Shedding the Last Layer: Mechanisms of Root Cap Cell Release
Abstract
1. Introduction
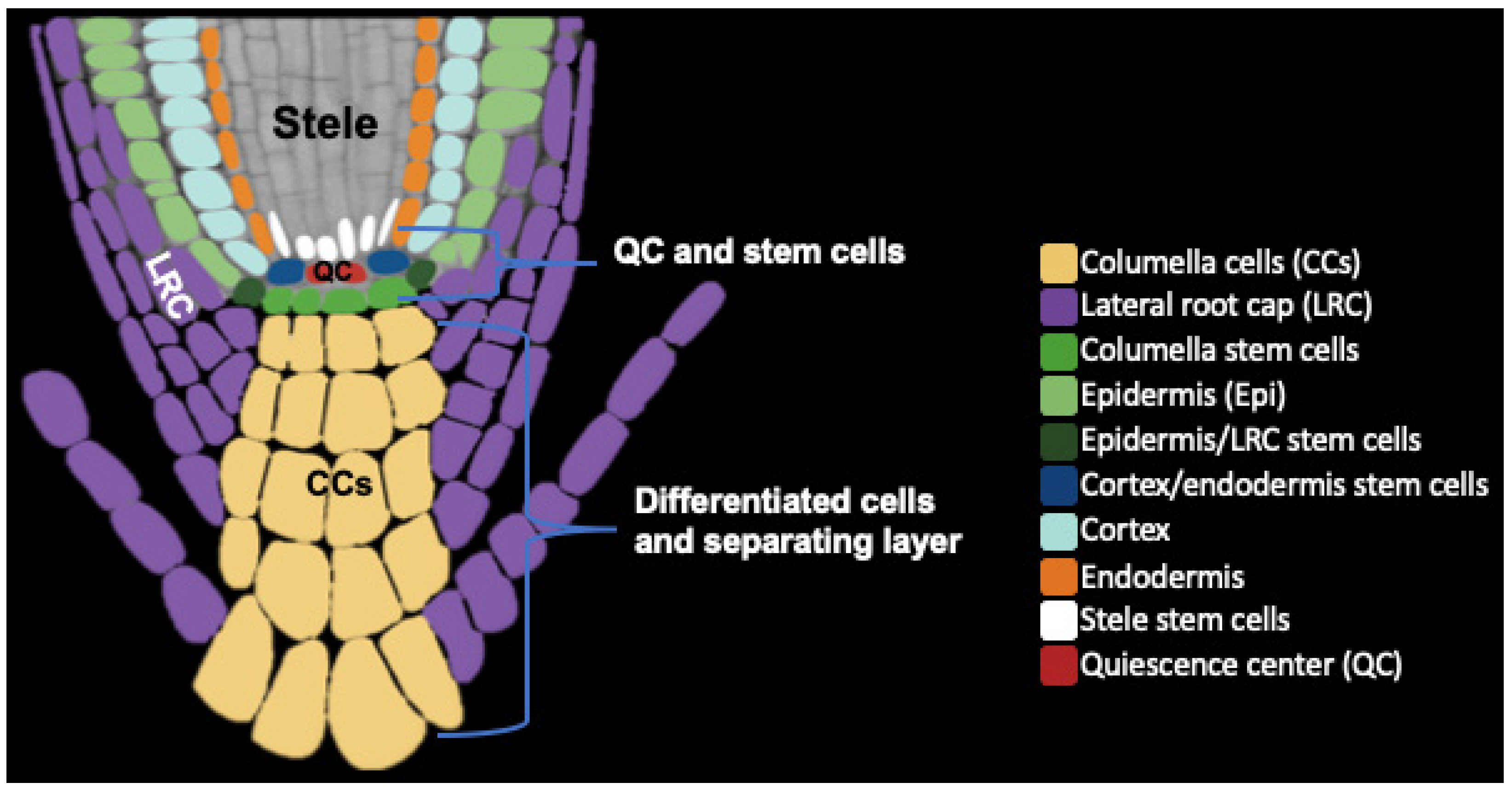
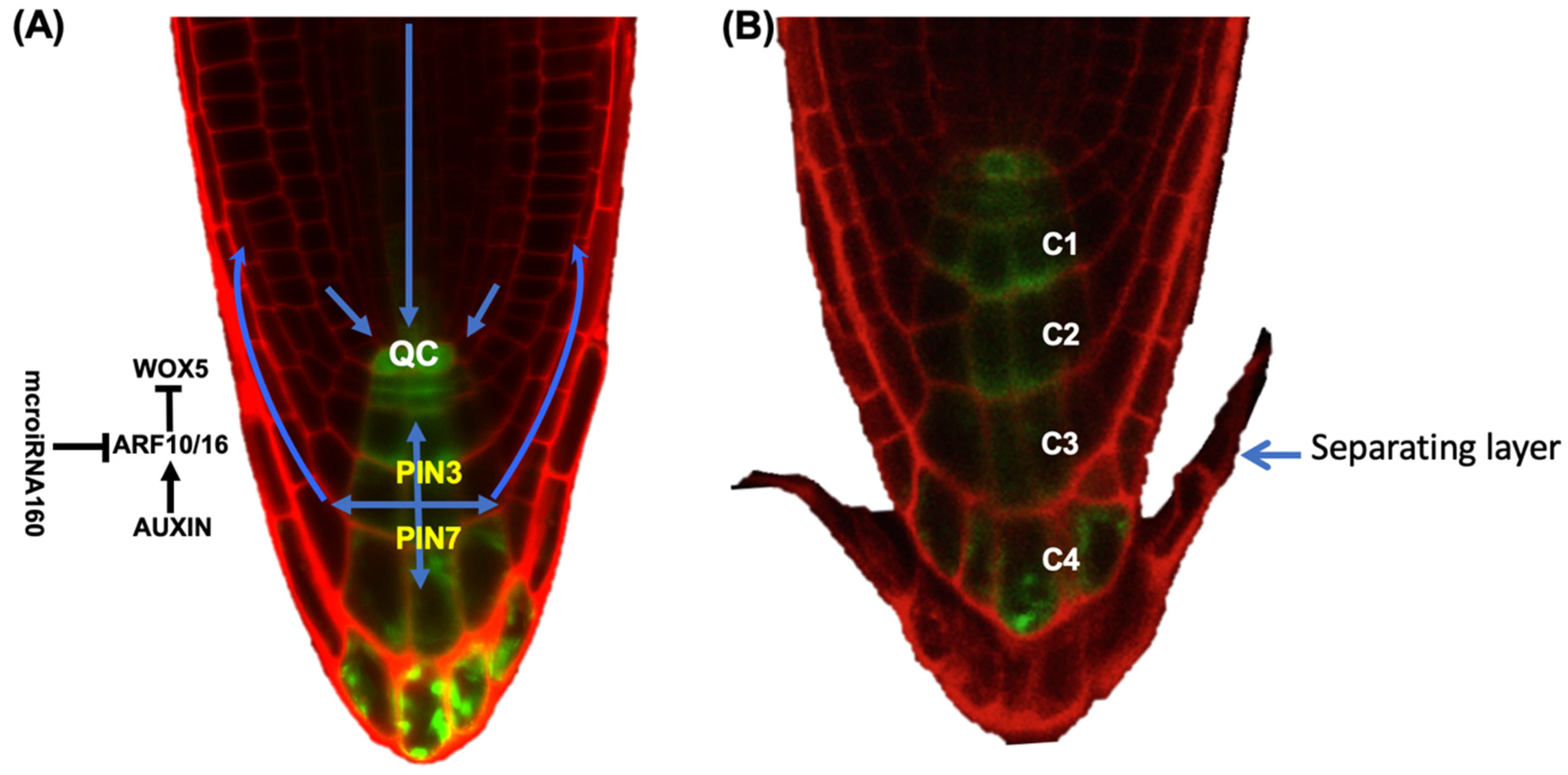
2. Root Cap Structure and BLC Release
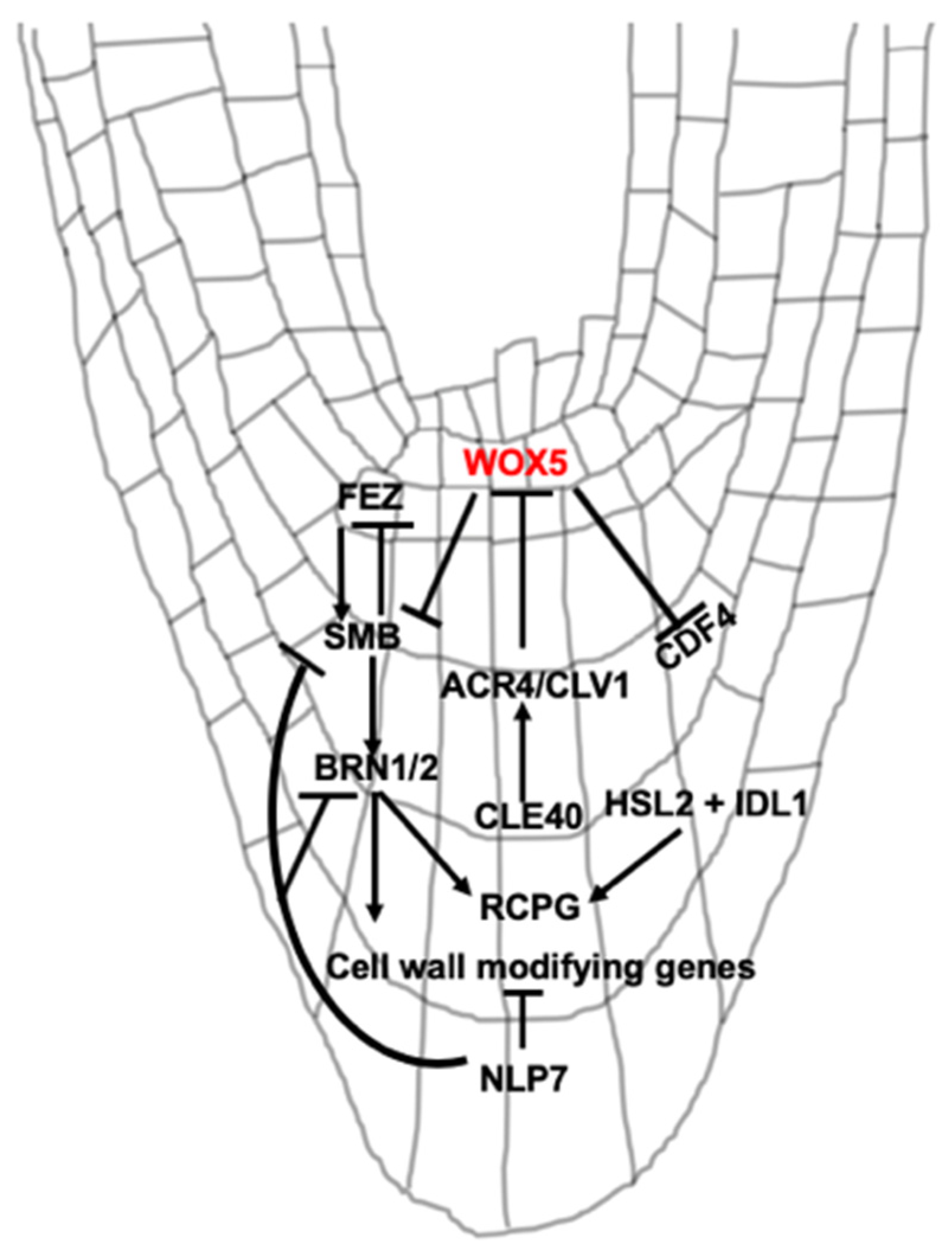
3. Signaling Peptide and Transcriptional Regulation of BLC Release
4. The Role of Hormones in BLC Release
5. Variations in BC Release Among Species
6. BC and BLC Protective Functions in Biotic and Abiotic Stresses
7. Conclusions and Future Perspectives
- How do external stresses impact the release of root cap cells and the molecular pathways that control release? How are such stresses communicated in the root cap such that root cap cell production and release remain linked during environmental stress?
- Do differences in either the timing or type (BLC vs BC) of release impact whole root growth and root architecture? Do BLC and BC release impact root cap protection differently?
- How are signals between the separating layer and stem cells communicated and what are additional signals?
- What are the mechanisms used by the auxin pathway to regulate cell production from CSCs and the release of BLCs?
- What other transcription factors are involved in BLC and BC release? Are there other hormones besides auxin involved in BLC release? Does WOX5 have an impact on BLC release?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knox, J.P. Cell adhesion, cell separation and plant morphogenesis. Plant J. 1992, 2, 137–141. [Google Scholar] [CrossRef]
- Jarvis, M.C.; Briggs, S.P.H.; Knox, J.P. Intercellular adhesion and cell separation in plants. Plant Cell Environ. 2003, 26, 977–989. [Google Scholar] [CrossRef]
- Strohm, A.K.; Baldwin, K.L.; Masson, P.H. Molecular mechanisms of root gravity sensing and signal transduction. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Tsugeki, R.; Fedoroff, N.V. Genetic ablation of root cap cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 12941–12946. [Google Scholar] [CrossRef]
- Massa, G.D.; Gilroy, S. Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J. 2003, 33, 435–445. [Google Scholar] [CrossRef]
- Barlow, P.W. The root cap: Cell dynamics, cell differentiation and cap function. J. Plant Growth Regul. 2002, 21, 261–286. [Google Scholar] [CrossRef]
- Fendrych, M.; Van Hautegem, T.; Van Durme, M.; Olvera-Carrillo, Y.; Huysmans, M.; Karimi, M.; Lippens, S.; Guérin, C.J.; Krebs, M.; Schumacher, K.; et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 2014, 24, 931–940. [Google Scholar] [CrossRef]
- Hawes, M.C.; Brigham, L.A.; Wen, F.; Woo, H.H.; Zhu, Y. FUNCTION OF ROOT BORDER CELLS IN PLANT HEALTH: Pioneers in the Rhizosphere. Annu. Rev. Phytopathol. 1998, 36, 311–327. [Google Scholar] [CrossRef]
- Hawes, M.C.; Lin, H.J. Correlation of pectolytic enzyme activity with the programmed release of cells from root caps of pea (Pisum sativum). Plant Physiol. 1990, 94, 1855–1859. [Google Scholar] [CrossRef]
- Driouich, A.; Durand, C.; Vicre-Gibouin, M. Formation and separation of root border cells. Trends Plant Sci. 2007, 12, 14–19. [Google Scholar] [CrossRef]
- Vermeer, J.; McCully, M.E. The rhizosphere in Zea: New insight into its structure and development. Planta 1982, 156, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Hawes, M.C.; Gunawardena, U.; Miyasaka, S.; Zhao, X. The role of root border cells in plant defense. Trends Plant Sci. 2000, 5, 128–133. [Google Scholar] [CrossRef]
- Hawes, M.C.; Bengough, G.; Cassab, G.; Ponce, G. Root caps and rhizosphere. J. Plant Growth Regul. 2002, 21, 352–367. [Google Scholar] [CrossRef]
- Vicré, M.; Santaella, C.; Blanchet, S.; Gateau, A.; Driouich, A. Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 2005, 138, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Plancot, B.; Santaella, C.; Jaber, R.; Kiefer-Meyer, M.C.; Follet-Gueye, M.L.; Leprince, J.; Gattin, I.; Souc, C.; Driouich, A.; Vicré-Gibouin, M. Deciphering the responses of root border-like cells of Arabidopsis and flax to pathogen-derived elicitors. Plant Physiol. 2013, 163, 1584–1597. [Google Scholar] [CrossRef] [PubMed]
- Driouich, A.; Follett-Gueye, M.; Vicre-Gibouin, M.; Hawes, M. Root border cells and secretions are critical elements in plant host defense. Curr. Opin. Plant Biol. 2013, 16, 489–495. [Google Scholar] [CrossRef]
- Driouich, A.; Durand, C.; Cannesan, M.A.; Percoco, G.; Vicré-Gibouin, M. Border cells versus border-like cells: Are they alike? J. Exp. Bot. 2010, 61, 3827–3831. [Google Scholar] [CrossRef]
- Hawes, M.; Allen, C.; Turgeon, B.G.; Curlango-Rivera, G.; Tran, T.M.; Huskey, D.A.; Xiong, Z. Root Border Cells and Their Role in Plant Defense. Annu. Rev. Phytopathol. 2016, 54, 143–161. [Google Scholar] [CrossRef]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.C.; De Winter, F.; Parizot, B.; Mertens, J.; Karimi, M.; Fendrych, M.; Nowack, M.K. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell 2018, 30, 2197–2213. [Google Scholar] [CrossRef]
- Maeda, K.; Kunieda, T.; Tamura, K.; Hatano, K.; Hara-Nishimura, I.; Shimada, T. Identification of periplasmic root-cap mucilage in developing columella cells of Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 1296–1303. [Google Scholar] [CrossRef]
- Cai, M.; Wang, N.; Xing, C.; Wang, F.; Wu, K.; Du, X. Immobilization of aluminum with mucilage secreted by root cap and root border cells is related to aluminum resistance in Glycine max L. Environ. Sci. Pollut. Res. 2013, 20, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Berhin, A.; de Bellis, D.; Franke, R.B.; Buono, R.A.; Nowack, M.K.; Nawrath, C. The Root Cap Cuticle: A Cell Wall Structure for Seedling Establishment and Lateral Root Formation. Cell 2019, 176, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, C.; Jin, X.; Grönlund, A.; Fischer, U. A Local Auxin Gradient Regulates Root Cap Self-Renewal and Size Homeostasis. Curr. Biol. 2018, 28, 2581–2587. [Google Scholar] [CrossRef]
- Shi, C.L.; von Wangenheim, D.; Herrmann, U.; Wildhagen, M.; Kulik, I.; Kopf, A.; Ishida, T.; Olsson, V.; Anker, M.K.; Albert, M.; et al. The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nat. Plants 2018, 4, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Mravec, J.; Guo, X.; Hansen, A.R.; Schückel, J.; Kračun, S.K.; Mikkelsen, M.D.; Mouille, G.; Johansen, I.E.; Ulvskov, P.; Domozych, D.S.; et al. Pea border cell maturation and release involve complex cell wall structural dynamics. Plant Physiol. 2017, 174, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.; Vicré-Gibouin, M.; Follet-Gueye, M.L.; Duponchel, L.; Moreau, M.; Lerouge, P.; Driouich, A. The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 2009, 150, 1411–1421. [Google Scholar] [CrossRef]
- Bouton, S.; Leboeuf, E.; Mouille, G.; Leydecker, M.-T.; Talbotec, J.; Granier, F.; Lahaye, M.; Höfte, H.; Truong, H.-N. QUASIMODO1 Encodes a Putative Membrane-Bound Glycosyltransferase Required for Normal Pectin Synthesis and Cell Adhesion in Arabidopsis. Plant Cell 2002, 14, 2577–2590. [Google Scholar] [CrossRef]
- Mravec, J.; Kračun, S.K.; Rydahl, M.G.; Westereng, B.; Miart, F.; Clausen, M.H.; Fangel, J.U.; Daugaard, M.; Van Cutsem, P.; De Fine Licht, H.H.; et al. Tracking Developmentally Regulated Post-Synthetic Processing of Homogalacturonan and Chitin Using Reciprocal Oligosaccharide Probes. Development 2014, 141, 4841–4850. [Google Scholar] [CrossRef]
- Stephenson, M.B.; Hawes, M.C. Correlation of Pectin Methylesterase Activity in Root Caps of Pea with Root Border Cell Separation. Plant Physiol. 1994, 106, 739–745. [Google Scholar] [CrossRef]
- Wen, F.; Zhu, Y.; Hawes, M.C. Effect of Pectin Methylesterase Gene Expression on Pea Root Development. Plant Cell 1999, 11, 1129–1140. [Google Scholar] [CrossRef]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef]
- Drisch, R.C.; Stahl, Y. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front. Plant Sci. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rahni, R.; Efroni, I.; Birnbaum, K.D. A Case for Distributed Control of Local Stem Cell Behavior in Plants. Dev. Cell 2016, 6, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Augstein, F.; Carlsbecker, A. Getting to the roots: A developmental genetic view of root anatomy and function from Arabidopsis to lycophytes. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Forzani, C.; Aichinger, E.; Sornay, E.; Willemsen, V.; Laux, T.; Dewitte, W.; Murray, J.A.H. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 2014, 24, 1939–1944. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef]
- Bennett, T.; van den Toorn, A.; Willemsen, V.; Scheres, B. Precise control of plant stem cell activity through parallel regulatory inputs. Development 2014, 141, 4055–4064. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A Signaling Module Controlling the Stem Cell Niche in Arabidopsis Root Meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef]
- Stahl, Y.; Grabowski, S.; Bleckmann, A.; Kühnemuth, R.; Weidtkamp-Peters, S.; Pinto, K.G.; Kirschner, G.K.; Schmid, J.B.; Wink, R.H.; Hülsewede, A.; et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 2013, 23, 362–371. [Google Scholar] [CrossRef]
- Berckmans, B.; Kirschner, G.; Gerlitz, N.; Stadler, R.; Simmon, R. CLE40 signalling regulates root stem cell fate. Plant Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Wink, R.H.; Simon, R. Mathematical modelling of WOX5-and CLE40-mediated columella stem cell homeostasis in Arabidopsis. J. Exp. Bot. 2015, 66, 5375–5384. [Google Scholar] [CrossRef] [PubMed]
- Pi, L.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-Derived WOX5 Signal Maintains Root Columella Stem Cells through Chromatin-Mediated Repression of CDF4 Expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, V.; Bauch, M.; Bennett, T.; Campilho, A.; Wolkenfelt, H.; Xu, J.; Haseloff, J.; Scheres, B. The NAC Domain Transcription Factors FEZ and SOMBRERO Control the Orientation of Cell Division Plane in Arabidopsis Root Stem Cells. Dev. Cell 2008, 15, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; van den Toorn, A.; Sanchez-Perez, G.F.; Campilho, A.; Willemsen, V.; Snel, B.; Scheres, B. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 2010, 22, 640–654. [Google Scholar] [CrossRef]
- Kamiya, M.; Higashio, S.Y.; Isomoto, A.; Kim, J.M.; Seki, M.; Miyashima, S.; Nakajima, K. Control of root cap maturation and cell detachment by BEARSKIN transcription factors in Arabidopsis. Development 2016, 143, 4063–4072. [Google Scholar] [CrossRef]
- Karve, R.; Suárez-Román, F.; Iyer-Pascuzzi, A.S. The transcription factor NIN-LIKE PROTEIN7 controls border-like cell release. Plant Physiol. 2016, 171, 2101–2111. [Google Scholar] [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef]
- Billou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Frimi, J.; Heldstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, L.; Hawes, M.C.; Rost, T.L. The production and release of living root cap border cells is a function of root apical meristem type in dicotyledonous angiosperm plants. Ann. Bot. 2006, 97, 917–923. [Google Scholar] [CrossRef]
- Mravec, J. Border cell release: Cell separation without cell wall degradation? Plant Signal. Behav. 2017, 12, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Koroney, A.S.; Plasson, C.; Pawlak, B.; Sidikou, R.; Driouich, A.; Menu-Bouaouiche, L.; Vicré-Gibouin, M. Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. Bot. 2016, 118, 797–808. [Google Scholar] [CrossRef]
- Brigham, L.A.; Woo, H.H.; Nicoll, S.M.; Hawes, M.C. Differential expression of proteins and mRNAs from border cells and root tips of pea. Plant Physiol. 1995, 109, 457–463. [Google Scholar] [CrossRef]
- Wen, F.; Vanetten, H.D.; Tsaprailis, G.; Hawes, M.C. Extracellular proteins in pea root tip and border cell exudates. Plant Physiol. 2007, 143, 773–783. [Google Scholar] [CrossRef]
- Cannesan, M.A.; Gangneux, C.; Lanoue, A.; Giron, D.; Laval, K.; Hawes, M.; Driouich, A.; Vicré-Gibouin, M. Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 2011, 108, 459–469. [Google Scholar] [CrossRef]
- Wen, F.; White, G.J.; Vanetten, H.D.; Xiong, Z.; Hawes, M.C. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 2009, 151, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.M.; MacIntyre, A.; Hawes, M.; Allen, C. Escaping Underground Nets: Extracellular DNases Degrade Plant Extracellular Traps and Contribute to Virulence of the Plant Pathogenic Bacterium Ralstonia solanacearum. PLoS Pathog. 2016, 12, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, U.; Hawes, M.C. Tissue specific localization of root infection by fungal pathogens: Role of root border cells. Mol. Plant-Microbe Interact. 2002, 15, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, U.; Rodriguez, M.; Straney, D.; Romeo, J.T.; VanEtten, H.D.; Hawes, M.C. Tissue-specific localization of pea root infection by Nectria haematococca. Mechanisms and consequences. Plant Physiol. 2005, 137, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Wang, W.; Curlango-Rivera, G.; Xiong, Z.; Lin, Z.; Huskey, D.A.; Hawes, M.C.; Vanetten, H.D.; Turgeon, B.G. A DNase from a fungal phytopathogen is a virulence factor likely deployed as counter defense against host-secreted extracellular dna. MBio 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Watson, B.S.; Bedair, M.F.; Urbanczyk-Wochniak, E.; Huhman, D.V.; Yang, D.S.; Allen, S.N.; Li, W.; Tang, Y.; Sumner, L.W. Integrated metabolomics and transcriptomics reveal enhanced specialized metabolism in Medicago truncatula root border cells. Plant Physiol. 2015, 167, 1699–1716. [Google Scholar] [CrossRef]
- Cannesan, M.A.; Durand, C.; Burel, C.; Gangneux, C.; Lerouge, P.; Ishii, T.; Laval, K.; Follet-Gueye, M.L.; Driouich, A.; Vicré-Gibouin, M. Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 2012, 159, 1658–1670. [Google Scholar] [CrossRef]
- Nagayama, T.; Nakamura, A.; Yamaji, N.; Satoh, S.; Furukawa, J.; Iwai, H. Changes in the Distribution of Pectin in Root Border Cells Under Aluminum Stress. Front. Plant Sci. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Yang, J.; Qu, M.; Fang, J.; Shen, R.F.; Feng, Y.M.; Liu, J.Y.; Bian, J.F.; Wu, L.S.; He, Y.M.; Yu, M. Alkali-soluble pectin is the primary target of aluminum immobilization in root border cells of pea (Pisum sativum). Front. Plant Sci. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Miyasaka, S.C.; Hawes, M.C. Possible Role of Root Border Cells in Detection and Avoidance of Aluminum Toxicity. Plant Physiol. 2001, 125, 1978–1987. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Gendi, X.; Song, J.; Wu, T.; Mei, X.; Liu, P. Effects of Iron Toxicity on the Morphological and Biological Characteristics of Rice Root Border Cells. J. Plant Nutr. 2017, 40, 332–343. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, N.; Iyer-Pascuzzi, A.S. Shedding the Last Layer: Mechanisms of Root Cap Cell Release. Plants 2020, 9, 308. https://doi.org/10.3390/plants9030308
Kumar N, Iyer-Pascuzzi AS. Shedding the Last Layer: Mechanisms of Root Cap Cell Release. Plants. 2020; 9(3):308. https://doi.org/10.3390/plants9030308
Chicago/Turabian StyleKumar, Narender, and Anjali S. Iyer-Pascuzzi. 2020. "Shedding the Last Layer: Mechanisms of Root Cap Cell Release" Plants 9, no. 3: 308. https://doi.org/10.3390/plants9030308
APA StyleKumar, N., & Iyer-Pascuzzi, A. S. (2020). Shedding the Last Layer: Mechanisms of Root Cap Cell Release. Plants, 9(3), 308. https://doi.org/10.3390/plants9030308





