Prevalent pH Controls the Capacity of Galdieria maxima to Use Ammonia and Nitrate as a Nitrogen Source
Abstract
1. Introduction
2. Material and Methods
2.1. Strains Cultivation
2.2. Phylogenetic Analysis
2.3. Experimental Procedure
2.4. Statistical Analysis
3. Results
3.1. Phylogenetic Analysis
3.2. Relationship between pH and Growth under Ammonium Source
3.3. Effect of pH and Nitrogen Source on Growth and Chlorophyll Content
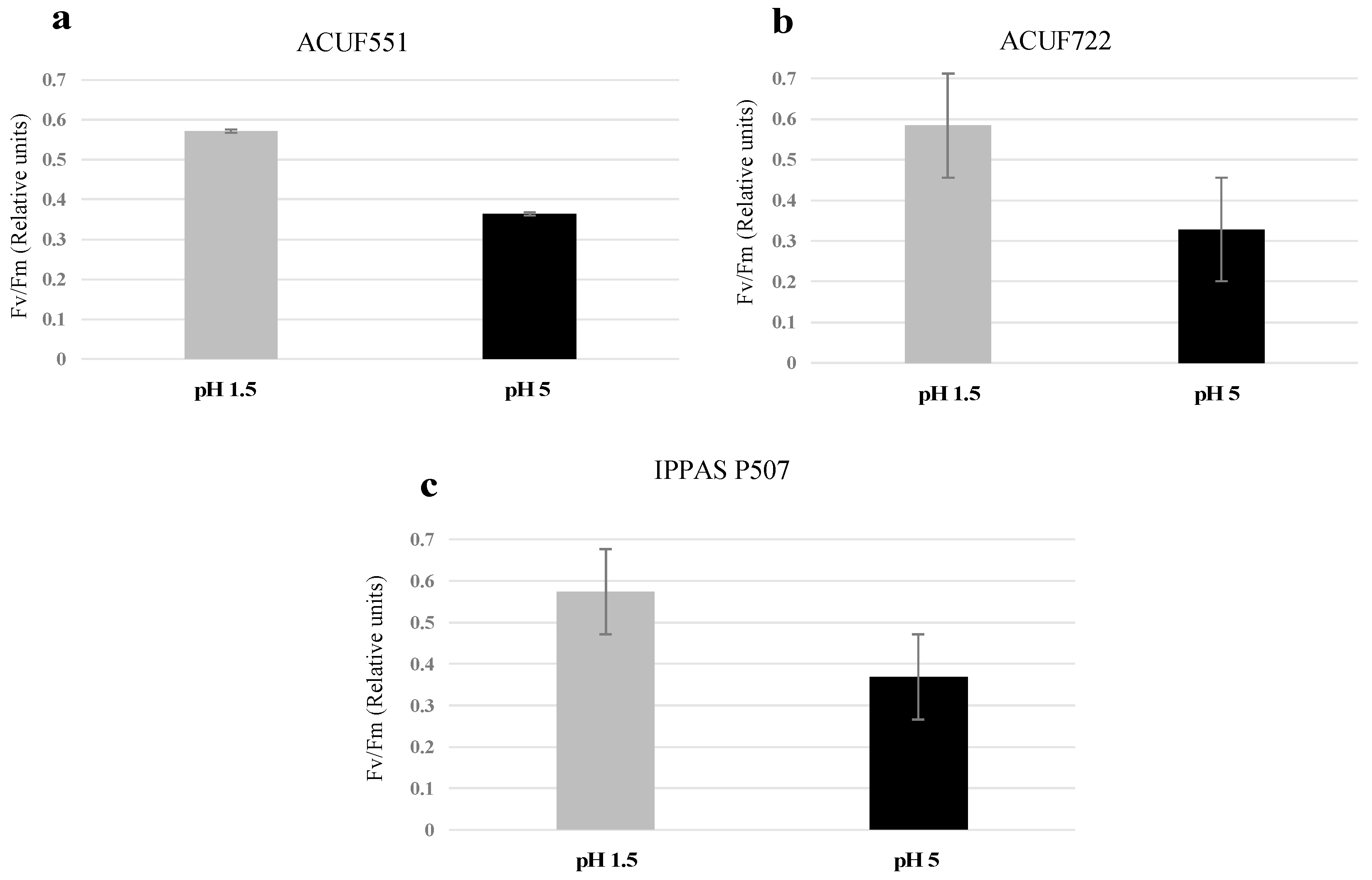
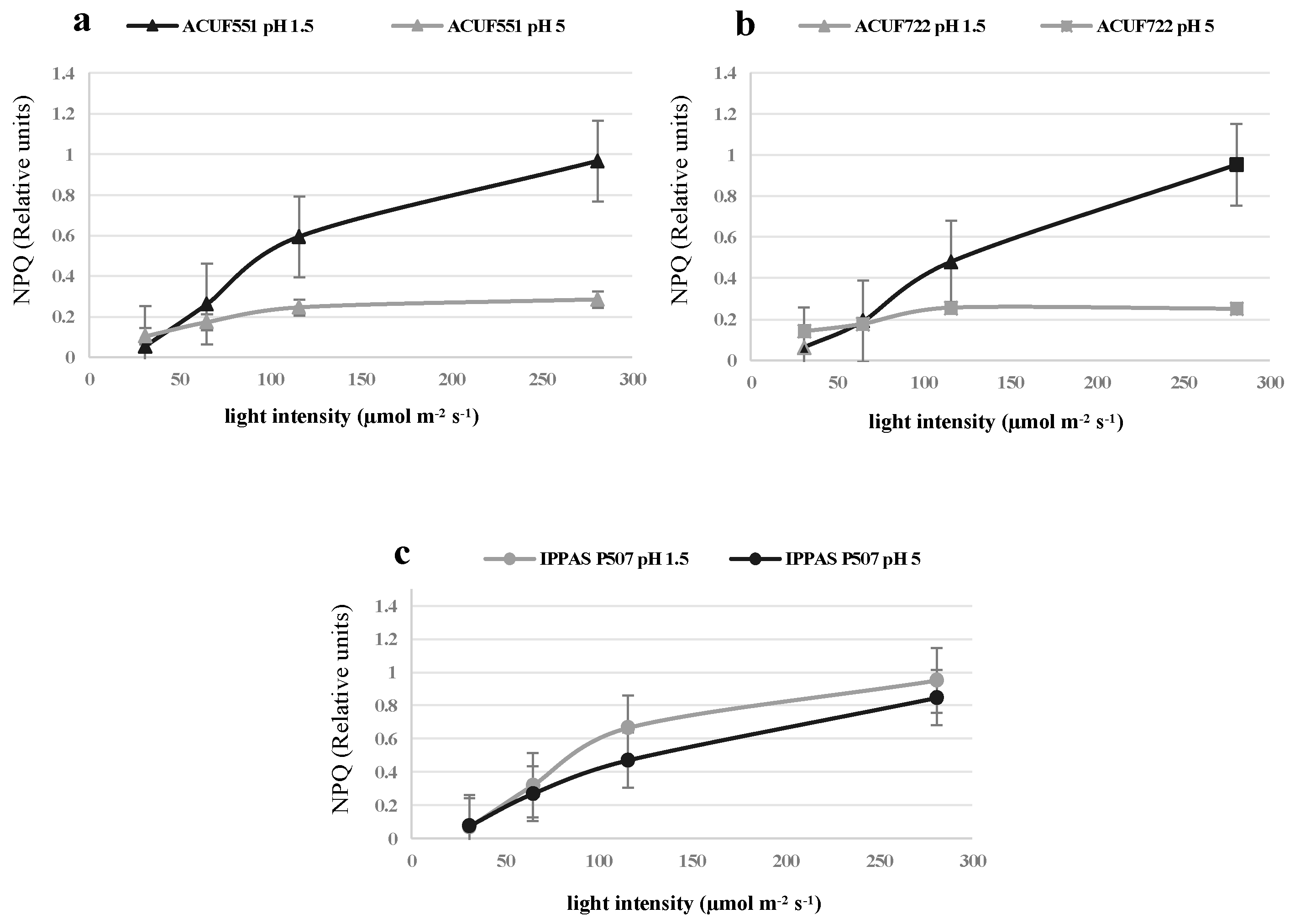
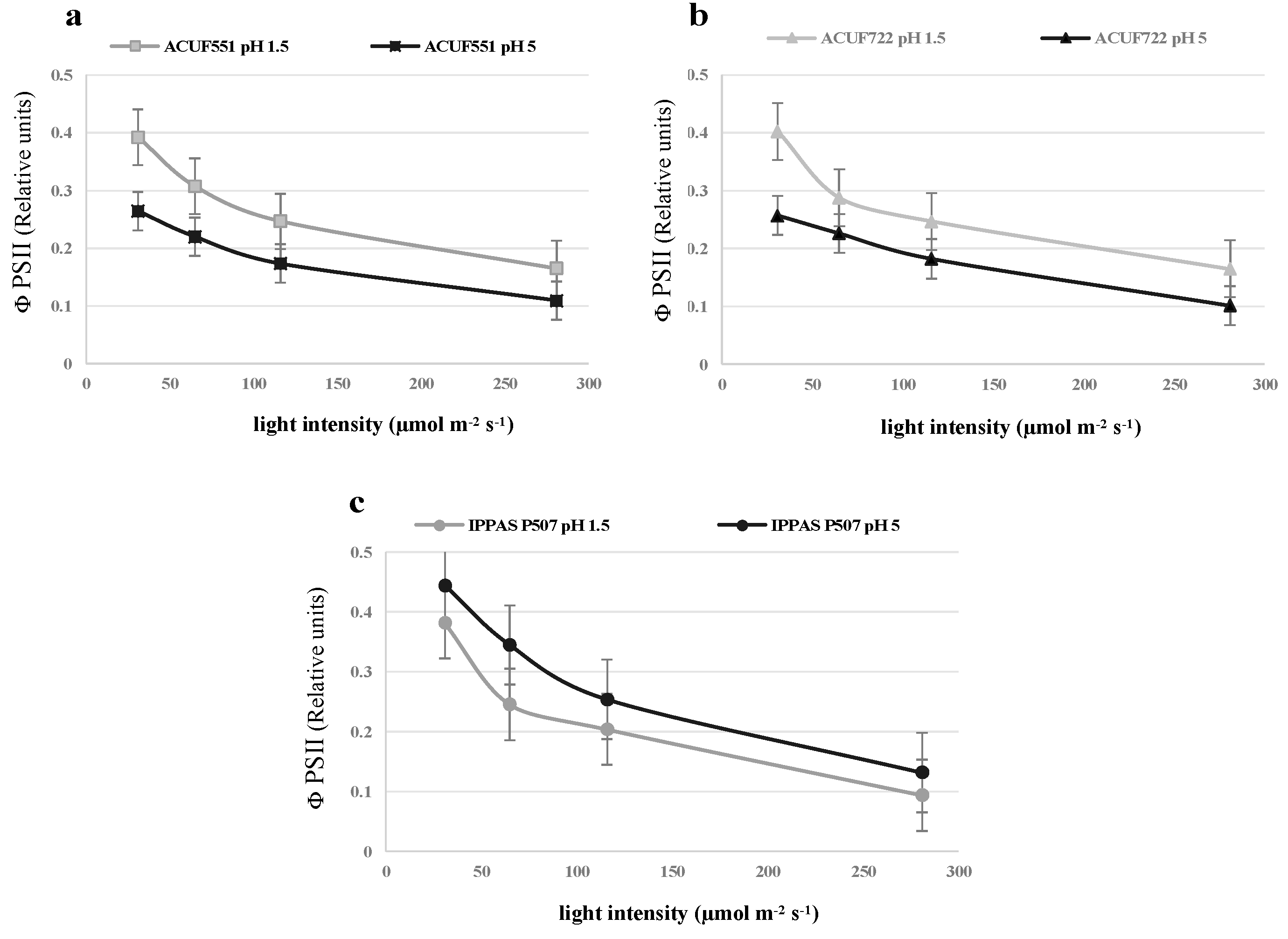
3.4. Effect of pH and Nitrogen Source on Photochemical Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dhakar, K.; Pandey, A. Wide pH range tolerance in extremophiles: Towards understanding an important phenomenon for future biotechnology. Appl. Microbiol. Biotechnol. 2016, 100, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Seckbach, J.; Oren, A.; Stan-Lotter, H. Polyextremophiles: Life under Multiple Forms of Stress; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Brock, T.D. The genus Cyanidium. In Thermophilic Microorganisms and Life at High Temperatures; Springer: New York, NY, USA, 1978; pp. 255–301. ISBN 1461262844. [Google Scholar]
- Albertano, P.; Pinto, G.; Pollio, A. Ecophysiology and Ultrastructure of an Acidophilic Species of Ochromonas (Chrysophyceae, Ochromonadales). Arch. für Protistenkd. 1994, 144, 75–82. [Google Scholar] [CrossRef]
- Schönknecht, G.; Chen, W.; Ternes, C.M.; Barbier, G.G.; Shrestha, R.P.; Stanke, M.; Bräutigam, A.; Baker, B.J.; Banfield, J.F.; Garavito, R.M.; et al. Gene Transfer from Bacteria and Archaea Facilitated Evolution of an Extremophilic Eukaryote. Science 2013, 339, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Ciniglia, C.; Yoon, H.S.; Pollio, A.; Pinto, G.; Bhattacharya, D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol. Ecol. 2004, 13, 1827–1838. [Google Scholar] [CrossRef]
- Toplin, J.A.; Norris, T.B.; Lehr, C.R.; McDermott, T.R.; Castenholz, R.W. Biogeographic and phylogenetic diversity of thermoacidophilic cyanidiales in Yellowstone National Park, Japan, and New Zealand. Appl. Environ. Microbiol. 2008, 74, 2822–2833. [Google Scholar] [CrossRef]
- Skorupa, D.J.; Reeb, V.; Castenholz, R.W.; Bhattacharya, D.; Mcdermott, T.R. Cyanidiales diversity in Yellowstone National Park. Lett. Appl. Microbiol. 2013, 57, 459–466. [Google Scholar] [CrossRef]
- Ciniglia, C.; Yang, E.C.; Pollio, A.; Pinto, G.; Iovinella, M.; Vitale, L.; Yoon, H.S. Cyanidiophyceae in Iceland: Plastid rbc L gene elucidates origin and dispersal of extremophilic Galdieria sulphuraria and G. maxima (Galdieriaceae, Rhodophyta). Phycologia 2014, 53, 542–551. [Google Scholar] [CrossRef]
- Hsieh, C.J.; Zhan, S.H.; Lin, Y.; Tang, S.L.; Liu, S.L. Analysis of rbcL sequences reveals the global biodiversity, community structure, and biogeographical pattern of thermoacidophilic red algae (Cyanidiales). J. Phycol. 2015, 51, 682–694. [Google Scholar] [CrossRef]
- Carfagna, S.; Landi, V.; Coraggio, F.; Salbitani, G.; Vona, V.; Pinto, G.; Pollio, A.; Ciniglia, C. Different characteristics of C-phycocyanin (C-PC) in two strains of the extremophilic Galdieria phlegrea. Algal Res. 2018, 31, 406–412. [Google Scholar] [CrossRef]
- Eren, A.; Iovinella, M.; Yoon, H.S.; Cennamo, P.; de Stefano, M.; de Castro, O.; Ciniglia, C. Genetic structure of Galdieria populations from Iceland. Polar Biol. 2018, 41, 1681–1691. [Google Scholar] [CrossRef]
- Iovinella, M.; Eren, A.; Pinto, G.; Pollio, A.; Davis, S.J.; Cennamo, P.; Ciniglia, C. Cryptic dispersal of Cyanidiophytina (Rhodophyta) in non-acidic environments from Turkey. Extremophiles 2018, 22, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Del Mondo, A.; Iovinella, M.; Petriccione, M.; Nunziata, A.; Davis, S.J.; Cioppa, D.; Ciniglia, C. A SPOTLIGHT ON RAD52 IN CYANIDIOPHYTINA (RHODOPHYTA): A RELIC IN ALGAL HERITAGE. Plants 2019, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Carfagna, S.; Bottone, C.; Cataletto, P.R.; Petriccione, M.; Pinto, G.; Salbitani, G.; Vona, V.; Pollio, A.; Ciniglia, C. Impact of sulfur starvation in autotrophic and heterotrophic cultures of the Extremophilic Microalga Galdieria Phlegrea (Cyanidiophyceae). Plant Cell Physiol. 2016, 57, 1890–1898. [Google Scholar] [CrossRef]
- Sentsova, Y.O. On the diversity of acido-thermophilic unicellular algae of the genus Galdieria (Rhodophyta, Cyanidiophyceae). Bot. J. 1991, 76, 69–79. [Google Scholar]
- Rigano, C.; Fuggi, A.; Rigano, V.D.M.; Aliotta, G. Studies on utilization of 2-ketoglutarate, glutamate and other amino acids by the unicellular alga Cyanidium caldarium. Arch. Microbiol. 1976, 107, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Rigano, C.; Aliotta, G.; Rigano Martino Di, V.; Fuggi, A.; Vona, V. Heterotrophic Growth Patterns in the Unicellular Alga Cyanidium Calderium. Arch. Microbiol. 1977, 113, 191–196. [Google Scholar] [CrossRef]
- Gross, W.; Schnarrenberger, C. Heterotrophic Growth of Two Strains of the Acido-Thermophilic Red Alga Galdieria sulphuraria. Plant Cell Physiol. 1995, 36, 633–638. [Google Scholar]
- Gross, W.; Oesterhelt, C. Ecophysiological studies on the red alga Galdieria sulphuraria isolated from southwest Iceland. Plant Biol. 1999, 1, 694–700. [Google Scholar] [CrossRef]
- Oesterhelt, C.; Schnarrenberger, C.; Gross, W. Characterization of a sugar/polyol uptake system in the red alga Galdieria sulphuraria. Eur. J. Phycol. 1999, 34, 271–277. [Google Scholar] [CrossRef]
- Lowell, C.; Castenholz, R.W. The lowering of external pH in confined environments by thermo-acidophilic algae (class: Cyanidiophyceae). Environ. Microbiol. Rep. 2013, 5, 660–664. [Google Scholar]
- Henkanatte-Gedera, S.M.; Selvaratnam, T.; Karbakhshravari, M.; Myint, M.; Nirmalakhandan, N.; Van Voorhies, W.; Lammers, P.J. Removal of dissolved organic carbon and nutrients from urban wastewaters by Galdieria sulphuraria: Laboratory to field scale demonstration. Algal Res. 2017, 24, 450–456. [Google Scholar] [CrossRef]
- Wang, J.; Curtis, W.R. Proton stoichiometric imbalance during algae photosynthetic growth on various nitrogen sources: Toward metabolic pH control. J. Appl. Phycol. 2016, 28, 43–52. [Google Scholar] [CrossRef]
- Allen, M.M.; Stanier, R.Y. Selective Isolation of Blue-green Algae from Water and Soil. J. Gen. Microbiol. 1968, 51, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Ciniglia, C.; Mastrobuoni, F.; Scortichini, M.; Petriccione, M. Oxidative damage and cell-programmed death induced in Zea mays L. by allelochemical stress. Ecotoxicology 2015, 24, 926–937. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinf. Appl. 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Juneau, P.; Green, B.R.; Harrison, P.J. Simulation of Pulse-Amplitude-Modulated (PAM) fluorescence: Limitations of some PAM-parameters in studying environmental stress effects. Photosynthetica 2005, 43, 75–83. [Google Scholar] [CrossRef]
- Hanelt, D.; Nultsch, W. Field Studies of Photoinhibition Show Non-Correlationsbetween Oxygen and Fluorescence Measurements in the Arctic Red Alga Palmaria palmata. J. Plant Physiol. 1995, 145, 31–38. [Google Scholar] [CrossRef]
- Carbone, D.A.; Gargano, I.; Pinto, G.; De Natale, A.; Pollio, A. Evaluating Microalgae Attachment to Surfaces: A first Approach towards a Laboratory Integrated Assessment. Chem. Eng. Trans. 2017, 57, 73–78. [Google Scholar]
- Carbone, D.A.; Gargano, I.; Olivieri, G.; Marzocchella, A.; Andreozzi, R.; Marotta, R.; Spasiano, D.; Pinto, G.; Pollio, A. LIGHT INTENSITIES MAXIMIZING PHOTOSYNTHESIS AND KINETICS OF PHOTOCHEMICAL STEPS IN Graesiella emersonii UNDER DIFFERENT CULTIVATION STRATEGIES. Environ. Eng. Manag. J. 2019, 18, 1519–1526. [Google Scholar]
- Schreiber, U.; Schliwa, U.; Bilger, W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 1986, 10, 51–62. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 31 January 2020).
- Scherholz, M.L.; Curtis, W.R. Achieving pH control in microalgal cultures through fed-batch addition of stoichiometrically-balanced growth media. BMC Biotechnol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Reysenbach, A.-L.; Shock, E. Merging Genomes with Geochemistry in Hydrothermal Ecosystems. Science 2002, 296, 1077–1082. [Google Scholar] [CrossRef]
- Imamura, S.; Terashita, M.; Ohnuma, M.; Maruyama, S.; Minoda, A.; Weber, A.P.M.; Inouye, T.; Sekine, Y.; Fujita, Y.; Omata, T.; et al. Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga cyanidioschyzon merolae: Genetic evidence for nitrite reduction by a sulfite reductase-like enzyme. Plant Cell Physiol. 2010, 51, 707–717. [Google Scholar] [CrossRef]
- Nozaki, H.; Takano, H.; Misumi, O.; Terasawa, K.; Matsuzaki, M.; Maruyama, S.; Nishida, K.; Yagisawa, F.; Yoshida, Y.; Fujiwara, T.; et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007, 5, 1–8. [Google Scholar] [CrossRef]
- Büchel, C.; Wilhelm, C. In vivo analysis of slow chlorophyll fluorescence induction kinetics in algae: Progress, problems and perspectives. Photochem. Photobiol. 1993, 58, 137–148. [Google Scholar] [CrossRef]
- Salleh, S.; McMinn, A. THE EFFECTS OF TEMPERATURE ON THE PHOTOSYNTHETIC PARAMETERS AND RECOVERY OF TWO TEMPERATE BENTHIC MICROALGAE, AMPHORA CF. COFFEAEFORMIS AND COCCONEIS CF. SUBLITTORALIS (BACILLARIOPHYCEAE)1. J. Phycol. 2011, 47, 1413–1424. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Schansker, G.; Brestic, M.; Bussotti, F.; Calatayud, A.; Ferroni, L.; Goltsev, V.; Guidi, L.; Jajoo, A.; Li, P.; et al. Frequently Asked Questions about Chlorophyll Fluorescence, the Sequel; Springer: Dordrecht, The Netherlands, 2017; Volume 132, ISBN 1112001603. [Google Scholar]
- Kalaji, H.M.; Schansker, G.; Ladle, R.J.; Goltsev, V.; Bosa, K.; Allakhverdiev, S.I.; Brestic, M.; Bussotti, F.; Calatayud, A.; Dąbrowski, P.; et al. Frequently asked questions about in vivo chlorophyll fluorescence: Practical issues. Photosynth. Res. 2014, 122, 121–158. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102. [Google Scholar] [CrossRef]
- Oesterhelt, C.; Schmälzlin, E.; Schmitt, J.M.; Lokstein, H. Regulation of photosynthesis in the unicellular acidophilic red alga Galdieria sulphuraria†. Plant J. 2007, 51, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Dhakar, K.; Sharma, A.; Priti, P.; Sati, P.; Kumar, B. Thermophilic bacteria that tolerate a wide temperature and pH range colonize the Soldhar (95 °C) and Ringigad (80 °C) hot springs of Uttarakhand, India. Ann. Microbiol. 2015, 65, 809–816. [Google Scholar] [CrossRef]
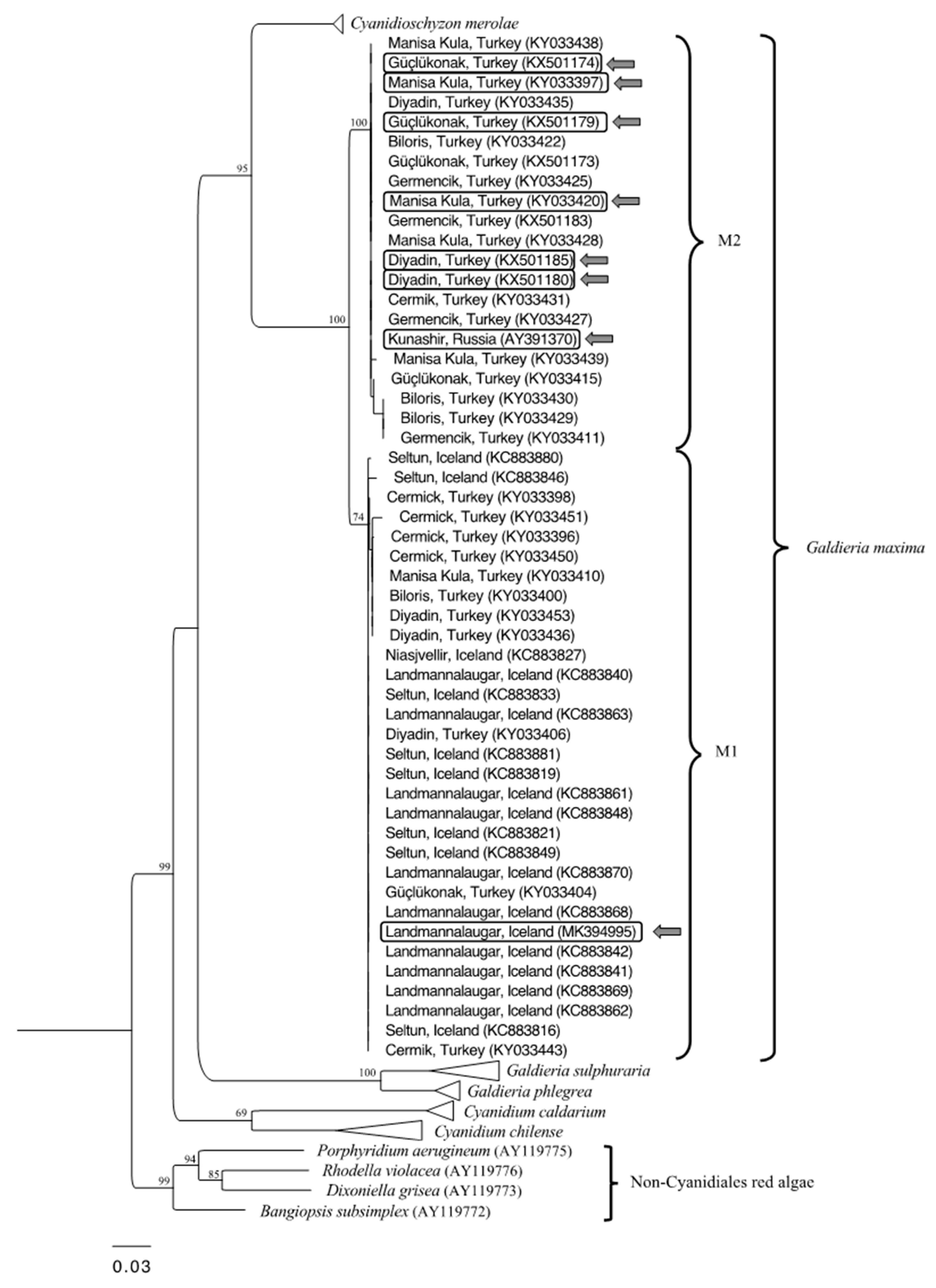
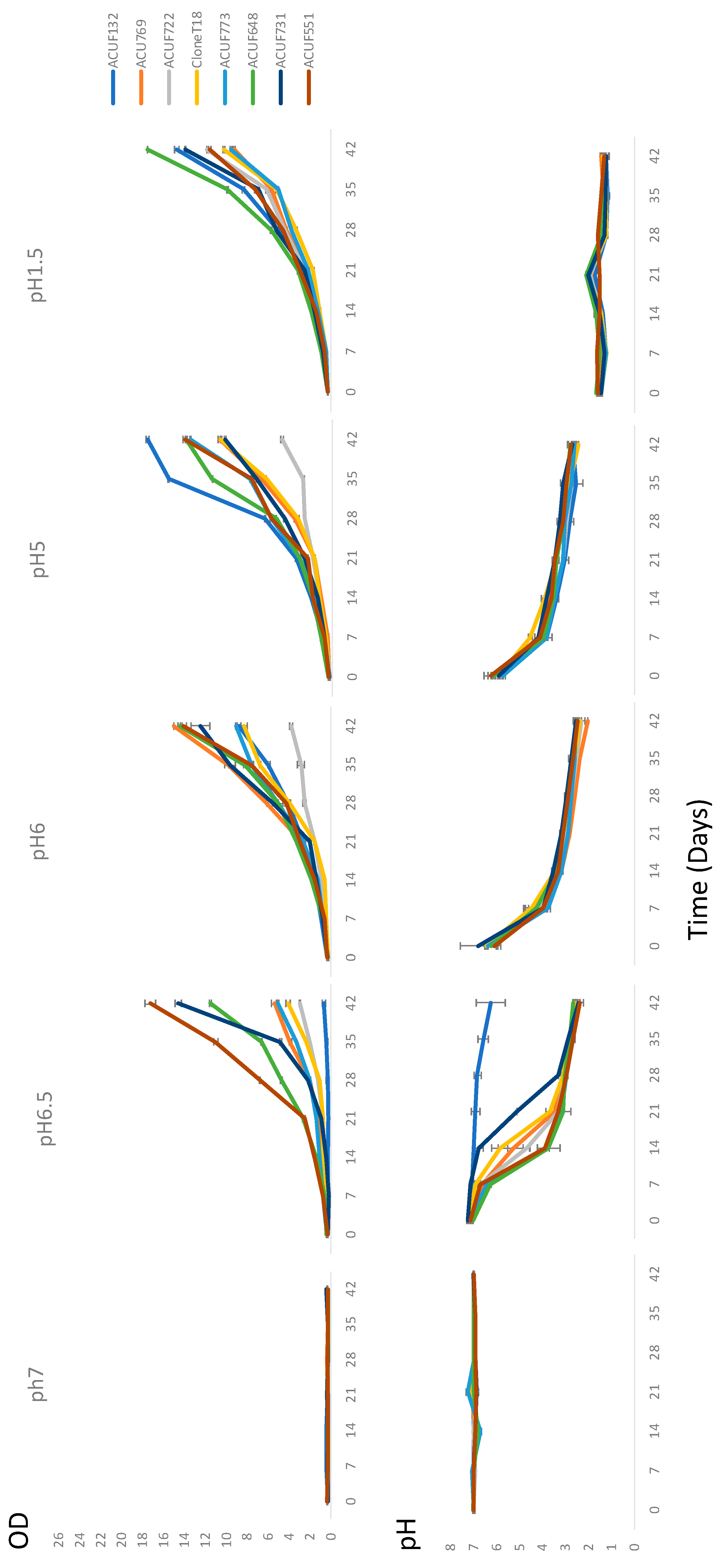
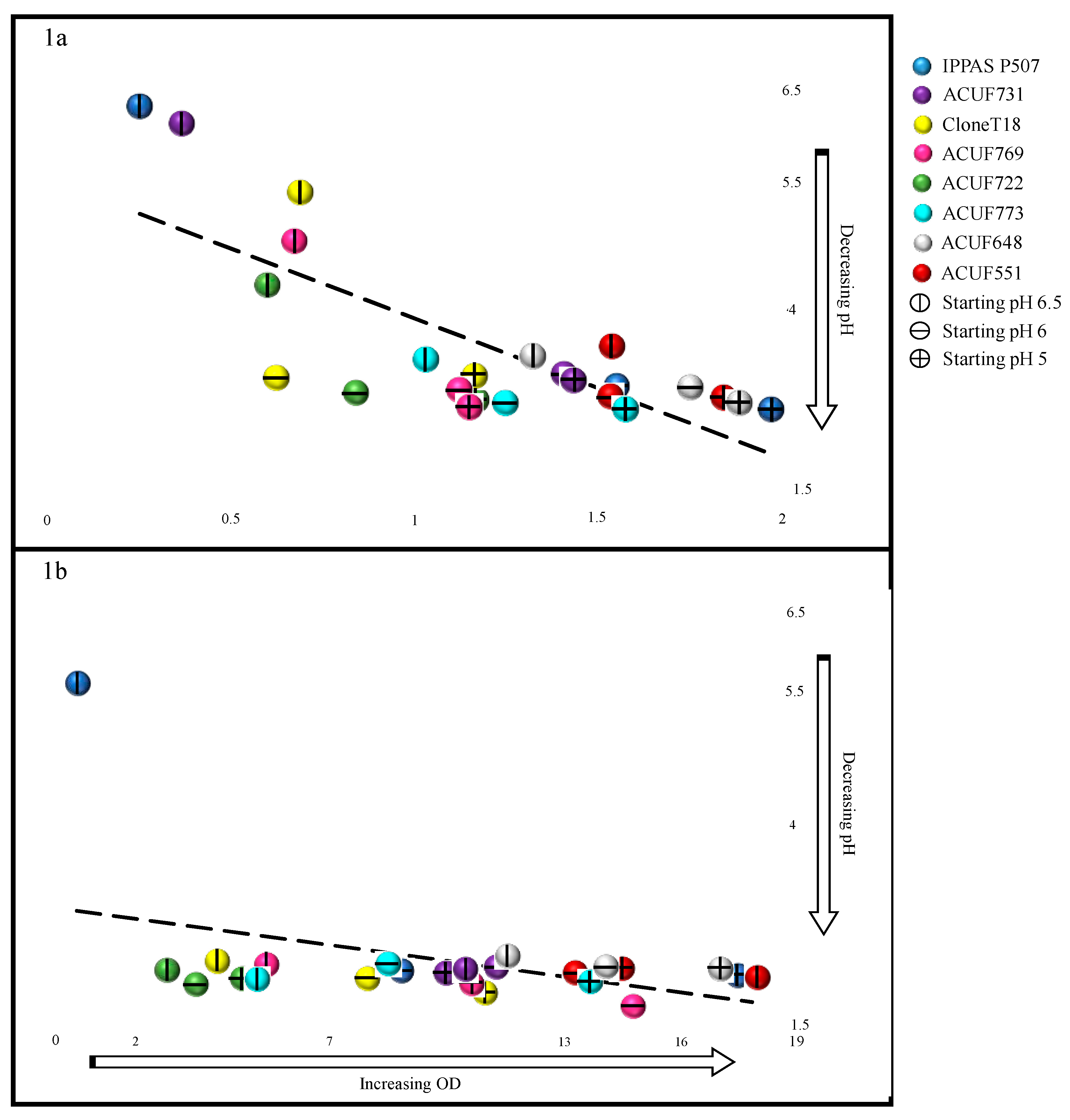
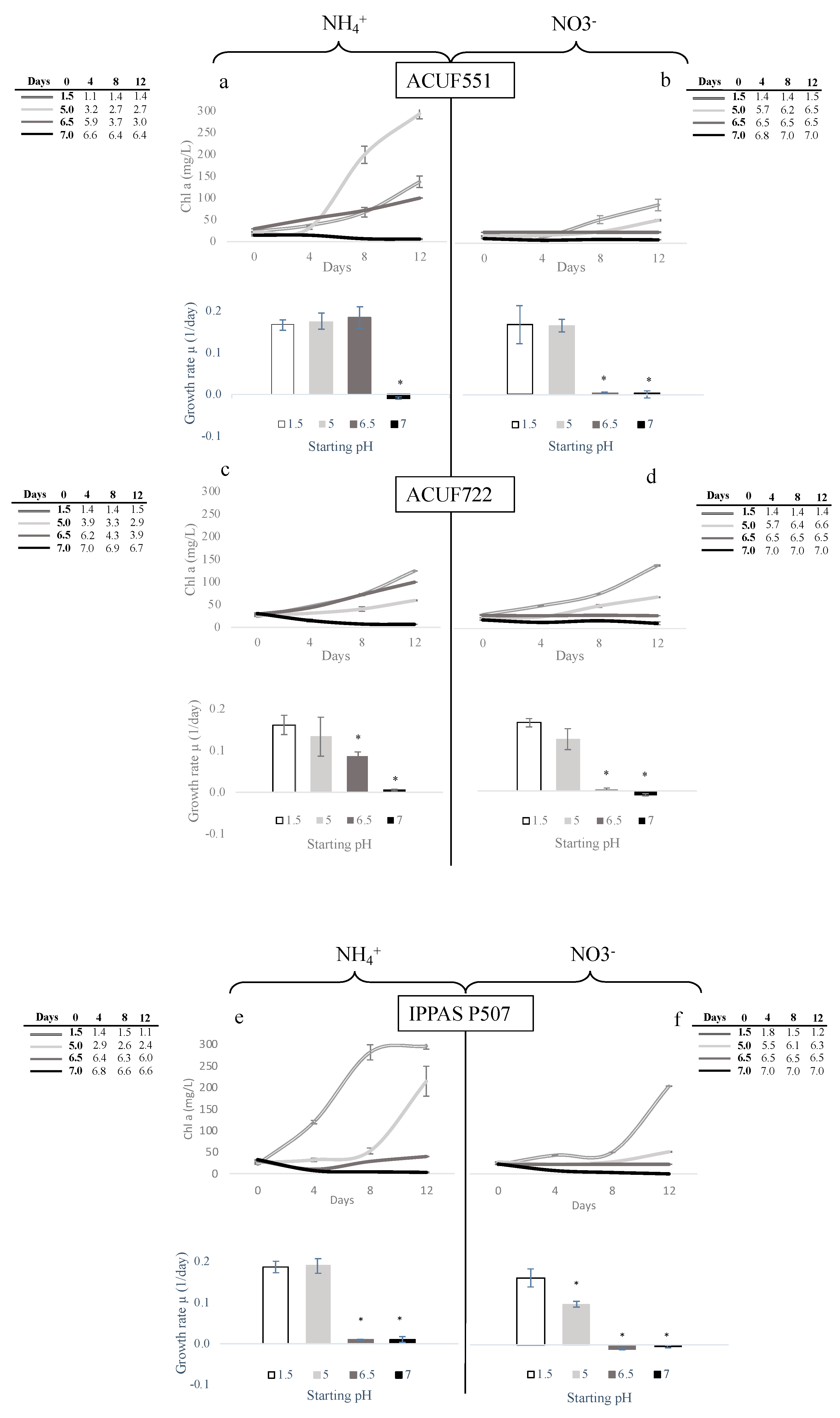
| Strains | Sampling Site | Temperature (°C) | pH |
|---|---|---|---|
| IPPAS_P507 | Kunashir (RUS) | 41.5 °C | 4 |
| ACUF769 | Güçlükonak (TURK) | 60 °C | 1 |
| ACUF722 | Güçlükonak (TURK) | 60 °C | 1 |
| CloneT18 | Diyadin (TURK) | 45 °C | 7 |
| ACUF773 | Diyadin (TURK) | 45 °C | 7 |
| ACUF648 | Kula, Manisa (TURK) | 41 °C | 7 |
| ACUF731 | Kula, Manisa (TURK) | 41 °C | 7 |
| ACUF551 | Landmannalaugar, Hekla (ICE) | 42 °C | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iovinella, M.; Carbone, D.A.; Cioppa, D.; Davis, S.J.; Innangi, M.; Esposito, S.; Ciniglia, C. Prevalent pH Controls the Capacity of Galdieria maxima to Use Ammonia and Nitrate as a Nitrogen Source. Plants 2020, 9, 232. https://doi.org/10.3390/plants9020232
Iovinella M, Carbone DA, Cioppa D, Davis SJ, Innangi M, Esposito S, Ciniglia C. Prevalent pH Controls the Capacity of Galdieria maxima to Use Ammonia and Nitrate as a Nitrogen Source. Plants. 2020; 9(2):232. https://doi.org/10.3390/plants9020232
Chicago/Turabian StyleIovinella, Manuela, Dora Allegra Carbone, Diana Cioppa, Seth J. Davis, Michele Innangi, Sabrina Esposito, and Claudia Ciniglia. 2020. "Prevalent pH Controls the Capacity of Galdieria maxima to Use Ammonia and Nitrate as a Nitrogen Source" Plants 9, no. 2: 232. https://doi.org/10.3390/plants9020232
APA StyleIovinella, M., Carbone, D. A., Cioppa, D., Davis, S. J., Innangi, M., Esposito, S., & Ciniglia, C. (2020). Prevalent pH Controls the Capacity of Galdieria maxima to Use Ammonia and Nitrate as a Nitrogen Source. Plants, 9(2), 232. https://doi.org/10.3390/plants9020232







