Assessment of Genetic Relationships between Streptocarpus x hybridus V. Parents and F1 Progenies Using SRAP Markers and FT-IR Spectroscopy
Abstract
1. Introduction
2. Results
2.1. Pollen Viability
2.2. Morphological Characterization and Genetic Relationships Between Parents and F1 Progenies
2.3. Assessment of Genetic Relationships based on SRAP Analysis
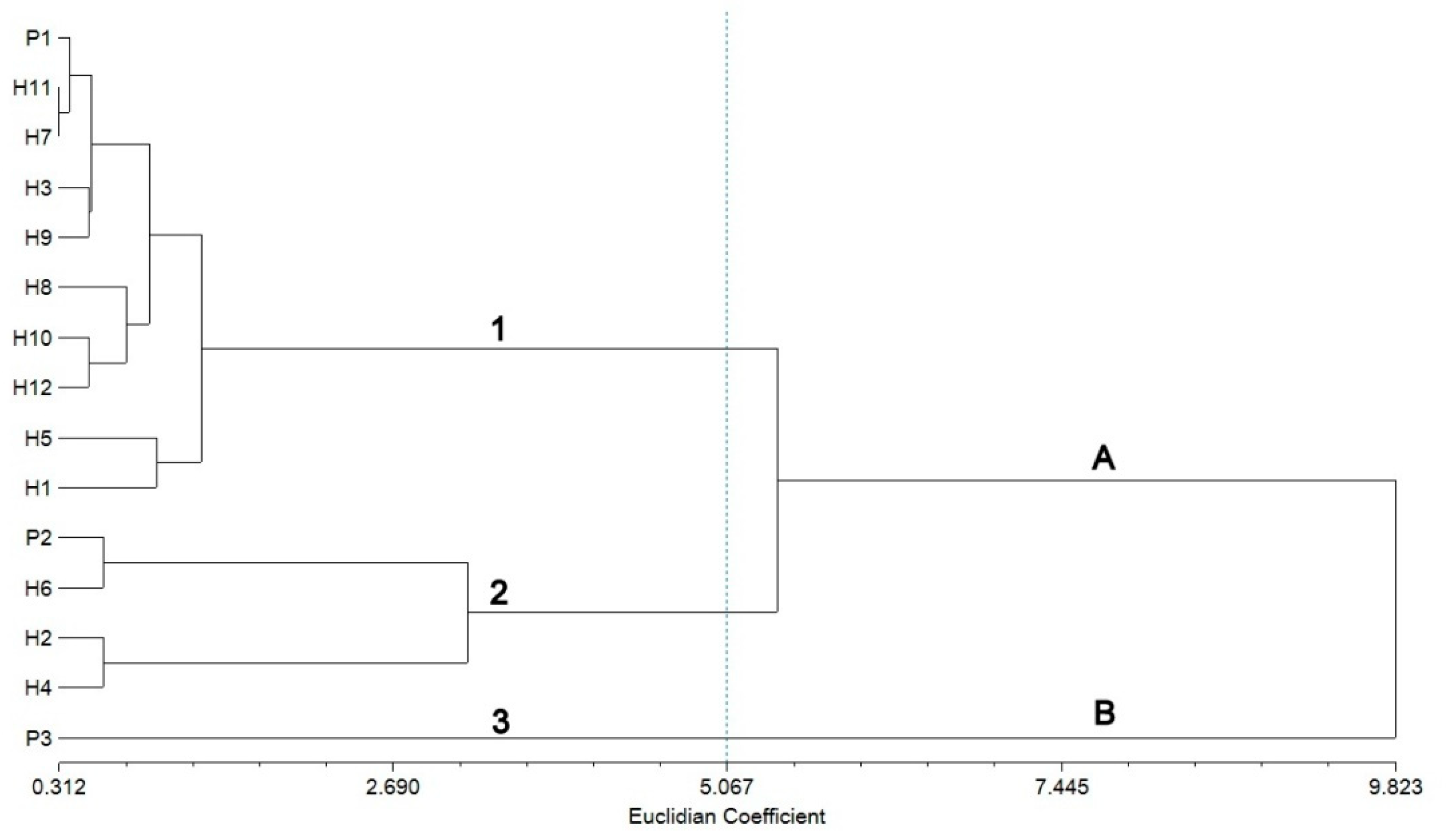
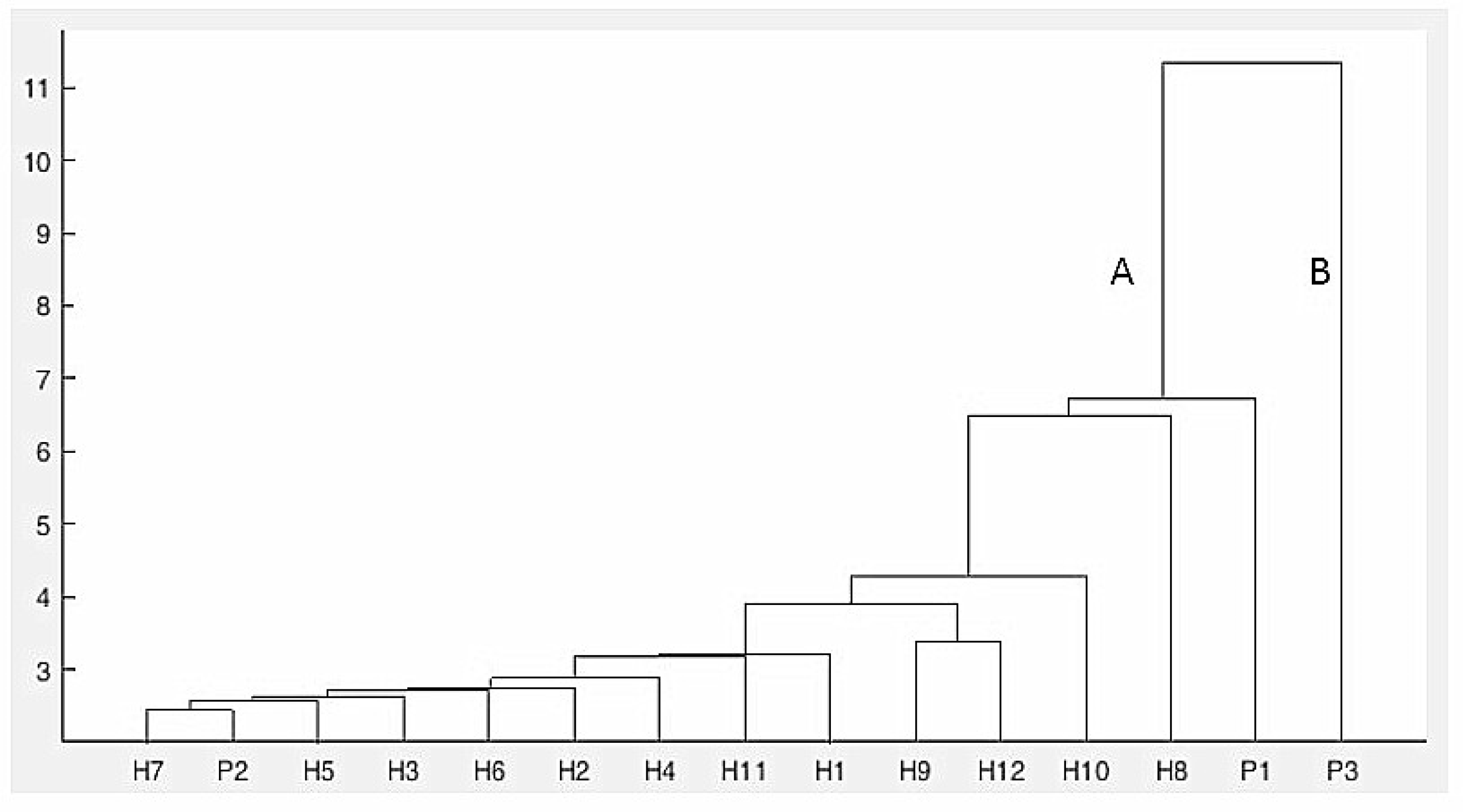
2.4. Cluster Analysis Revealed by SRAP Markers
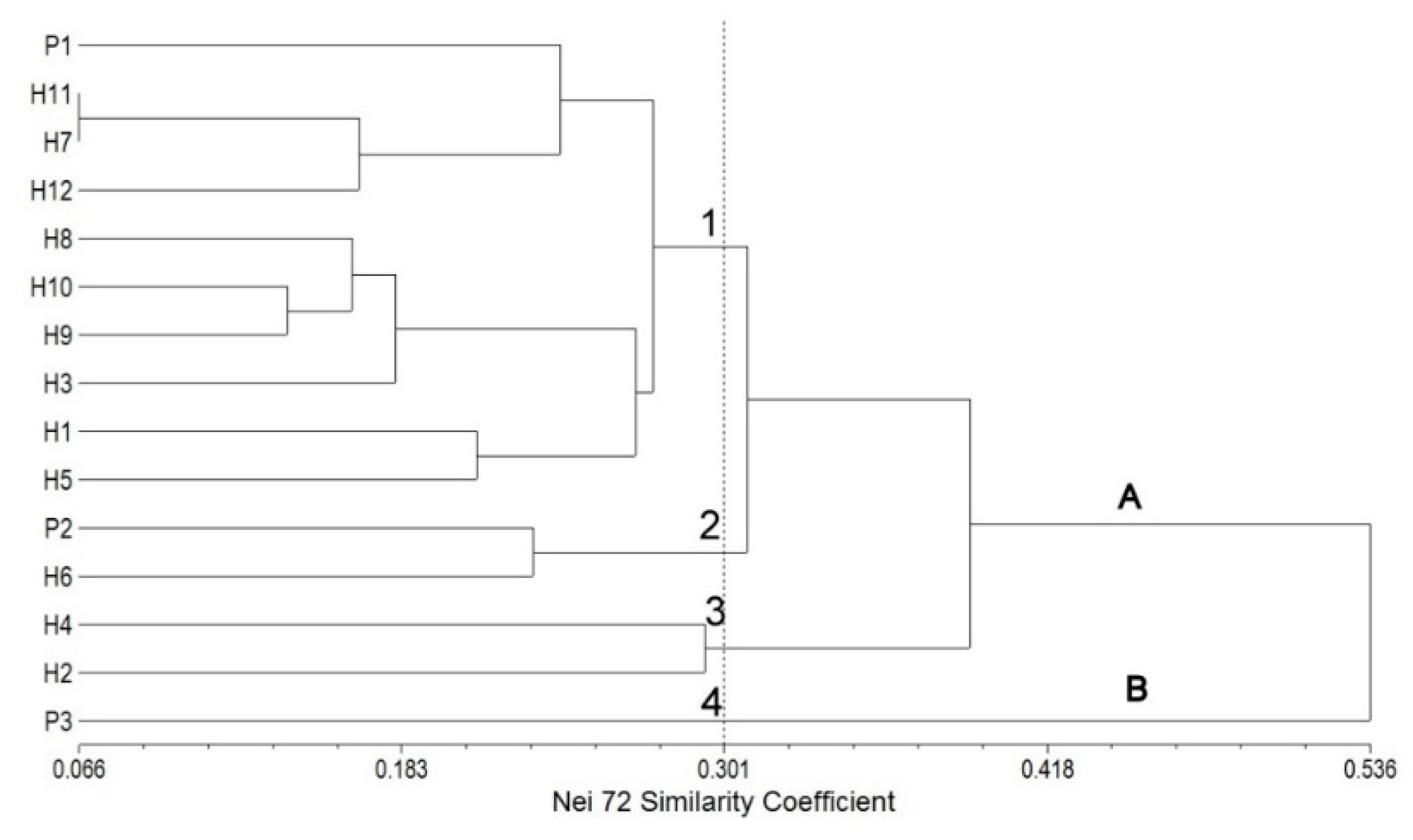
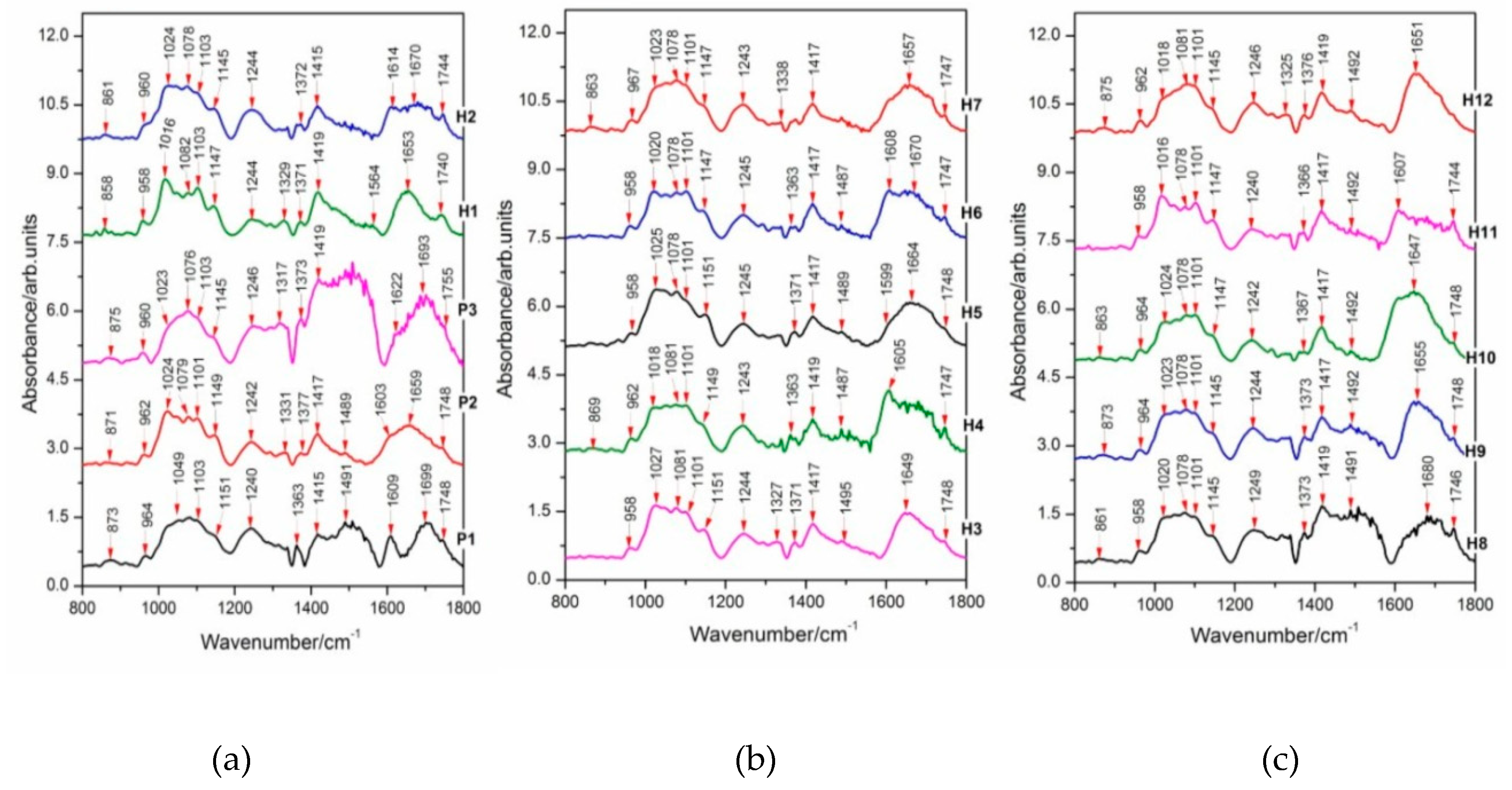
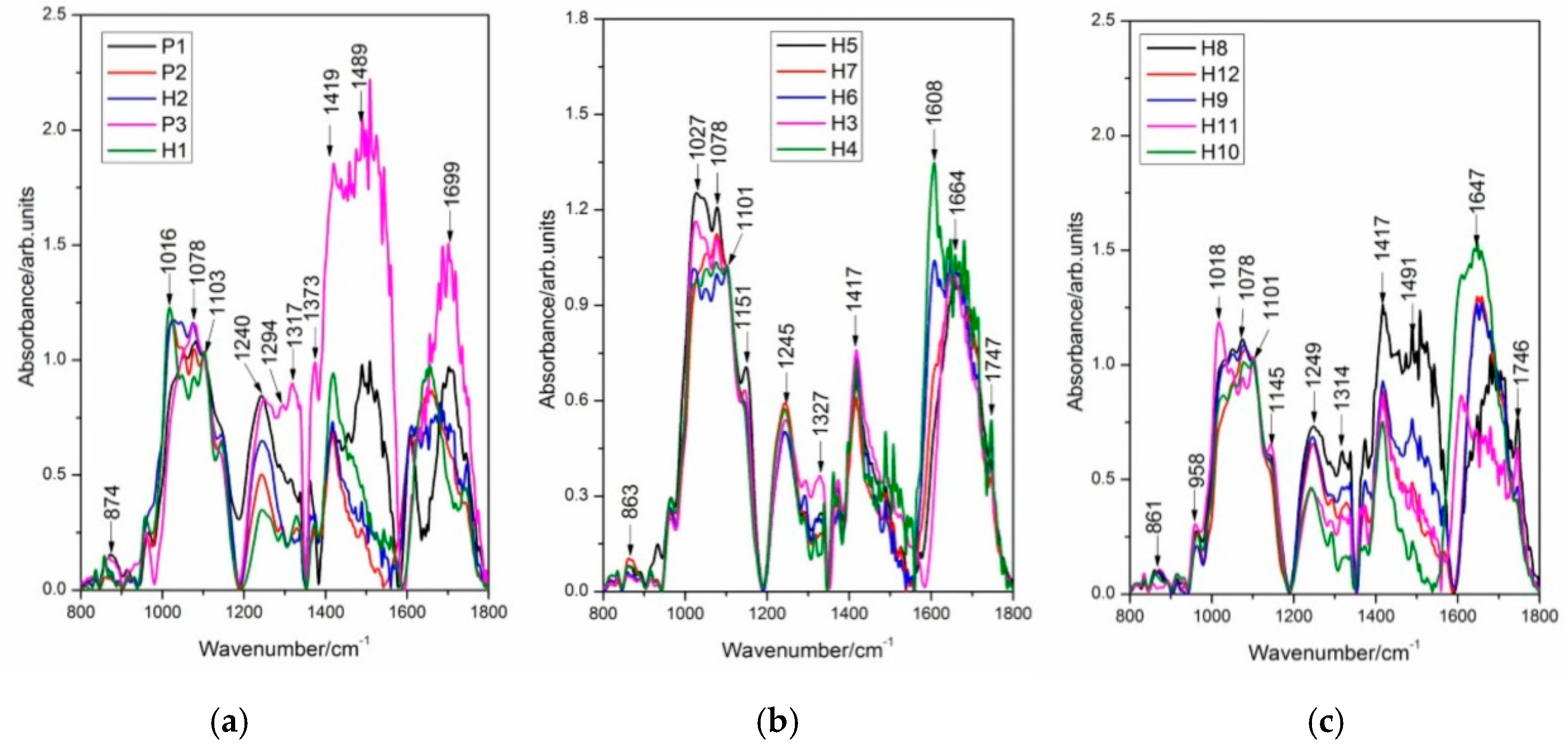
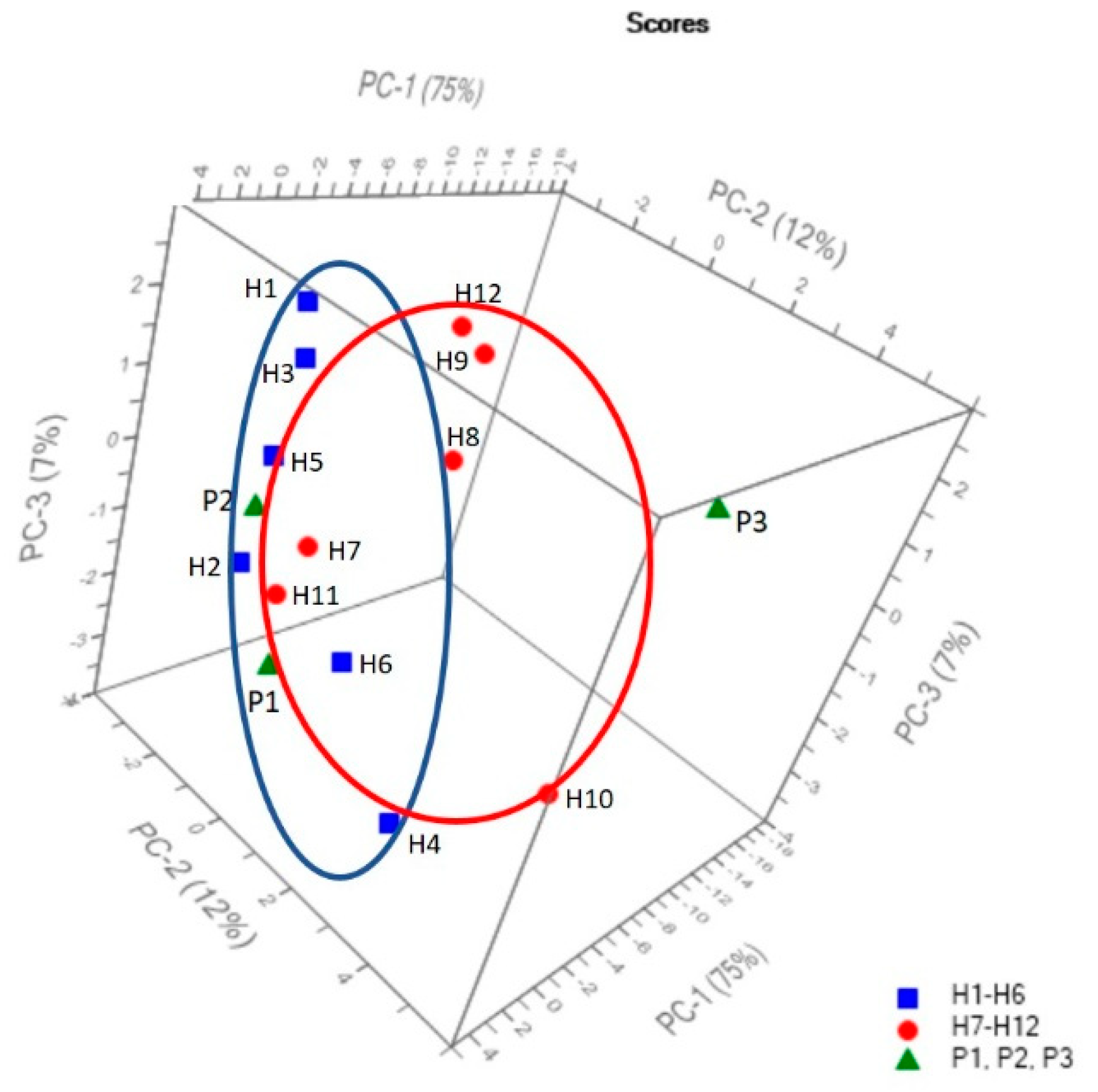
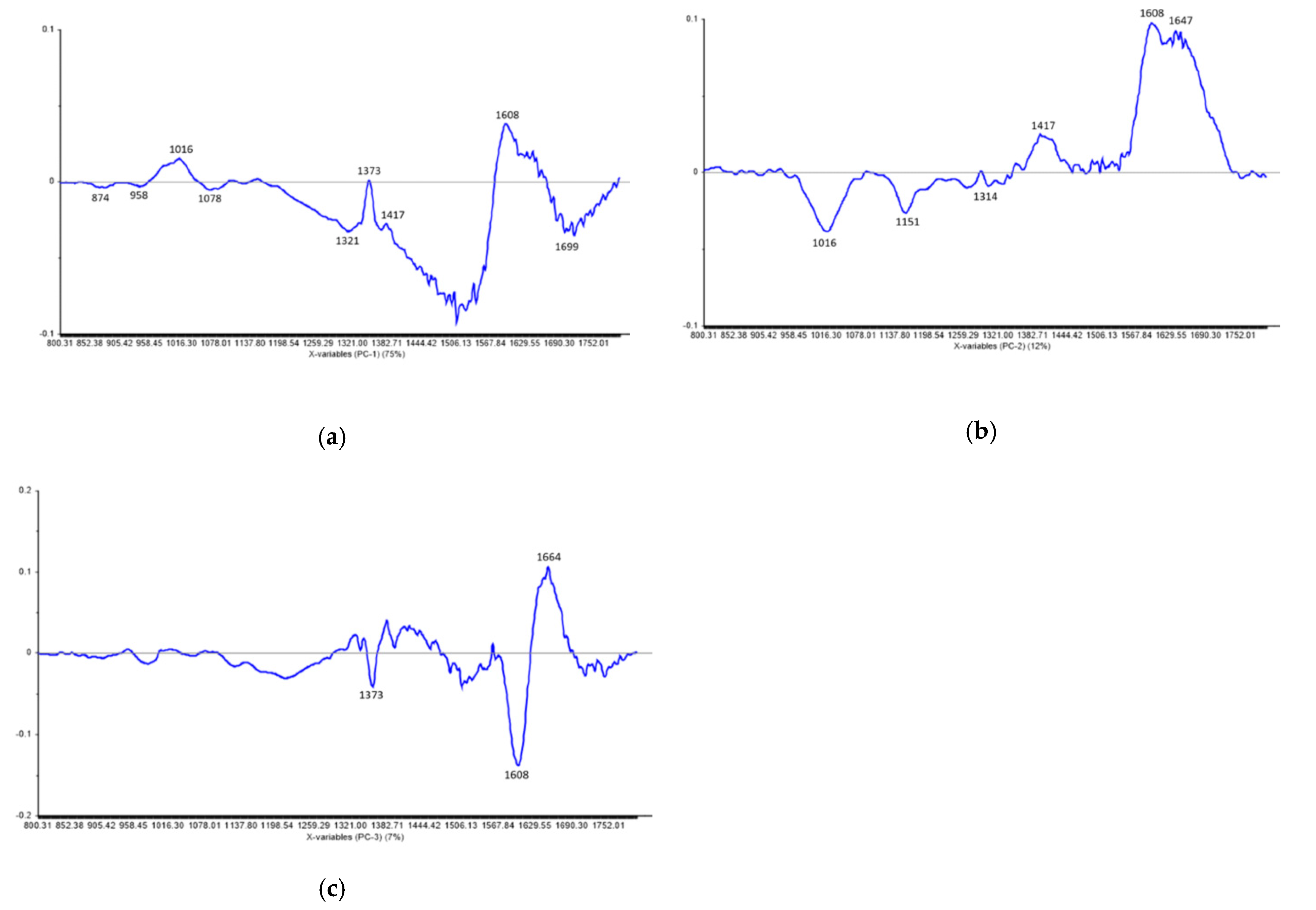
2.5. FT-IR Spectral Data Combined with Multivariate Analysis
2.6. Comparison of Molecular Marker Systems
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Pollen Viability Test
4.3. Cross Pollination Between Parents and F1 Progeny Selection
4.4. DNA Isolation
4.5. SRAP Analyses
4.6. FT-IR Spectroscopy Measurements
4.7. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chaudhury, A.; Power, J.B.; Davey, M.R. High frequency direct plant regeneration from leaf and petals of Cape Primrose (Streptocarpus). J. Crop Sci. Biotechnol 2010, 13, 107–112. [Google Scholar] [CrossRef]
- Hârţa, M.; Clapa, D.; Borsai, O.; Rusu, M.C.; Kelemen, C.; Pop, R.; Pamfil, D. Micropropagation and Assessment of Genetic Stability of Acclimatized Streptocarpus x hybridus Voss Plantlets Using RAPD Markers. Bull. UASVM Hortic. 2018, 75, 154–162. [Google Scholar] [CrossRef]
- Dibley, R. Streptocarpus, 2nd ed.Dibleys Nurseries: Wales, UK, 2008; pp. 8–10. [Google Scholar]
- Röper, A.C.; Orabi, J.; Lütken, H.; Christensen, B.; Skou, A.M.T.; Müller, R. Phenotypic and genotypic analysis of newly obtained interspecific hybrids in the Campanula genus. PLoS ONE 2015, 10, e0137537. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Nishii, K.; Barber, S.; Hackett, C.; Kidner, C.A.; Gharbi, K.; Nagano, A.J.; Iwamoto, A.; Möller, M. A first genetic map in the genus Streptocarpus generated with RAD sequencing based SNP markers. S. Afr. J. Bot. 2018, 117, 158–168. [Google Scholar] [CrossRef]
- Crino, P.; Tavazza, R.; Munoz, N.A.R.; Nisini, P.T.; Saccardo, F.; Ancora, G.; Pagnotta, M.A. Recovery, morphological and molecular characterization of globe artichoke ‘Romanesco’landraces. Genet. Resour. Crop Evol. 2008, 55, 823–833. [Google Scholar] [CrossRef]
- Kobayashi, N.; Akabane, M.; Handa, T.; Takayanagi, K. Inheritance of morphological characters and RAPD markers in intersubgeneric hybrids of azalea (Rhododendron kiusianum Makino x R. indicum (L.) (Sweet) x R. japonicum (A. Gray) suringer f. flavum Nakai. J. Japan Soc. Hort. Sci. 1996, 65, 45–53. [Google Scholar] [CrossRef][Green Version]
- Yan, Z.; Denneboom, C.; Hattendorf, A.; Dolstra, O.; Debener, T.; Stam, P.; Visser, P.B. Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theor. Appl. Genet 2005, 110, 766–777. [Google Scholar] [CrossRef]
- Rout, G.R.; Mohapatra, A. Use of molecular markers in ornamental plants: A critical reappraisal. Eur. J. Hort. Sci. 2006, 71, 53–68. [Google Scholar]
- Fu, X.P.; Ning, G.G.; Gao, L.P.; Bao, M.Z. Genetic diversity of Dianthus accessions as assessed using two molecular marker systems (SRAPs and ISSRs) and morphological traits. Sci. Hortic. 2008, 3, 263–270. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; Chang, Q.; Liu, P. Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 2011, 177, 15–24. [Google Scholar] [CrossRef]
- Yang, M.; Han, Y.; Xu, L.; Zhao, J.; Liu, Y. Comparative analysis of genetic diversity of lotus (Nelumbo) using SSR and SRAP markers. Sci. Hortic. 2012, 142, 185–195. [Google Scholar] [CrossRef]
- Longhi, S.; Giongo, L.; Buti, M.; Surbanovski, N.; Viola, R.; Velasco, R.; Ward, J.A.; Sargent, D.J. Molecular genetics and genomics of the Rosoideae: State of the art and future perspectives. Hortic. Res. 2014, 1, 1. [Google Scholar] [CrossRef] [PubMed]
- Turchetto, C.; Segatto, A.L.; Beduschi, J.; Bonatto, S.L.; Freitas, L.B. Genetic differentiation and hybrid identification using microsatellite markers in closely related wild species. AoB Plants 2015, 7, plv084. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Suzuki, T.; Nakatsuka, T.; Dohra, H.; Yamagishi, M.; Matsuyama, K.; Matsuura, H. RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom. 2016, 17, 611. [Google Scholar] [CrossRef]
- Jia, S.; Yan, Z.; Wang, Y.; Wei, Y.; Xie, Z.; Zhang, F. Genetic diversity and relatedness among ornamental purslane (Portulaca, L.) accessions unraveled by SRAP markers. 3 Biotech 2017, 7, 241. [Google Scholar] [CrossRef]
- de Oliveira Belo, G.; Souza, M.M.; Silva, G.S.; Lavinscky, M.P. Hybrids of Passiflora: P. gardneri versus P. gibertii, confirmation of paternity, morphological and cytogenetic characterization. Euphytica 2018, 214, 2. [Google Scholar] [CrossRef]
- Hao, Q.; Liu, Z.A.; Shu, Q.Y.; Zhang, R.; De Rick, J.; Wang, L.S. Studies on Paeonia cultivars and hybrids identification based on SRAP analysis. Hereditas 2008, 145, 38–47. [Google Scholar] [CrossRef]
- Aneja, B.; Yadav, N.R.; Chawla, V.; Yadav, R.C. Sequence-related amplified polymorphism (SRAP) molecular marker system and its applications in crop improvement. Mol. Breed. 2012, 30, 1635–1648. [Google Scholar] [CrossRef]
- Robarts, D.W.H.; Wolfe, A.D. Sequence-related amplified polymorphism (SRAP) markers: A potential resource for studies in plant molecular biology. Appl. Plant Sci. 2014, 2, 1400017. [Google Scholar] [CrossRef]
- Nahm, S.H.; Yang, S.G.; Kim, S.W.; Min, B.W. Rapid discrimination of F1 hybrid seeds from their parental lines and selection of protein-rich corn lines for silage corn breeding using FT-IR spectroscopy combined by multivariate analysis. J. Crop Sci. Biotechnol. 2015, 18, 161–169. [Google Scholar] [CrossRef]
- Conrad, A.O.; Bonello, P. Application of Infrared and Raman Spectroscopy for the Identification of Disease Resistant Trees. Front. Plant Sci. 2016, 6, 1152. [Google Scholar] [CrossRef] [PubMed]
- Ştefan, R.; Muntean, C.M.; Tripon, C.; Halmagyi, A.; Valimareanu, S. UV degradation of genomic DNA from in vitro grown plant species: A Fourier transform infrared spectroscopic assessment. Polym. Degrad. Stab. 2014, 108, 35–40. [Google Scholar] [CrossRef]
- Song, S.Y.; Jie, E.Y.; Ahn, M.S.; Lee, I.H.; Nou, I.S.; Min, B.W.; Kim, S.W. Fourier transform infrared (FT-IR) spectroscopy of genomic DNA to discriminate F1 progenies from their paternal lineage of Chinese cabbage (Brassica rapa subsp. pekinensis). Mol. Breed. 2014, 33, 453–464. [Google Scholar] [CrossRef]
- Hughes, M.; Möller, M.; Bellstedt, D.U.; Edwards, T.J.; Woodheas, M. EST and random genomic nuclear microsatellite markers for Streptocarpus. Mol. Ecol. Notes 2004, 4, 36–40. [Google Scholar] [CrossRef]
- Afkhami-Sarvestani, R.; Serek, M.; Winkelmann, T. Interspecific crosses within the Streptocarpus subgenus Streptocarpella and intergeneric crosses between Streptocarpella and Saintpaulia ionantha genotypes. Sci. Hortic. 2012, 148, 215–222. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, S.; Liu, J.; Zhao, Y.; Liu, J. Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE 2017, 12, e0177508. [Google Scholar] [CrossRef]
- Riek, J.D.; Calsyn, E.; Everaert, I.; Bockstaele, E.V.; Loose, M.D. AFLP based alternatives for the assessment of Distinctness, Uniformity and Stability of sugar beet varieties. Theor. Appl. Genet. 2001, 103, 1254–1265. [Google Scholar] [CrossRef]
- Pop, I.F.; Vicol, A.C.; Botu, M.; Raica, P.A.; Vahdati, K.; Pamfil, D. Relationships of walnut cultivars in a germplasm collection: Comparative analysis of phenotypic and molecular data. Sci. Hortic. 2013, 153, 124–135. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 949. [Google Scholar] [CrossRef]
- Muntean, C.M.; Ştefan, R.; Bindea, M.; Cozma, V. Fourier transform infrared spectroscopy of DNA from Borrelia burgdorferi sensu lato and Ixodes ricinus ticks. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 110, 185–192. [Google Scholar] [CrossRef]
- Taillandier, E.; Liquier, J. Vibrational spectroscopy of nucleic acids. In Handbook of Vibrational Spectroscopy Applications in Life, Pharmaceutical and Natural Sciences; Chalmers, J.M., Griffiths, P.R., Eds.; John Wiley & Sons Ltd.: Chichester, UK, 2002; pp. 3465–3480. [Google Scholar]
- Geinguenaud, F.; Calandrini, V.; Teixeira, J.; Mayer, C.; Liquier, J.; Lavelle, C.; Arluison, V. Conformational transition of DNA bound to Hfq probed by infrared spectroscopy. Phys. Chem. 2011, 13, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Rauch, C.; Pichler, A.; Trieb, M.; Wellensohn, B.; Liedt, K.R.; Mayer, E. Z-DNA’s Conformer Substrates Revealed by FT-IR Difference Spectroscopy of Nonoriented Left-Handed Double Helical Poly (dG-dC). J. Biomol. Struct. Dyn. 2005, 22, 595–614. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef] [PubMed]
- Taboury, J.A.; Liquier, J.; Taillandier, E. Characterization of DNA structures by infrared spectroscopy: Double helical forms of poly(dG-dC)•poly(dG-dC), poly(dD8G-dC)•poly(dD8G-dC), and poly(dG-dm5C)•poly(dG-dm5C). Can. J. Chem. 1985, 63, 1904–1909. [Google Scholar] [CrossRef]
- Pevsner, A.; Diem, M. Infrared Spectroscopic Studies of Major Cellular Components. Part II: The Effect of Hydration on the Spectra of Nucleic Acids. Appl. Spectrosc. 2001, 55, 1502–1505. [Google Scholar] [CrossRef]
- Andrushchenko, V.; Van De Sande, J.H.; Wieser, H. Vibrational circular dichroism and IR absorption of DNA complexes with Cu2+ ions. Biopolymers 2003, 72, 374–390. [Google Scholar] [CrossRef]
- Sfihi, H.; Liquier, J.; Urpi, L.; Verdaguer, N.; Subirana, J.A.; Igolen, J.; Taillandier, E. A and Z canonical conformations in d (CnGCGn) crystals characterized by microFTIR and microRaman spectroscopies. Biopolymers 1993, 33, 1715–1723. [Google Scholar] [CrossRef]
- Taillandier, E.; Liquier, J. Infrared spectroscopy of DNA. Methods Enzymol. 1992, 211, 307–335. [Google Scholar]
- Taillandier, E.; Liquier, J.; Ghomi, M. Conformational transitions of nucleic acids studied by IR and Raman spectroscopies. J. Mol. Struct. 1989, 214, 85–211. [Google Scholar] [CrossRef]
- Taillandier, E.; Peticolas, W.L.; Adam, S.; Huynh Dinh, T.; Igolen, J. Polymorphism of the d(CCCGCGGG)2 double helix studied by FT-i.r. spectroscopy Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 1990, 46, 107–112. [Google Scholar] [CrossRef]
- Arakawa, H.; Neault, J.F.; Tajmir-Riahi, H.A. Silver (I) complexes with DNA and RNA studied by Fourier transform infrared spectroscopy and capillary electrophoresis. Biophys. J. 2001, 81, 1580–1587. [Google Scholar] [CrossRef]
- Adnet, F.; Liquier, J.; Taillandier, E.; Singh, M.P.; Rao, K.E.; Lown, J.W. FTIR study of specific binding interactions between DNA minor groove binding ligands and polynucleotides. J. Biomol. Struct. Dyn. 1992, 10, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Lema-Rumińska, J.; Zalewska, M. Changes in flower colour among Lady Group of Chrysanthemum× grandiflorum/Ramat./Kitam. as a result of mutation breeding. Folia Hortic. 2005, 17, 61–72. [Google Scholar]
- Gitonga, V.W.; Stolker, R.; Ribot, S.; Keizer, P.; Koning-Boucoiran, C.F.S.; Krens, F.A. Inheritance of determinants of flower colour in tetraploid roses. In Proceedings of the XXIII International Eucarpia Symposium, Section Ornamentals: Colourful Breeding and Genetics, Leiden, The Netherlands, 31 August 2009. [Google Scholar]
- Aros, D.; Suazo, M.; Rivas, C.; Zapata, P.; Ubeda, C.; Bridge, M. Molecular and morphological characterization of new interspecific hybrids of alstroemeria originated from A. caryophylleae scented lines. Euphytica 2019, 215, 93. [Google Scholar] [CrossRef]
- Richards, A.J. Plant Breeding Systems; George Allen & Unwin: London, UK, 1986; pp. 381–458. [Google Scholar]
- Oehlkers, F. Cytoplasmic inheritance in the genus Streptocarpus Lindley. Adv. Genet. 1964, 12, 329–370. [Google Scholar]
- Xuan, C.; Guo, H.; Xue, D.; Liu, J. Identification of Zoysia hybrids by SRAP analysis. Mol. Plant Breed. 2008, 6, 1233–1238. [Google Scholar]
- Mishra, M.K.; Suresh, N.; Bhat, A.M.; Suryaprakash, N.; Kumar, S.; Kumar, A.; Jayarama, A. Genetic molecular analysis of Coffea Arabica (Rubiaceae) hybrids using SRAP markers. Rev. Biol. Trop. 2011, 59, 607–617. [Google Scholar]
- Letellier, R.; Ghomi, M.; Taillandier, E. Interpretation of DNA vibration modes: IV—A single-helical approach to assign the phosphate-backbone contribution to the vibrational spectra in A and B conformations. J. Biomol. Struct. Dyn. 1989, 6, 755–768. [Google Scholar] [CrossRef]
- Lindqvist, M.; Gräslund, A. An FTIR and CD study of the structural effects of G-tract length and sequence context on DNA conformation in solution. J. Mol. Biol. 2001, 314, 423–432. [Google Scholar] [CrossRef]
- Muntean, C.M.; Halmagyi, A.; Puia, M.D.; Pavel, I. FT-Raman of genomic DNA from plant tissues. J. Spectrosc. 2009, 23, 59–70. [Google Scholar] [CrossRef]
- Cantor, M.; Stana, D.; Pop, I. Streptocarpus -flowering pot plant -propagation and culture. Not. Bot. Horti. Agrobot. Cluj Napoca 2004, 32, 15–19. [Google Scholar] [CrossRef]
- Lázaro, A.; Totland, Ø. The influence of floral symmetry, dependence on pollinators and pollination generalization on flower size variation. Ann. Bot. 2014, 114, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Guang-Ning, Z.; Weeden, F.N.F.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Pop, R.; Ardelean, M.; Pamfil, D.; Gaboreanu, I.M. The efficiency of different DNA isolation and purification in ten cultivars of Vitis vinifera. Bul. USAMV CN (ZB) 2003, 59, 259–261. [Google Scholar]
- Bodea, M.; Pamfil, D.; Pop, R.; Sisea, R.C. DNA isolation from desiccated leaf material from plum tree (Prunus domestica L.) molecular analysis. Bul. UASVM CN (H) 2016, 1, 1–2. [Google Scholar] [CrossRef]
- Wang, S.; Yang, W.; Shen, H. Genetic diversity in Apium graveolens and related species revealed by SRAP and SSR markers. Sci. Hortic. 2011, 129, 1–8. [Google Scholar] [CrossRef]
- Sneath, P.; Sokal, R.R. Numerical taxonomy. In The Principles and Practice of Numerical Classification, 1st ed.; Freeman, W.H., Ed.; Taylor & Francis: San Francisco, CA, USA, 1973; pp. 39–42. [Google Scholar]
- Mgendi, M.G.; Manoko, M.K.; Nyomora, A.M. Genetic diversity between cultivated and non-cultivated Moringa oleifera Lam. provenances assessed by RAPD markers. J. Cell Mol. Biol. 2010, 8, 95–102. [Google Scholar]
- Ward, J. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]

| Sample no. | Parents and F1 Progenies | Color | RHSCC Code |
|---|---|---|---|
| 1 | P1 (“Black Panther”) | dark violet, blue violet | 83B, N88A |
| 2 | P2 (“Slumber Song”) | purple pink, yellow | N73A, 4A |
| 3 | P3 (“Snow White”) | white | 155B |
| 4 | H1 | violet | N82B |
| 5 | H2 | pink violet | N77B |
| 6 | H3 | pink purple, blue violet | 72A, N88A |
| 7 | H4 | violet | N82D |
| 8 | H5 | dark violet, violet | 83B, 82B |
| 9 | H6 | purple pink, yellow, white | N74C, 6A, 155B |
| 10 | H7 | blue violet | N88C |
| 11 | H8 | pink violet, white | N77B, 155A |
| 12 | H9 | violet | N89A |
| 13 | H10 | blue violet, dark violet | N88C, 83B |
| 14 | H11 | blue violet, violet | N88C, 82B |
| 15 | H12 | blue violet, dark pink violet | N88A, N77A |
| Primer Combination | Number of Total Bands | Size Range of Bands (bp) | Number of Polymorphic Bands | % of Polymorphism | PIC |
|---|---|---|---|---|---|
| Me8-Em6 | 11 | 487–1553 | 10 | 90.90 | 0.42 |
| Me8-Em2 | 9 | 686–1949 | 8 | 88.88 | 0.34 |
| Me4-Em8 | 13 | 109–1502 | 12 | 92.30 | 0.45 |
| Me1-Em8 | 7 | 104–523 | 7 | 100 | 0.35 |
| Me6-Em1 | 15 | 237–1697 | 14 | 93.33 | 0.39 |
| Me6-Em8 | 11 | 211–1348 | 11 | 100 | 0.37 |
| Me1-Em2 | 16 | 175–1712 | 15 | 93.75 | 0.48 |
| Me4-Em2 | 14 | 219–1013 | 13 | 92.85 | 0.46 |
| Total | 96 | 90 | - | - | |
| Mean | 12 ± 0.02 | 11.25 ± 0.06 | 93.75 ± 0.26 | 0.40 ± 0.07 |
| Type of SRAP Marker | Band Presence/ Absence in ♀ Parent | Band Presence/ Absence in ♂ Parent | Band Presence/ Absence in F1 Hybrid | Total Number of Occurrences for Each Type of SRAP Marker | |
|---|---|---|---|---|---|
| P1 × P2 | P1 × P3 | ||||
| I | + | + | + | 96 | 59 |
| II | + | − | + | 16 | 58 |
| III | − | + | + | 31 | 19 |
| Total | 143 | 136 | |||
| Streptocarpus Parents and Hybrids | Tentative Assignment 1 | References | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | H1 | H2 | H3 | H4 | H5 | H6 | ||
| 873 | 871 | 858 | 861 | - | 869 | - | - | bk, deoxyribose, possible A-form | [31,32,33,34] |
| 964 | 962 | 958 | 960 | 958 | 962 | 958 | 958 | Furanose-phosphate skeletal motions | [35] |
| - | 1024 | 1016 | 1024 | 1027 | 1018 | 1025 | 1020 | Deoxyribose | [36] |
| - | 1079 | 1082 | 1078 | 1081 | 1081 | 1078 | 1078 | υsPO2− | [31,37,38] |
| 1103 | 1101 | 1103 | 1103 | 1101 | 1101 | 1101 | 1101 | υsPO2− | [37,38] |
| 1151 | 1149 | 1147 | 1145 | 1151 | 1149 | 1151 | 1147 | Deoxyribose, C3‘-endo/anti, A-form | [39] |
| 1240 | 1242 | 1244 | 1244 | 1244 | 1243 | 1245 | 1245 | υaPO2−, possible A-form | [31,32,40] |
| 1363 | 1377 | 1371 | 1372 | 1371 | 1363 | 1371 | 1363 | dA, dG (C2′-endo/anti) | [32,39,41] |
| 1415 | 1417 | 1419 | 1415 | 1417 | 1419 | 1417 | 1417 | C2′-endo/anti sugar pucker (B-form) | [32,39] |
| 1491 | 1489 | - | - | 1495 | 1487 | 1489 | 1487 | Base ring modes | [42] |
| 1609 | 1603 | - | 1614 | - | 1605 | 1599 | 1608 | dA, possible C=C, C=N | [31,43,44] |
| - | 1659 | 1653 | 1670 | 1649 | - | 1664 | 1670 | dT (C=O) | [32] |
| Streptocarpus Parents and Hybrids | Tentative Assignment 1 | References | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | P3 | H7 | H8 | H9 | H10 | H11 | H12 | ||
| 873 | 875 | 863 | 861 | 873 | 863 | - | 875 | bk, deoxyribose, possible A-form | [31,32,33,34] |
| 964 | 960 | 967 | 958 | 964 | 964 | 958 | 962 | Furanose-phosphate skeletal motions | [35] |
| - | 1023 | 1023 | 1020 | 1023 | 1024 | 1016 | 1018 | Deoxyribose | [36] |
| 1049 | 1076 | 1078 | 1078 | 1078 | 1078 | 1078 | 1081 | υsPO2− | [31,37,38] |
| 1103 | 1103 | 1101 | 1101 | 1101 | 1101 | 1101 | 1101 | υsPO2− | [37,38] |
| 1151 | 1145 | 1147 | 1145 | 1145 | 1147 | 1147 | 1145 | Deoxyribose, C3‘-endo/anti, A-form | [39] |
| 1240 | 1246 | 1243 | 1249 | 1244 | 1242 | 1240 | 1246 | υaPO2−, possible A-form | [31,32,40] |
| 1363 | 1373 | - | 1373 | 1373 | 1367 | 1366 | 1376 | dA, dG (C2′-endo/anti) | [32,39,41] |
| 1415 | 1419 | 1417 | 1419 | 1417 | 1417 | 1417 | 1419 | C2′-endo/anti sugar pucker (B-form) | [32,39] |
| 1491 | - | - | 1491 | 1492 | 1492 | 1492 | 1492 | Base ring modes | [42] |
| 1609 | 1622 | - | - | - | - | 1607 | - | dA, possible C=C, C=N | [31,43,44] |
| - | - | 1657 | - | 1655 | 1647 | - | 1651 | dT (C=O) | [32] |
| Primer | Sequences 3′-5′ | Primer | Sequences 5′-3′ |
|---|---|---|---|
| Me*1 | TGAGTCCAAACCGGATA | Em3 | GACTGCGTACGAATTGAC |
| Me4 | TGAGTCCAAACCGGACC | Em4 | GACTGCGTACGAATTTGA |
| Me6 | TGAGTCCAAACCGGTAA | Em5 | GACTGCGTACGAATTAAC |
| Me8 | TGAGTCCAAACCGGTGC | Em6 | GACTGCGTACGAATTGCA |
| Em**1 | GACTGCGTACGAATTAAT | Em7 | GACTGCGTACGAATTCAA |
| Em2 | GACTGCGTACGAATTTGC | Em8 | GACTGCGTACGAATTCTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hârţa, M.; Borsai, O.; Muntean, C.M.; Dina, N.E.; Fǎlǎmaş, A.; Olar, L.E.; Szabo, K.; Pamfil, D.; Ştefan, R. Assessment of Genetic Relationships between Streptocarpus x hybridus V. Parents and F1 Progenies Using SRAP Markers and FT-IR Spectroscopy. Plants 2020, 9, 160. https://doi.org/10.3390/plants9020160
Hârţa M, Borsai O, Muntean CM, Dina NE, Fǎlǎmaş A, Olar LE, Szabo K, Pamfil D, Ştefan R. Assessment of Genetic Relationships between Streptocarpus x hybridus V. Parents and F1 Progenies Using SRAP Markers and FT-IR Spectroscopy. Plants. 2020; 9(2):160. https://doi.org/10.3390/plants9020160
Chicago/Turabian StyleHârţa, Monica, Orsolya Borsai, Cristina M. Muntean, Nicoleta E. Dina, Alexandra Fǎlǎmaş, Loredana Elena Olar, Katalin Szabo, Doru Pamfil, and Răzvan Ştefan. 2020. "Assessment of Genetic Relationships between Streptocarpus x hybridus V. Parents and F1 Progenies Using SRAP Markers and FT-IR Spectroscopy" Plants 9, no. 2: 160. https://doi.org/10.3390/plants9020160
APA StyleHârţa, M., Borsai, O., Muntean, C. M., Dina, N. E., Fǎlǎmaş, A., Olar, L. E., Szabo, K., Pamfil, D., & Ştefan, R. (2020). Assessment of Genetic Relationships between Streptocarpus x hybridus V. Parents and F1 Progenies Using SRAP Markers and FT-IR Spectroscopy. Plants, 9(2), 160. https://doi.org/10.3390/plants9020160









