Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley
Abstract
1. Introduction
2. Results and Discussion
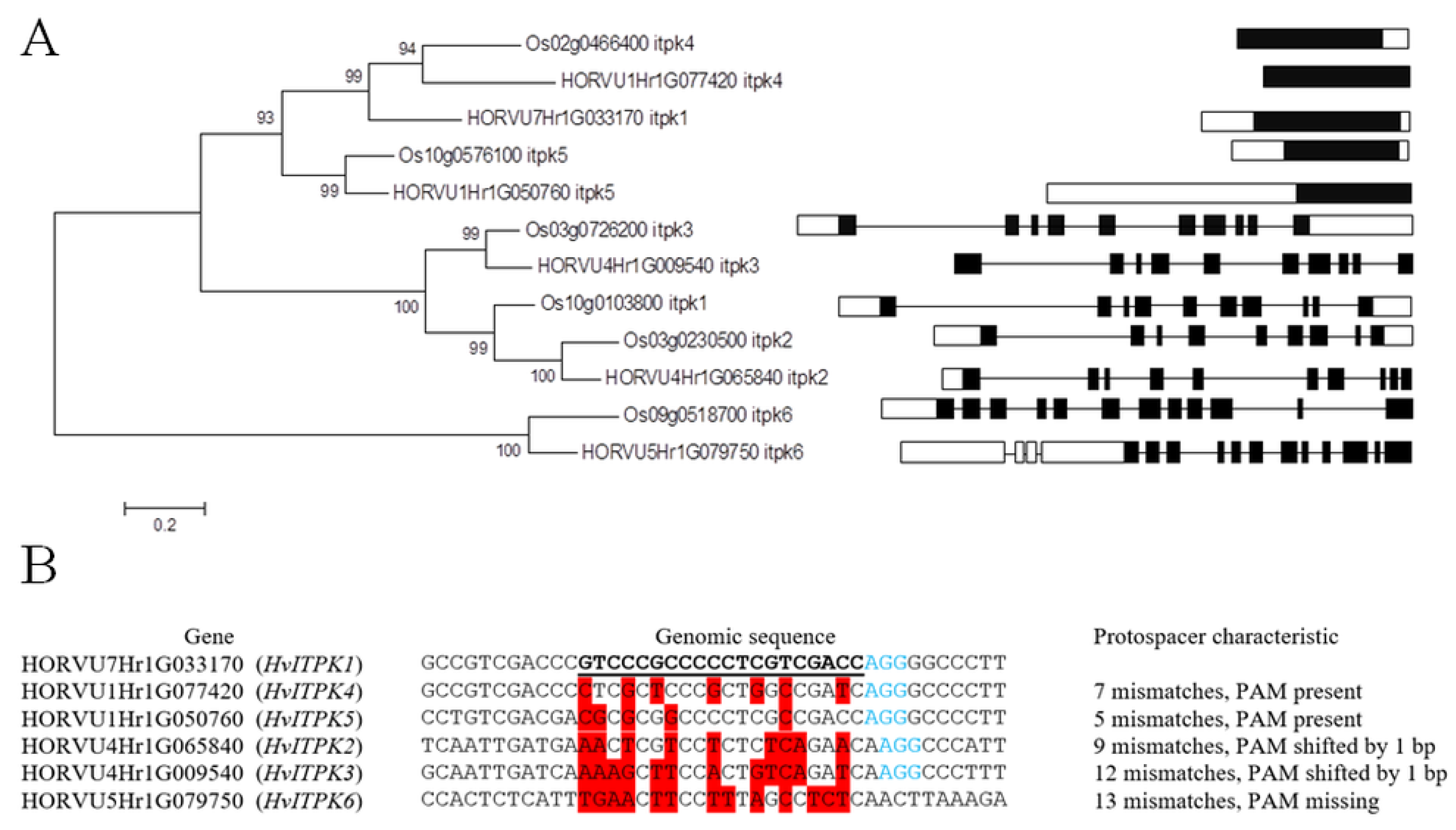
2.1. Target Gene Analysis
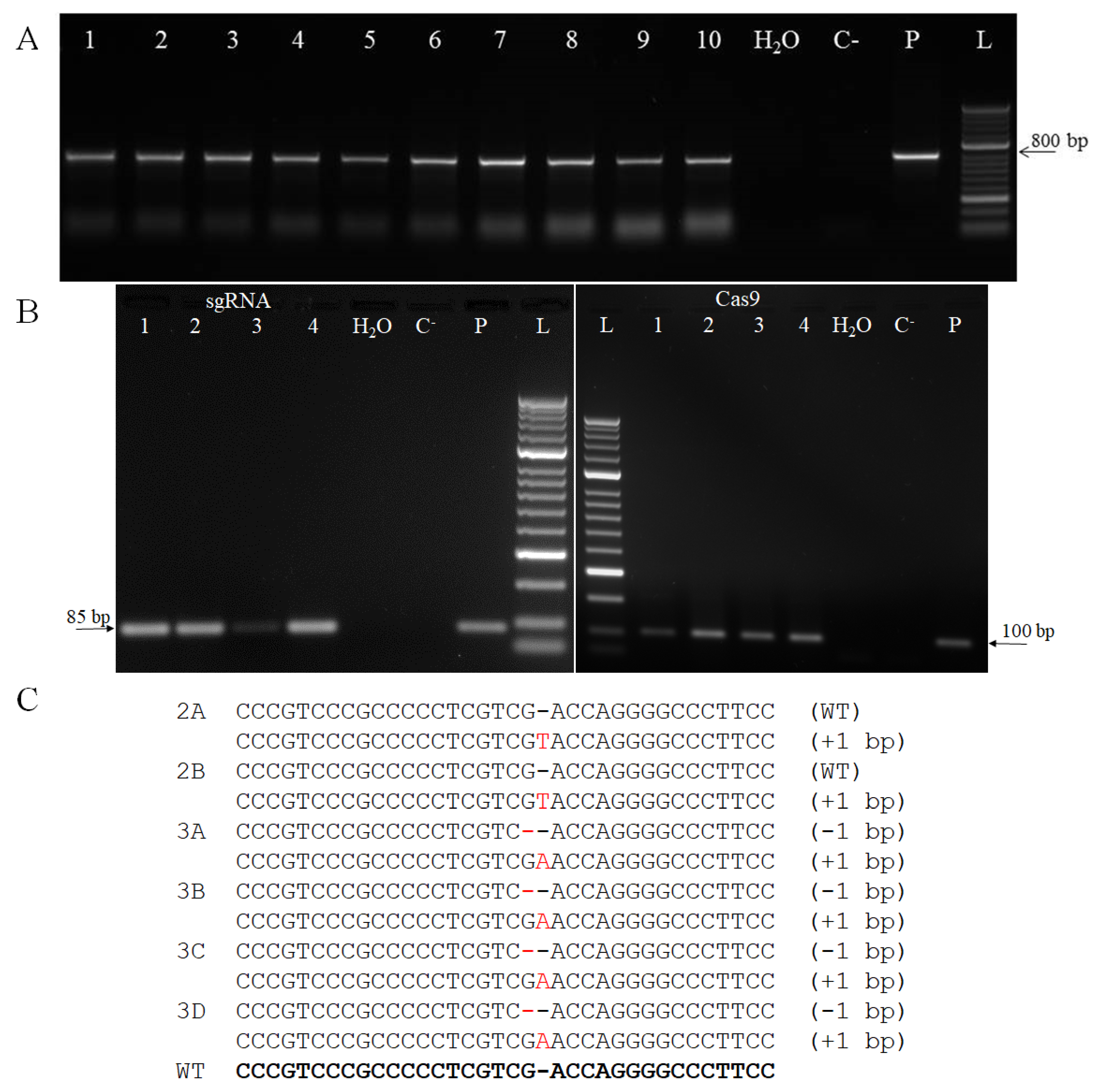
2.2. Barley Transformation
2.3. Phosphate Analysis
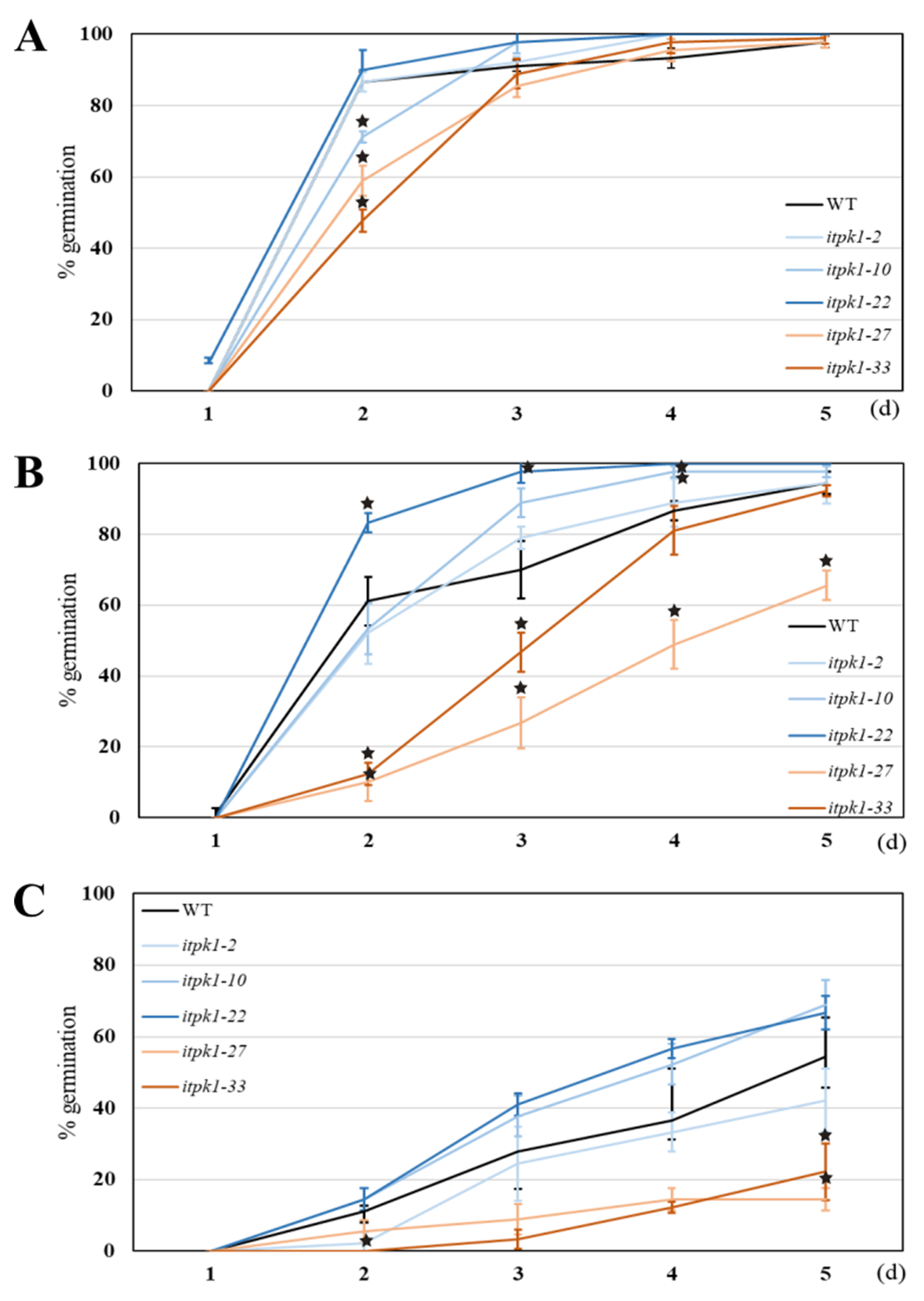
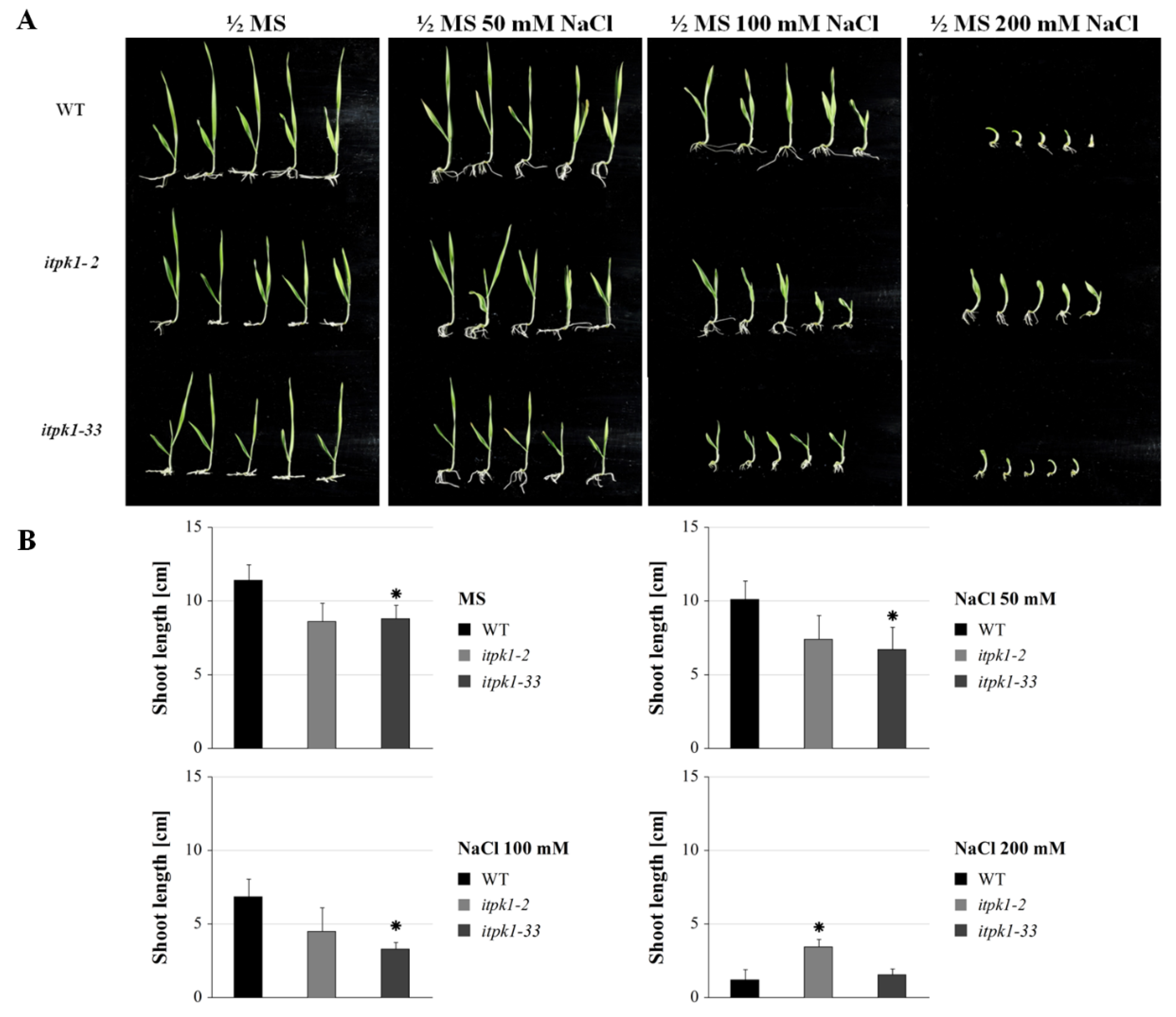
2.4. Abiotic Stress Assay
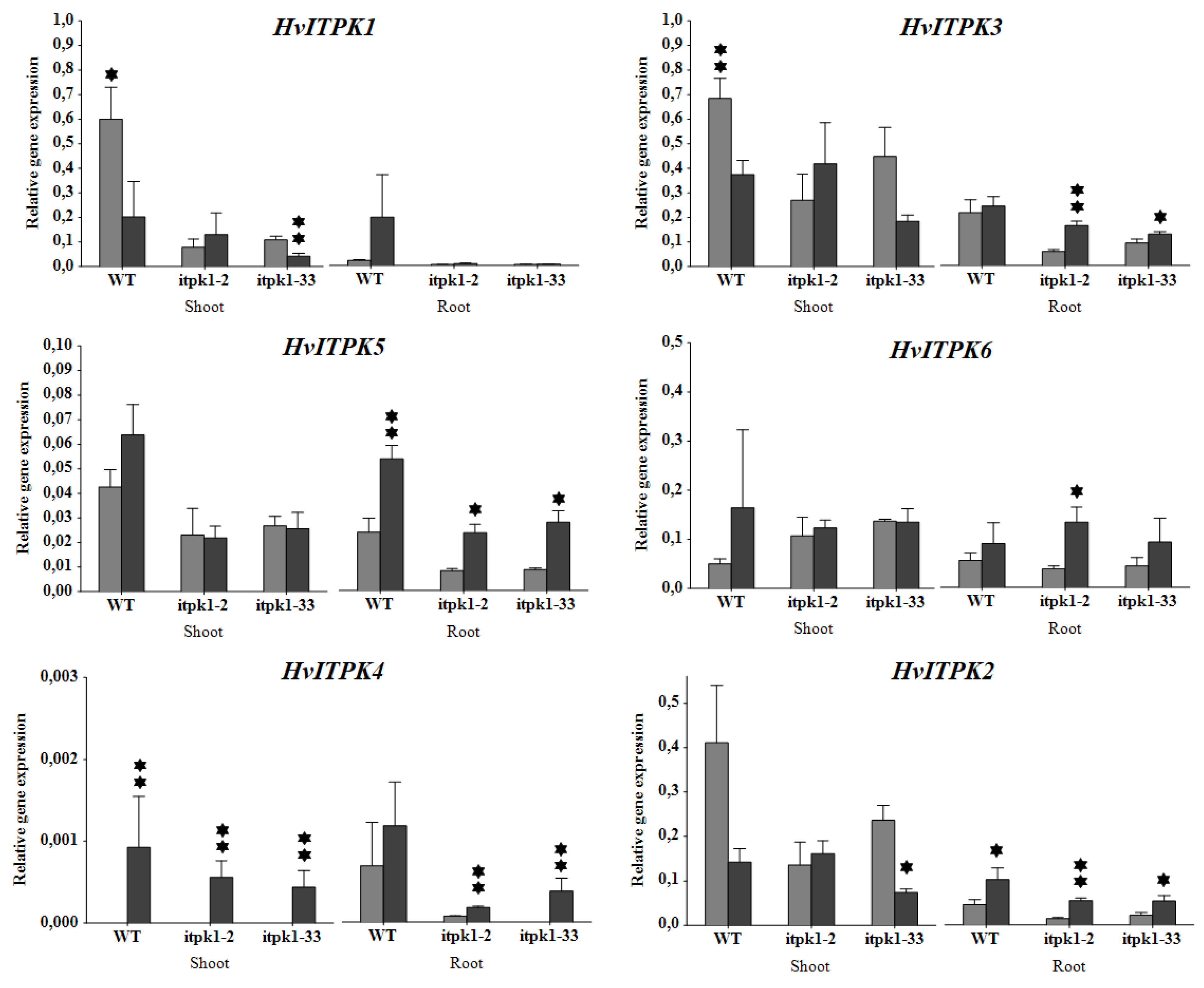
2.5. Transcription Profiling of ITPK Genes under Salinity Stress
3. Materials and Methods
3.1. Design and Cloning of Protospacer
3.2. Barley Transformation
3.3. Detection of the Cas9 and sgRNA Transcripts
3.4. Genotyping of Primary Regenerants and their Progeny
3.5. Seed Phosphate Analysis
3.6. Abiotic Stress Experiments
3.6.1. Seedling Growth Assay
3.6.2. Germination Assay
3.7. Expression Profiling of Barley ITPK Genes
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cosgrove, D. Inositolhexakisphosphates. In Inositol Phosphates: Their Chemistry, Biochemistry and Physiology; Cosgrove, D., Ed.; Elsevier Scientific Publishing Company: New York, NY, USA, 1980; pp. 26–43. ISBN 0444418741. [Google Scholar]
- Raboy, V. Forward genetics studies of seed phytic acid. Isr. J. Plant Sci. 2007, 55, 171–181. [Google Scholar] [CrossRef]
- Suzuki, M.; Tanaka, K.; Kuwano, M.; Yoshida, K.T. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): Implications for the phytic acid biosynthetic pathway. Gene 2007, 405, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.J.G.; Divecha, N.; Van Den Ende, H.; Musgrave, A.; Munnik, T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta 1999, 208, 294–298. [Google Scholar] [CrossRef]
- Warkentin, T.D.; Delgerjav, O.; Arganosa, G.; Rehman, A.U.; Bett, K.E.; Anbessa, Y.; Rossnagel, B.; Raboy, V. Development and characterization of low-phytate pea. Crop Sci. 2012, 52, 74–78. [Google Scholar] [CrossRef]
- Sakai, H.; Iwai, T.; Matsubara, C.; Usui, Y.; Okamura, M. A decrease in phytic acid content substantially affects the distribution of mineral elements within rice seeds. Plant Sci. 2015, 238, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Liu, Y.; Liu, Y.; Tan, Y.; Huang, J.; Shu, Q. Mutation of inositol 1,3,4-trisphosphate 5/6-kinase6 impairs plant growth and phytic acid synthesis in rice. Plants 2019, 8, 114. [Google Scholar] [CrossRef]
- Harada, A.; Sakai, T.; Okada, K. phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc. Natl. Acad. Sci. USA 2003, 100, 8583–8588. [Google Scholar] [CrossRef]
- Franklin-Tong, V.E.; Drøbak, B.K.; Allan, A.C.; Watkins, P.A.C.; Trewavas, A.J. Growth of pollen tubes of Papaver rhoeas is regulated by a slow-moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell 1996, 8, 1305–1321. [Google Scholar] [CrossRef]
- Yuan, F.-J.; Zhao, H.-J.; Ren, X.-L.; Zhu, S.-L.; Fu, X.-J.; Shu, Q.-Y. Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 2007, 115, 945–957. [Google Scholar] [CrossRef]
- Wilcox, J.R.; Premachandra, G.S.; Young, K.A.; Raboy, V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 2000, 40, 1601–1605. [Google Scholar] [CrossRef]
- Larson, S.R.; Raboy, V. Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: Correspondence with a low phytic acid mutation. Theor. Appl. Genet. 1999, 99, 27–36. [Google Scholar] [CrossRef]
- Larson, S.R.; Rutger, J.N.; Young, K.A.; Raboy, V. Isolation and Genetic Mapping of a Non-Lethal Rice (Oryza sativa L.) Low Phytic Acid Mutation. Crop Sci. 2000, 40, 1397–1405. [Google Scholar] [CrossRef]
- Guttieri, M.; Bowen, D.; Dorsch, J.A.; Raboy, V.; Souza, E. Identification and characterization of a low phytic acid wheat. Crop Sci. 2004, 44, 418–424. [Google Scholar] [CrossRef]
- Larson, S.R.; Young, K.A.; Cook, A.; Blake, T.K.; Raboy, V. Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor. Appl. Genet. 1998, 97, 141–146. [Google Scholar] [CrossRef]
- Jang, D.A.; Fadel, J.G.; Klasing, K.C.; Mireles, A.J.; Ernst, R.A.; Young, K.A.; Cook, A.; Raboy, V. Evaluation of low-phytate corn and barley on broiler chick performance. Poult. Sci. 2003, 82, 1914–1924. [Google Scholar] [CrossRef]
- Veum, T.L.; Ledoux, D.R.; Raboy, V. Low-phytate barley cultivars improve the utilization of phosphorus, calcium, nitrogen, energy, and dry matter in diets fed to young swine. J. Anim. Sci. 2007, 85, 961–971. [Google Scholar] [CrossRef][Green Version]
- Thacker, P.A.; Rossnagel, B.G.; Raboy, V. Phosphorus digestibility in low-phytate barley fed to finishing pigs. Can. J. Anim. Sci. 2003, 83, 101–104. [Google Scholar] [CrossRef]
- Leytem, A.B.; Taylor, J.B.; Raboy, V.; Plumstead, P.W. Dietary low-phytate mutant-M 955 barley grain alters phytate degradation and mineral digestion in sheep fed high-grain diets. Anim. Feed Sci. Technol. 2007, 138, 13–28. [Google Scholar] [CrossRef]
- Poulsen, H.D.; Johansen, K.S.; Hatzack, F.; Boisen, S.; Rasmussen, S.K. Nutritional value of low-phytate barley evaluated in rats. Acta Agric. Scand. Sect. A Anim. Sci. 2001, 51, 53–58. [Google Scholar] [CrossRef]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Pilu, R.; Panzeri, D.; Gavazzi, G.; Rasmussen, S.K.; Consonni, G.; Nielsen, E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241). Theor. Appl. Genet. 2003, 107, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Bregitzer, P.; Raboy, V. Effects of four independent low-phytate mutations in barley (Hordeum vulgare L.) on seed phosphorus characteristics and malting quality. Cereal Chem. 2006, 83, 460–464. [Google Scholar] [CrossRef]
- Kuo, H.; Chang, T.; Chiang, S.; Wang, W.; Charng, Y.; Chiou, T. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J. 2014, 80, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V.; Peterson, K.; Jackson, C.; Marshall, J.M.; Hu, G.; Saneoka, H.; Bregitzer, P. A substantial fraction of barley (Hordeum vulgare L.) low phytic acid mutations have little or no effect on yield across diverse production environments. Plants 2015, 4, 225–239. [Google Scholar] [CrossRef]
- Brinch-Pedersen, H.; Hatzack, F.; Stöger, E.; Arcalis, E.; Pontopidan, K.; Holm, P.B. Heat-stable phytases in transgenic wheat (Triticum aestivum L.): Deposition pattern, thermostability, and phytate hydrolysis. J. Agric. Food Chem. 2006, 54, 4624–4632. [Google Scholar] [CrossRef]
- Abid, N.; Khatoon, A.; Maqbool, A.; Irfan, M.; Bashir, A.; Asif, I.; Shahid, M.; Saeed, A.; Brinch-Pedersen, H.; Malik, K.A. Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transgenic Res. 2017, 26, 109–122. [Google Scholar] [CrossRef]
- Holme, I.B.; Dionisio, G.; Brinch-Pedersen, H.; Wendt, T.; Madsen, C.K.; Vincze, E.; Holm, P.B. Cisgenic barley with improved phytase activity. Plant Biotechnol. J. 2012, 10, 237–247. [Google Scholar] [CrossRef]
- Du, H.; Liu, L.; You, L.; Yang, M.; Yubing, H.; Li, X.; Xiong, L. Characterization of an inositol 1,3,4-trisphosphate 5/6-kinase gene that is essential for drought and salt stress responses in rice. Plant Mol. Biol. 2011, 77, 547–563. [Google Scholar] [CrossRef]
- Marathe, A.; Krishnan, V.; Vinutha, T.; Dahuja, A.; Jolly, M.; Sachdev, A. Exploring the role of Inositol 1,3,4-trisphosphate 5/6 kinase-2 (GmITPK2) as a dehydration and salinity stress regulator in Glycine max (L.) Merr. through heterologous expression in E. Coli. Plant Physiol. Biochem. 2018, 123, 331–341. [Google Scholar] [CrossRef]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Loewus, F.A.; Murthy, P.P.N. Myo-Inositol metabolism in plants. Plant Sci. 2000, 150, 1–19. [Google Scholar] [CrossRef]
- Aggarwal, S.; Shukla, V.; Bhati, K.K.; Kaur, M.; Sharma, S.; Singh, A.; Mantri, S.; Pandey, A.K. Hormonal regulation and expression profiles of wheat genes involved during phytic acid biosynthesis pathway. Plants 2015, 4, 298–319. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Chen, Æ.Q.; Wang, Æ.X. OsITL1 gene encoding an inositol 1,3,4-trisphosphate 5/6-kinase is a negative regulator of osmotic stress signaling. Biotechnol. Lett. 2008, 30, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 756–761. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82:1–e82:11. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA —Guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–822. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, D.J.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, 1–12. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2015, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Keith Joung, J. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef]
- Bo, L.; Jia-jie, W.; Dao-lin, F. Constructing the barley model for genetic transformation in Triticeae. J. Integr. Agric. 2015, 14, 453–468. [Google Scholar] [CrossRef]
- Hamada, H.; Liu, Y.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Abid, G.; Silue, S.; Muhovski, Y.; Jacquemin, J.M.; Toussaint, A.; Baudoin, J.P. Role of myo-inositol phosphate synthase and sucrose synthase genes in plant seed development. Gene 2009, 439, 1–10. [Google Scholar] [CrossRef]
- Bhati, K.K.; Aggarwal, S.; Sharma, S.; Mantri, S.; Singh, S.P.; Bhalla, S.; Kaur, J.; Tiwari, S.; Roy, J.K.; Tuli, R.; et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 2014, 224, 74–85. [Google Scholar] [CrossRef]
- Shi, J.; Hongyu, W.; Wu, Y.; Hazebroek, J.; Meeley, R.B.; Ertl, D.S. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003, 131, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhang, H.; Lou, D.; Yu, D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Serhantova, V.; Ehrenbergerova, J.; Ohnoutkova, L. Callus induction and regeneration efficiency of spring barley cultivars registered in the Czech Republic. Plant Soil Environ. 2004, 50, 456–462. [Google Scholar] [CrossRef]
- Harwood, W.A. A protocol for high-throughput Agrobacterium-mediated barley transformation. In Cereal Genomics: Methods and Protocols; Henry, R.J., Furtado, A., Eds.; Humana Press: Totowa, NJ, USA, 2014; pp. 251–260. ISBN 978-1-62703-715-0. [Google Scholar]
- Holme, I.B.; Wendt, T.; Gil, J.; Deleuran, L.C.; Starker, C.G.; Voytas, D.F.; Brinch-Pedersen, H. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 2017, 95, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Kapusi, E.; Corcuera-Gómez, M.; Melnik, S.; Stoger, E. Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef]
- Li, M.; Hensel, G.; Mascher, M.; Melzer, M.; Budhagatapalli, N.; Rutten, T.; Himmelbach, A.; Beier, S.; Korzun, V.; Kumlehn, J.; et al. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 2019, 31, 1430–1445. [Google Scholar] [CrossRef]
- Holubová, K.; Hensel, G.; Vojta, P.; Tarkowski, P.; Gene, H. Modification of Barley Plant Productivity Through Regulation of Cytokinin Content by Reverse–Genetics Approaches Preparation of Constructs for Silencing. Front. Plant Sci. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Sánchez-León, S.; Gil-Humanes, J.; Ozuna, C.V.; Sousa, C.; Voytas, D.F.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Jang, G.; Lee, S.; Um, T.; Hyun Chang, S.; Yong Lee, H.; Chung, P.J.; Kim, J.-K.; Do Choi, Y. Genetic chimerism of CRISPR/Cas9-mediated rice mutants. Plant Biotechnol. Rep. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- Hatzack, F.; Johansen, K.S.; Rasmussen, S.K. Nutritionally relevant parameters in low-phytate barley (Hordeum vulgare L.) grain mutants. J. Agric. Food Chem. 2000, 48, 6074–6080. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy, P.P.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Tai, T.H. Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genomics 2011, 286, 119–133. [Google Scholar] [CrossRef]
- Nagy, R.; Grob, H.; Weder, B.; Green, P.; Klein, M.; Frelet-Barrand, A.; Schjoerring, J.K.; Brearley, C.; Martinoia, E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporterinvolved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622. [Google Scholar] [CrossRef]
- Panzeri, D.; Cassani, E.; Doria, E.; Tagliabue, G.; Forti, L.; Campion, B.; Bollini, R.; Brearley, C.A.; Pilu, R.; Nielsen, E.; et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011, 191, 70–83. [Google Scholar] [CrossRef]
- Stiles, A.; Qian, X.; Shears, S.; Grabau, E. Metabolic and signaling properties of an Itpk gene family in Glycine max. FEBS Lett. 2008, 582, 1853–1858. [Google Scholar] [CrossRef]
- Marathe, A.; Krishnan, V.; Mahajan, M.M.; Thimmegowda, V.; Dahuja, A.; Jolly, M.; Praveen, S.; Sachdev, A. Characterization and molecular modeling of Inositol 1,3,4 tris phosphate 5/6 kinase-2 from Glycine max (L.) Merr.: Comprehending its evolutionary conservancy at functional level. 3 Biotech 2018, 8, 50. [Google Scholar] [CrossRef]
- Forster, B.P.; Pakniyat, H.; Macaulay, M.; Matheson, W.; Phillips, M.S.; Thomas, W.T.B.; Powell, W. Variation in the leaf sodium content of the Hordeum vulgare (Barley) cultivar Maythorpe and its derived mutant cv. Golden promise. Heredity (Edinb). 1994, 73, 249–253. [Google Scholar] [CrossRef]
- Kaur, H.; Verma, P.; Petla, B.P.; Rao, V.; Saxena, S.C.; Majee, M. Ectopic expression of the ABA-inducible dehydration-responsive chickpea l-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in Arabidopsis enhances tolerance to salinity and dehydration stress. Planta 2013, 237, 321–335. [Google Scholar] [CrossRef]
- Smits, A.H.; Ziebell, F.; Joberty, G.; Zinn, N.; Mueller, W.F.; Clauder-Münster, S.; Eberhard, D.; Fälth Savitski, M.; Grandi, P.; Jakob, P.; et al. Biological plasticity rescues target activity in CRISPR knock outs. Nat. Methods 2019, 16, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Hu, H.; Kai, J.; Traw, M.B.; Yang, S.; Zhang, X. Different knockout genotypes of OsIAA23 in rice using CRISPR/Cas9 generating different phenotypes. Plant Mol. Biol. 2019, 100, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.-G. DSDecode: A Web-Based Tool for Decoding of Sequencing Chromatograms for Genotyping of Targeted Mutations. Mol. Plant 2018, 8, 1431–1433. [Google Scholar] [CrossRef]
- Vaculova, K.; Balounova, M.; Sedlackova, I.; Kvasnicka, F.; Mikulikova, R.; Belakova, S.; Benesova, K.; Pouch, M.; Ehrenbergerova, J. Metodika Prebreedingu Ječmene Jarního s Diferencovaným Obsahem Přirozených Škodlivých Látek v Zrně Pro Šlechtění Odrůd Nesladovnického Typu, 1st ed.; Agrotest Fyto, S.R.O.: Kromeriz, Czech Republic, 2011; pp. 12–15. ISBN 978-80-87555-00-2. [Google Scholar]
- Chen, P.S.; Toribara, T.Y.; Warner, H. Microdetermination of Phosphorus. Anal. Chem. 1956, 28, 1756–1758. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Skubacz, A.; Kurowska, M.; Slota, M.; Swiergolik, D.; Szarejko, I. Methods for the Simple and Reliable Assessment of Barley Sensitivity to Abiotic Stresses During Early Development. In Barley: Methods and Protocols; Harwood, W.A., Ed.; Springer Science + Business Media: New York, NY, USA, 2019; pp. 127–151. ISBN 978-1-4939-8942-3. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlčko, T.; Ohnoutková, L. Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants 2020, 9, 195. https://doi.org/10.3390/plants9020195
Vlčko T, Ohnoutková L. Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants. 2020; 9(2):195. https://doi.org/10.3390/plants9020195
Chicago/Turabian StyleVlčko, Tomáš, and Ludmila Ohnoutková. 2020. "Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley" Plants 9, no. 2: 195. https://doi.org/10.3390/plants9020195
APA StyleVlčko, T., & Ohnoutková, L. (2020). Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants, 9(2), 195. https://doi.org/10.3390/plants9020195



