosman, B. Reduced phloem uptake of Myzus persicae on an aphid resistant pepper accession. BMC Plant Biol. 2018, 18, 138. [Google Scholar] [CrossRef] [PubMed]
- Scheer, J.M.; Ryan, C.A. The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 2002, 99, 9585–9590. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.S.; Jones, D.A.; Keddie, J.S.; Thomas, C.M.; Harrison, K.; Jones, J.D. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 1996, 84, 451–459. [Google Scholar] [CrossRef]
- Jones, D.A.; Brading, P.; Dixon, M.; Hammond-Kosack, K.; Harrison, K.; Hatzixanthis, K.; Parniske, M.; Piedras, P.; Torres, M.; Tang, S.; et al. Molecular, genetic and physiological analysis of Cladosporium resistance gene function in tomato. Symp. Soc. Exp. Biol. 1998, 51, 111–113. [Google Scholar] [PubMed]
- Thomas, C.M.; Dixon, M.S.; Parniske, M.; Golstein, C.; Jones, J.D. Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998, 353, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Jones, D.A.; Parniske, M.; Harrison, K.; Balint-Kurti, P.J.; Hatzixanthis, K.; Jones, J.D. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 1997, 9, 2209–2224. [Google Scholar] [PubMed]
- Takken, F.L.; Thomas, C.M.; Joosten, M.H.; Golstein, C.; Westerink, N.; Hille, J.; Nijkamp, H.J.; De Wit, P.J.; Jones, J.D. A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 1999, 20, 279–288. [Google Scholar] [CrossRef]
- Takken, F.L.; Schipper, D.; Nijkamp, H.J.; Hille, J. Identification and Ds-tagged isolation of a new gene at the Cf-4 locus of tomato involved in disease resistance to Cladosporium fulvum race 5. Plant J. 1998, 14, 401–411. [Google Scholar] [CrossRef]
- Dixon, M.S.; Hatzixanthis, K.; Jones, D.A.; Harrison, K.; Jones, J.D. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 1998, 10, 1915–1925. [Google Scholar] [CrossRef]
- Grushtskaia, Z.E.; Lemesh, V.A.; Poliksenova, V.D.; Khotyleva, L.V. Cloning of the Cf-6 tomato leaf mould resistance locus using SSR markers. Genetika 2007, 43, 1511–1516. [Google Scholar]
- Jones, D.A.; Thomas, C.M.; Hammond-Kosack, K.E.; Balint-Kurti, P.J.; Jones, J.D. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Harrison, K.; Jones, J.D. Developmentally regulated cell death on expression of the fungal avirulence gene Avr9 in tomato seedlings carrying the disease-resistance gene Cf-9. Proc. Natl. Acad. Sci. USA 1994, 91, 10445–10449. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Tang, S.; Harrison, K.; Jones, J.D. The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product avr 9. Plant Cell 1998, 10, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- van der Hoorn, R.A.; Wulff, B.B.; Rivas, S.; Durrant, M.C.; van der Ploeg, A.; de Wit, P.J.; Jones, J.D. Structure-Function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 2005, 17, 1000–1015. [Google Scholar] [CrossRef]
- Parniske, M.; Wulff, B.B.; Bonnema, G.; Thomas, C.M.; Jones, D.A.; Jones, J.D. Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol. Plant Microbe Interact. 1999, 12, 93–102. [Google Scholar] [CrossRef]
- Laugé, R.; Joosten, M.H.; Haanstra, J.P.; Goodwin, P.H.; Lindhout, P.; De Wit, P.J. Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proc. Natl. Acad. Sci. USA 1998, 95, 9014–9018. [Google Scholar] [CrossRef]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef]
- Bar, M.; Sharfman, M.; Ron, M.; Avni, A. BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 2010, 63, 791–800. [Google Scholar] [CrossRef]
- Catanzariti, A.M.; Do, H.T.; Bru, P.; de Sain, M.; Thatcher, L.F.; Rep, M.; Jones, D.A. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 2017, 89, 1195–1209. [Google Scholar] [CrossRef]
- Gonzalez-Cendales, Y.; Catanzariti, A.M.; Baker, B.; Mcgrath, D.J.; Jones, D.A. Identification of I-7 expands the repertoire of genes for resistance to Fusarium wilt in tomato to three resistance gene classes. Mol. Plant Pathol. 2016, 17, 448–463. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Sun, Y.; Wang, H.; Qi, J.; Wan, B.; Ye, W.; Lin, Y.; Shao, Y.; Dong, S.; et al. Leucine-Rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Kawchuk, L.M.; Hachey, J.; Lynch, D.R.; Kulcsar, F.; van Rooijen, G.; Waterer, D.R.; Robertson, A.; Kokko, E.; Byers, R.; Howard, R.J.; et al. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 6511–6515. [Google Scholar] [CrossRef] [PubMed]
- Fradin, E.F.; Zhang, Z.; Juarez Ayala, J.C.; Castroverde, C.D.M.; Nazar, R.N.; Robb, J.; Liu, C.-M.; Thomma, B.P.H.J. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009, 150, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Nazar, R.N.; Xu, X.; Kurosky, A.; Robb, J. Antagonistic function of the Ve R-genes in tomato. Plant Mol. Biol. 2018, 98, 67–79. [Google Scholar] [CrossRef] [PubMed]
- van Ooijen, G.; van den Burg, H.A.; Cornelissen, B.J.; Takken, F.L. Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 2007, 45, 43–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, M.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364. [Google Scholar] [CrossRef]
- Horsefield, S.; Burdett, H.; Zhang, X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.; Gilley, J.; Lai, J.-S.; Rank, M.X.; et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Osborne Nishimura, E.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD. Science 2019, 365, 799–803. [Google Scholar] [CrossRef]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.; Takken, F.L.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Di, X.; Cao, L.; Hughes, R.K.; Tintor, N.; Banfield, M.J.; Takken, F.L.W. Structure-Function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 2017, 216, 897–914. [Google Scholar] [CrossRef]
- Biju, V.C.; Fokkens, L.; Houterman, P.M.; Rep, M.; Cornelissen, B.J.C. Multiple evolutionary trajectories have led to the emergence of races in Fusarium oxysporum f. sp. lycopersici. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jiang, L.; Bai, B.; Zhao, W.; Chen, X.; Li, J.; Liu, Y.; Chen, Z.; Wang, B.; Wang, C.; et al. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Zhu, M.; Huang, S.; Zhang, W.; Dinesh-Kumar, S.P.; Tao, X. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant 2019, 12, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 2008, 132, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Köhn, B.A.; Dedi, C.; Baulcombe, D.C. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 1995, 8, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Ritter, E.; Debener, T.; Barone, A.; Salamini, F.; Gebhardt, C. RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX). Mol. Gen. Genet. 1991, 227, 81–85. [Google Scholar] [CrossRef]
- Santa Cruz, S.; Baulcombe, D. Analysis of potato virus X coat protein genes in relation to resistance conferred by the genes Nx, Nb and Rx1 of potato. J. Gen. Virol. 1995, 76 Pt 8, 2057–2061. [Google Scholar] [CrossRef]
- Slootweg, E.J.; Spiridon, L.N.; Roosien, J.; Butterbach, P.; Pomp, R.; Westerhof, L.; Wilbers, R.; Bakker, E.; Bakker, J.; Petrescu, A.J.; et al. Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 2013, 162, 1510–1528. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lin, N.C.; Martin, G.B. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 2002, 109, 589–598. [Google Scholar] [CrossRef]
- Martin, G.B.; Brommonschenkel, S.H.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-Based cloning of a protein kinase gene conferring disease resistance in tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef]
- Salmeron, J.M.; Oldroyd, G.E.; Rommens, C.M.; Scofield, S.R.; Kim, H.S.; Lavelle, D.T.; Dahlbeck, D.; Staskawicz, B.J. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 1996, 86, 123–133. [Google Scholar] [CrossRef]
- Tang, X.; Frederick, R.D.; Zhou, J.; Halterman, D.A.; Jia, Y.; Martin, G.B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 1996, 274, 2060–2063. [Google Scholar] [CrossRef] [PubMed]
- Ntoukakis, V.; Balmuth, A.L.; Mucyn, T.S.; Gutierrez, J.R.; Jones, A.M.; Rathjen, J.P. The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.J.; Andriotis, V.M.; Durrant, M.C.; Rathjen, J.P. A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 2004, 16, 2809–2821. [Google Scholar] [CrossRef]
- Xing, W.; Zou, Y.; Liu, Q.; Liu, J.; Luo, X.; Huang, Q.; Chen, S.; Zhu, L.; Bi, R.; Hao, Q.; et al. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 2007, 449, 243–247. [Google Scholar] [CrossRef]
- Mucyn, T.S.; Clemente, A.; Andriotis, V.M.E.; Balmuth, A.L.; Oldroyd, G.E.D.; Staskawicz, B.J.; Rathjen, J.P. The tomato NBARC-LRR protein Prf interacts with Pto kinase In Vivo to regulate specific plant immunity. Plant Cell 2006, 18, 2792–2806. [Google Scholar] [CrossRef]
- Ntoukakis, V.; Saur, I.M.; Conlan, B.; Rathjen, J.P. The changing of the guard: The Pto/Prf receptor complex of tomato and pathogen recognition. Curr. Opin. Plant Biol. 2014, 20, 69–74. [Google Scholar] [CrossRef]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Shirasu, K.; Guerois, R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 2010, 35, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Shirasu, K. The HSP90 complex of plants. Biochim. Biophys. Acta 2012, 1823, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Kearney, B.; Staskawicz, B.J. Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature 1990, 346, 385–386. [Google Scholar] [CrossRef]
- Leister, R.T.; Dahlbeck, D.; Day, B.; Li, Y.; Chesnokova, O.; Staskawicz, B.J. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 2005, 17, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Han, S.W.; Hwang, B.K. Molecular functions of Xanthomonas type III effector AvrBsT and its plant interactors in cell death and defense signaling. Planta 2017, 245, 237–253. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, G.; Lal, N.K.; Nagalakshmi, U.; Li, Y.; Zheng, W.; Huang, P.J.; Branon, T.C.; Ting, A.Y.; Walley, J.W.; et al. TurboID-Based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 2019, 10, 3252. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Seong, K.; Thomazella, D.P.T.; Kim, J.R.; Pham, J.; Seo, E.; Cho, M.J.; Schultink, A.; Staskawicz, B.J. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in. Proc. Natl. Acad. Sci. USA 2018, 115, E10979–E10987. [Google Scholar] [CrossRef]
- Sacco, M.A.; Mansoor, S.; Moffett, P. A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 2007, 52, 82–93. [Google Scholar] [CrossRef]
- Tameling, W.I.; Baulcombe, D.C. Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 2007, 19, 1682–1694. [Google Scholar] [CrossRef]
- Tameling, W.I.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.; Goverse, A.; Shirasu, K.; Joosten, M.H. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 2010, 22, 4176–4194. [Google Scholar] [CrossRef]
- Slootweg, E.; Roosien, J.; Spiridon, L.N.; Petrescu, A.J.; Tameling, W.; Joosten, M.; Pomp, R.; van Schaik, C.; Dees, R.; Borst, J.W.; et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 2010, 22, 4195–4215. [Google Scholar] [CrossRef]
- Cao, L.; Blekemolen, M.C.; Tintor, N.; Cornelissen, B.J.C.; Takken, F.L.W. The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 2018, 11, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.J.; Palsson, L.O.; Takken, F.L.W.; Goverse, A.; et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J. Biol. Chem. 2018, 293, 3218–3233. [Google Scholar] [CrossRef] [PubMed]
- Swords, K.M.; Dahlbeck, D.; Kearney, B.; Roy, M.; Staskawicz, B.J. Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2. J. Bacteriol. 1996, 178, 4661–4669. [Google Scholar] [CrossRef]
- Tai, T.H.; Dahlbeck, D.; Clark, E.T.; Gajiwala, P.; Pasion, R.; Whalen, M.C.; Stall, R.E.; Staskawicz, B.J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 1999, 96, 14153–14158. [Google Scholar] [CrossRef]
- van der Vossen, E.A.; van der Voort, J.N.; Kanyuka, K.; Bendahmane, A.; Sandbrink, H.; Baulcombe, D.C.; Bakker, J.; Stiekema, W.J.; Klein-Lankhorst, R.M. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 2000, 23, 567–576. [Google Scholar] [CrossRef]
- Ernst, K.; Kumar, A.; Kriseleit, D.; Kloos, D.U.; Phillips, M.S.; Ganal, M.W. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 2002, 31, 127–136. [Google Scholar] [CrossRef]
- Ori, N.; Eshed, Y.; Paran, I.; Presting, G.; Aviv, D.; Tanksley, S.; Zamir, D.; Fluhr, R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 1997, 9, 521–532. [Google Scholar] [CrossRef]
- Simons, G.; Groenendijk, J.; Wijbrandi, J.; Reijans, M.; Groenen, J.; Diergaarde, P.; Van der Lee, T.; Bleeker, M.; Onstenk, J.; de Both, M.; et al. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 1998, 10, 1055–1068. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef]
- Vos, P.; Simons, G.; Jesse, T.; Wijbrandi, J.; Heinen, L.; Hogers, R.; Frijters, A.; Groenendijk, J.; Diergaarde, P.; Reijans, M.; et al. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 1998, 16, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Abramovitch, R.B.; Kim, Y.J.; Chen, S.; Dickman, M.B.; Martin, G.B. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003, 22, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Ronald, P.C.; Salmeron, J.M.; Carland, F.M.; Staskawicz, B.J. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 1992, 174, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- Ballvora, A.; Ercolano, M.R.; Weiss, J.; Meksem, K.; Bormann, C.A.; Oberhagemann, P.; Salamini, F.; Gebhardt, C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002, 30, 361–371. [Google Scholar] [CrossRef] [PubMed]
- van der Lee, T.; Testa, A.; Robold, A.; van ‘t Klooster, J.; Govers, F. High-Density genetic linkage maps of Phytophthora infestans reveal trisomic progeny and chromosomal rearrangements. Genetics 2004, 167, 1643–1661. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; van der Vossen, E.A.; Kuang, H.; Vleeshouwers, V.G.; Zhang, N.; Borm, T.J.; van Eck, H.J.; Baker, B.; Jacobsen, E.; Visser, R.G. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005, 42, 251–261. [Google Scholar] [CrossRef]
- Armstrong, M.R.; Whisson, S.C.; Pritchard, L.; Bos, J.I.; Venter, E.; Avrova, A.O.; Rehmany, A.P.; Böhme, U.; Brooks, K.; Cherevach, I.; et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 2005, 102, 7766–7771. [Google Scholar] [CrossRef]
- Vossen, J.H.; van Arkel, G.; Bergervoet, M.; Jo, K.R.; Jacobsen, E.; Visser, R.G. The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor. Appl. Genet. 2016, 129, 1785–1796. [Google Scholar] [CrossRef]
- Song, J.; Bradeen, J.M.; Naess, S.K.; Raasch, J.A.; Wielgus, S.M.; Haberlach, G.T.; Liu, J.; Kuang, H.; Austin-Phillips, S.; Buell, C.R.; et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. USA 2003, 100, 9128–9133. [Google Scholar] [CrossRef]
- van der Vossen, E.; Sikkema, A.; Hekkert, B.; Gros, J.; Stevens, P.; Muskens, M.; Wouters, D.; Pereira, A.; Stiekema, W.; Allefs, S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003, 36, 867–882. [Google Scholar] [CrossRef]
- Oh, S.K.; Young, C.; Lee, M.; Oliva, R.; Bozkurt, T.O.; Cano, L.M.; Win, J.; Bos, J.I.; Liu, H.Y.; van Damme, M.; et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 2009, 21, 2928–2947. [Google Scholar] [CrossRef] [PubMed]
- van der Vossen, E.A.G.; Gros, J.; Sikkema, A.; Muskens, M.; Wouters, D.; Wolters, P.; Pereira, A.; Allefs, S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 2005, 44, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Querci, M.; Kanyuka, K.; Baulcombe, D.C. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 2000, 21, 73–81. [Google Scholar] [CrossRef]
- Brommonschenkel, S.H.; Frary, A.; Tanksley, S.D. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormelink, R. The Tomato spotted wilt virus cell-to-cell movement protein (NSM ) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef]
- Calder, V.L.; Palukaitis, P. Nucleotide sequence analysis of the movement genes of resistance breaking strains of tomato mosaic virus. J. Gen. Virol. 1992, 73 Pt 1, 165–168. [Google Scholar] [CrossRef]
- Lanfermeijer, F.C.; Warmink, J.; Hille, J. The products of the broken Tm-2 and the durable Tm-2(2) resistance genes from tomato differ in four amino acids. J. Exp. Bot. 2005, 56, 2925–2933. [Google Scholar] [CrossRef]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.; de Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1037–1049. [Google Scholar] [CrossRef]
- Yoon, M.; Rikkerink, E.H.A. Rpa1 mediates an immune response to avrRpm1Psa and confers resistance against Pseudomonas syringae pv. actinidiae. Plant J. 2019. [Google Scholar] [CrossRef]
- Bonas, U.; Conrads-Strauch, J.; Balbo, I. Resistance in tomato to Xanthomonas campestris pv vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol. Gen. Genet. 1993, 238, 261–269. [Google Scholar] [CrossRef]
- Kay, S.; Boch, J.; Bonas, U. Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant Microbe Interact. 2005, 18, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; Ballvora, A.; Gürlebeck, D.; Peart, J.; Baulcombe, D.; Ganal, M.; Baker, B.; Bonas, U.; Lahaye, T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004, 37, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Paal, J.; Henselewski, H.; Muth, J.; Meksem, K.; Menéndez, C.M.; Salamini, F.; Ballvora, A.; Gebhardt, C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004, 38, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Erickson, F.L.; Holzberg, S.; Calderon-Urrea, A.; Handley, V.; Axtell, M.; Corr, C.; Baker, B. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 1999, 18, 67–75. [Google Scholar] [CrossRef]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Schultink, A.; Qi, T.; Lee, A.; Steinbrenner, A.D.; Staskawicz, B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017, 92, 787–795. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network properties of robust immunity in plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef]
- Kadota, Y.; Liebrand, T.W.H.; Goto, Y.; Sklenar, J.; Derbyshire, P.; Menke, F.L.H.; Torres, M.A.; Molina, A.; Zipfel, C.; Coaker, G.; et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector-and PAMP-triggered immunity in plants. New Phytol. 2019, 221, 2160–2175. [Google Scholar] [CrossRef]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef] [PubMed]
- van der Burgh, A.M.; Joosten, M.H.A.J. Plant immunity: Thinking outside and inside the box. Trends Plant Sci. 2019, 24, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Leibman-Markus, M.; Pizarro, L.; Schuster, S.; Lin, Z.J.D.; Gershony, O.; Bar, M.; Coaker, G.; Avni, A. The intracellular nucleotide-binding leucine-rich repeat receptor (SlNRC4a) enhances immune signalling elicited by extracellular perception. Plant Cell Environ. 2018, 41, 2313–2327. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Peng, H.C.; Li, B.; Atamian, H.S.; Takken, F.L.; Kaloshian, I. The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant J. 2011, 67, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Gabriëls, S.H.; Vossen, J.H.; Ekengren, S.K.; van Ooijen, G.; Abd-El-Haliem, A.M.; van den Berg, G.C.; Rainey, D.Y.; Martin, G.B.; Takken, F.L.; de Wit, P.J.; et al. An NB-LRR protein required for HR signalling mediated by both extra-and intracellular resistance proteins. Plant J. 2007, 50, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Belhaj, K.; Bozkurt, T.O.; Birk, M.S.; Kamoun, S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 2016, 209, 1344–1352. [Google Scholar] [CrossRef]
- Qi, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 2011, 12, 702–708. [Google Scholar] [CrossRef]
- Kud, J.; Wang, W.; Gross, R.; Fan, Y.; Huang, L.; Yuan, Y.; Gray, A.; Duarte, A.; Kuhl, J.C.; Caplan, A.; et al. The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 2019, 15, e1007720. [Google Scholar] [CrossRef]
- Jacob, F.; Kracher, B.; Mine, A.; Seyfferth, C.; Blanvillain-Baufumé, S.; Parker, J.E.; Tsuda, K.; Schulze-Lefert, P.; Maekawa, T. A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol. 2018, 217, 1667–1680. [Google Scholar] [CrossRef]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 286. [Google Scholar] [CrossRef]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef] [PubMed]
- Boschi, F.; Schvartzman, C.; Murchio, S.; Ferreira, V.; Siri, M.I.; Galván, G.A.; Smoker, M.; Stransfeld, L.; Zipfel, C.; Vilaró, F.L.; et al. Enhanced bacterial wilt resistance in potato through expression of Arabidopsis EFR and introgression of quantitative resistance from Solanum commersonii. Front. Plant Sci. 2017, 8, 1642. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Han, M.; Blanco-Portales, R.; Song, W.; Weide, R.; Guo, L.Y.; van der Vossen, E.A.; Govers, F. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol. J. 2014, 12, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.Y.; Yeh, Y.H.; Liu, A.C.; Cheng, C.P.; Zimmerli, L. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014, 79, 243–255. [Google Scholar] [CrossRef]
- Kunwar, S.; Iriarte, F.; Fan, Q.; Evaristo da Silva, E.; Ritchie, L.; Nguyen, N.S.; Freeman, J.H.; Stall, R.E.; Jones, J.B.; Minsavage, G.V.; et al. Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 2018, 108, 1402–1411. [Google Scholar] [CrossRef]
- Lacombe, S.; Bangratz, M.; Vignols, F.; Brugidou, C. The rice yellow mottle virus P1 protein exhibits dual functions to suppress and activate gene silencing. Plant J. 2010, 61, 371–382. [Google Scholar] [CrossRef]
- Hao, G.; Pitino, M.; Duan, Y.; Stover, E. Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from Nicotiana benthamiana. Mol. Plant Microbe Interact. 2016, 29, 132–142. [Google Scholar] [CrossRef]
- Jehle, A.K.; Lipschis, M.; Albert, M.; Fallahzadeh-Mamaghani, V.; Fürst, U.; Mueller, K.; Felix, G. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 2013, 25, 2330–2340. [Google Scholar] [CrossRef]
- Jehle, A.K.; Fürst, U.; Lipschis, M.; Albert, M.; Felix, G. Perception of the novel MAMP eMax from different Xanthomonas species requires the Arabidopsis receptor-like protein ReMAX and the receptor kinase SOBIR. Plant Signal. Behav. 2013, 8, e27408. [Google Scholar] [CrossRef]
- Wu, J.; Reca, I.B.; Spinelli, F.; Lironi, D.; De Lorenzo, G.; Poltronieri, P.; Cervone, F.; Joosten, M.H.A.J.; Ferrari, S.; Brutus, A. An EFR-Cf-9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1-and BAK1-dependent recognition of elf18. Mol. Plant Pathol. 2019, 20, 751–764. [Google Scholar] [CrossRef]
- Hillmer, R.A.; Tsuda, K.; Rallapalli, G.; Asai, S.; Truman, W.; Papke, M.D.; Sakakibara, H.; Jones, J.D.G.; Myers, C.L.; Katagiri, F. The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 2017, 13, e1006639. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-Spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Greene, G.H.; Yoo, H.; Liu, L.; Marqués, J.; Motley, J.; Dong, X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane trafficking in plant immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef]
- Yang, F.; Kimberlin, A.N.; Elowsky, C.G.; Liu, Y.; Gonzalez-Solis, A.; Cahoon, E.B.; Alfano, J.R. A plant immune receptor degraded by selective autophagy. Mol. Plant 2019, 12, 113–123. [Google Scholar] [CrossRef]
- Li, P.; Day, B. Battlefield cytoskeleton: Turning the tide on plant immunity. Mol. Plant Microbe Interact. 2019, 32, 25–34. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Anfoka, G.; Moshe, A.; Fridman, L.; Amrani, L.; Rotem, O.; Kolot, M.; Zeidan, M.; Czosnek, H.; Gorovits, R. Tomato yellow leaf curl virus infection mitigates the heat stress response of plants grown at high temperatures. Sci. Rep. 2016, 6, 19715. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 10. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef]
- Roberts, R.; Mainiero, S.; Powell, A.F.; Liu, A.E.; Shi, K.; Hind, S.R.; Strickler, S.R.; Collmer, A.; Martin, G.B. Natural variation for unusual host responses and flagellin-mediated immunity against Pseudomonas syringae in genetically diverse tomato accessions. New Phytol. 2019, 223, 447–461. [Google Scholar] [CrossRef] [PubMed]
| Receptor | Type | Species | Ligand | Ligand Source | Reference |
|---|---|---|---|---|---|
| CSPR | RLK | Nicotiana benthamiana | csp22 (cold shock protein) | Pathogen (various bacterial species) | Saur et al. [56] |
| CORE | RLK | Solanum lycopersicum | csp22 (cold shock protein) | Pathogen (various bacterial species) | Wang et al. [16] |
| FLS2 | RLK | Solanum lycopersicum | flg22, flg15 (flagellin) | Pathogen (various bacterial species) | Robatzek et al. [7] |
| FLS3 | RLK | Solanum lycopersicum | flgII-28 (flagellin) | Pathogen (various bacterial species) | Hind et al. [6] |
| I-3 | RLK | Solanum lycopersicum | Avr3 | Pathogen (Fusarium oxysporum) | Catanzariti et al. [57] |
| LRPK1 | RLK | Solanum tuberosum | ? | Pathogen (Phytophthora infestans) | Wang et al. [58] |
| Nt-Sd-RLK | RLK | Nicotiana tabacum | LPS (lipopolysaccharide)? | Pathogen (bacteria) | Sanabria et al. [10] |
| PEPR1 | RLK | Solanum lycopersicum | SlPep6 | Host | Lori et al. [59] |
| PSKR1 | RLK | Solanum lycopersicum | PSK (phytosulfokine) | Host | Zhang et al. [60] |
| PSKR2 | RLK | Solanum lycopersicum | PSK (phytosulfokine) | Host | Zhang et al. [60] |
| Rmprp-1 | RLK | Capsicum annuum | ? | Pathogen (Myzus persicae) | Sun et al. [61] |
| SlLYK1 | RLK | Solanum lycopersicum | Chitin? | ? | Liao et al. [9] |
| SR160 | RLK | Solanum lycopersicum | ? | ? | Scheer and Ryan [62] |
| SYR1 | RLK | Solanum lycopersicum | systemin | Host | Wang et al [8] |
| SYR2 | RLK | Solanum lycopersicum | systemin | Host | Wang et al [8] |
| Cf-2 | RLP | Solanum lycopersicum | Avr2 | Pathogen (Cladosporium fulvum) | Dixon et al. [63]; Jones et al. [64]; Thomas et al. [65] |
| Cf-4 | RLP | Solanum lycopersicum | Avr4 | Pathogen (C. fulvum) | Thomas et al. [66]; Jones et al. [64]; Thomas et al. [65] |
| Cf-4A (Hcr9-4E) | RLP | Solanum lycopersicum | Avr4E | Pathogen (C. fulvum) | Takken et al. [67]; Takken et al., 1998 [68] |
| Cf-5 | RLP | Solanum lycopersicum | Avr5 | Pathogen (C. fulvum) | Dixon et al. [69]; Jones et al. [64]; Thomas et al. [65] |
| Cf-6 | RLP | Solanum lycopersicum | ? | Pathogen (C. fulvum) | Grushtskaia et al. [70] |
| Cf-9 | RLP | Solanum lycopersicum | Avr9 | Pathogen (C. fulvum) | Jones et al., 1994 [71]; Hammond-Kossack et al. [72]; Hammond-Kossack et al. [73]; van der Hoorn et al. [74] |
| Cf-9B (Hcr9-9B) | RLP | Solanum lycopersicum | ? | Pathogen (C. fulvum) | Parniske et al. [75] |
| Cf-ECP2 | RLP | Solanum lycopersicum | ECP2 | Pathogen (C. fulvum) | Laugé et al. [76] |
| CuRe1 | RLP | Solanum lycopersicum | ? | Parasitic plant (Cuscuta reflexa) | Hegenauer et al. [21] |
| EIX1 | RLP | Solanum lycopersicum | Ethylene-inducing xylanase (EIX) | Pathogen (Trichoderma spp.) | Ron and Avni [77]; Bar et al. [78] |
| EIX2 | RLP | Solanum lycopersicum | Ethylene-inducing xylanase (EIX) | Pathogen (Trichoderma spp.) | Ron and Avni [77] |
| ELR | RLP | Solanum tuberosum | INF1 elicitin | Pathogen (P. infestans) | Du et al. [19] |
| I | RLP | Solanum lycopersicum | Avr1 | Pathogen (F. oxysporum) | Catanzariti et al. [79] |
| I-7 | RLP | Solanum lycopersicum | ? | Pathogen (F. oxysporum) | Gonzalez-Cendales et al. [80] |
| RXEG1 | RLP | Nicotiana benthamiana | XEG1 (glycoside hydrolase 12 protein) | Pathogen (P. sojae) | Wang et al. [81] |
| Ve1 | RLP | Solanum lycopersicum | Ave1 | Pathogen (Verticillium dahliae, V. albo-atrum) | Kawchuk et al. [82]; Fradin et al. [83]; Castroverde et al. [37] |
| Ve2 | RLP | Solanum lycopersicum | ? | Pathogen (V. dahliae, V. albo-atrum) | Kawchuk et al. [82]; Nazar et al. [84] |
| Receptor | Type | Species | Ligand | Ligand Source | Reference |
|---|---|---|---|---|---|
| Bs2 | CNL | Capsicum annuum | AvrBs2 | Xanthomonas campestris | Swords et al. [124]; Tai et al. [125] |
| Gpa2 (Rxh1) | CNL | Solanum tuberosum | RBP-1 | Globodera pallida | van der Vossen et al. [126] |
| Hero | CNL | Solanum lycopersicum | Globodera rostochiensis, G. pallida | Ernst et al. [127] | |
| I-2 | CNL | Solanum lycopersicum | Avr2 | Fusarium oxysporum | Ori et al. [128] Simons et al. [129] |
| Mi-1.2 | CNL | Solanum lycopersicum | Meloidogyne incognita, M. arenaria, M. javanica, Bemisia tabacci | Milligan et al. [130] Vos et al. [131] | |
| Prf | CNL | Solanum lycopersicum | AvrPto, AvrPtoB | Pseudomonas syringae | Salmeron et al. [101]; Abramovitch et al. [132]; Ronald et al. [133] |
| R1 | CNL | Solanum tuberosum | Avr1 | Phytophthora infestans | Ballvora et al. [134]; van der Lee et al. [135] |
| R3a | CNL | Solanum tuberosum | Avr3a | Phytophthora infestans | Huang et al. [136]; Armstrong et al. [137] |
| R8 | CNL | Solanum tuberosum | Avr8 | Phytophthora infestans | Vossen et al. [138] |
| Rpi-blb1 (RB) | CNL | Solanum bulbocastanum | Avrblb1 | Phytophthora infestans | Song et al. [139]; van der Vossen et al. [140]; Oh et al. [141] |
| Rpi-blb2 | CNL | Solanum bulbocastanum | Avrblb2 | Phytophthora infestans | van der Vossen et al. [142]; Oh et al. [141] |
| Rx1 | CNL | Solanum tuberosum | CP | Potato virus x | Bendahmane et al. [95] |
| Rx2 | CNL | Solanum acaule | CP | Potato virus x | Bendahmane et al. [143] |
| Sw-5 | CNL | Solanum lycopersicum | Tospovirus | Brommonschenkel et al. [144] | |
| Sw-5b | CNL | Solanum tuberosum | NSm (viral movement protein) | Tospovirus | Hallwass et al. [145]; Zhu et al. [92] |
| Tm-2 | CNL | Solanum lycopersicum | MP | Tomato mosaic virus | Calder et al. [146]; Lanfermeijer et al. [147] |
| Tm-2-2 | CNL | Solanum lycopersicum | MP | Tomato mosaic virus | Calder et al. [146]; Lanfermeijer et al. [148] |
| Rpa1 | CNL | Nicotiana tabacum | AvrRpm1psa | Pseudomonas syringae pv. actinidiae | Yoon and Rikkerink [149] |
| Bs4 | TNL | Solanum lycopersicum | AvrBs4, Hax4 | Xanthomonas campestris | Bonas et al. [150]; Kay et al. [151]; Schornack et al. [152] |
| Gro1-4 | TNL | Solanum tuberosum | Globodera rostochiensis | Paal et al. [153] | |
| N | TNL | Nicotiana tabacum | Helicase | Tobacco mosaic virus | Erickson et al. [154]; Whitham et al. [155] |
| Roq1 | TNL | Nicotiana benthamiana | XopQ HopQ1 | Xanthomonas campestris Pseudomonas syringae | Schultink et al. [156] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Castroverde, C.D.M. Diversity, Function and Regulation of Cell Surface and Intracellular Immune Receptors in Solanaceae. Plants 2020, 9, 434. https://doi.org/10.3390/plants9040434
Kim JH, Castroverde CDM. Diversity, Function and Regulation of Cell Surface and Intracellular Immune Receptors in Solanaceae. Plants. 2020; 9(4):434. https://doi.org/10.3390/plants9040434
Chicago/Turabian StyleKim, Jong Hum, and Christian Danve M. Castroverde. 2020. "Diversity, Function and Regulation of Cell Surface and Intracellular Immune Receptors in Solanaceae" Plants 9, no. 4: 434. https://doi.org/10.3390/plants9040434
APA StyleKim, J. H., & Castroverde, C. D. M. (2020). Diversity, Function and Regulation of Cell Surface and Intracellular Immune Receptors in Solanaceae. Plants, 9(4), 434. https://doi.org/10.3390/plants9040434





