HTS-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine
Abstract
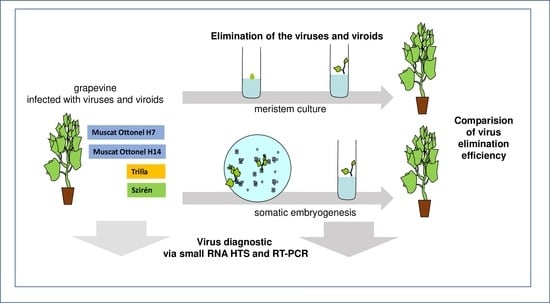
1. Introduction
2. Results
2.1. Virus Diagnostics of the Mother Plants
2.2. Virus Elimination Using Somatic Embryogenesis
2.3. Virus Elimination Using a Meristem Culture
2.4. Comparison of the Efficiency of the Sanitation Methods
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Grapevine In Vitro Cultures
4.3. Meristem Cultures
4.4. Somatic Embryogenesis
4.5. Pipeline for Data Evaluation of the HTS Results (Bioinformatics)
4.6. Validation of Predicted Viral Diagnostics by RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martelli, G. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Meng, B.; Martelli, G.P.; Golino, D.A.; Fuchs, M. Grapevine viruses: Molecular Biology, Diagnostics and Management; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Perrin, M.; Gertz, C.; Masson, J.E. High efficiency initiation of regenerable embryogenic callus from anther filaments of 19-grapevine genotypes grown worldwide. Plant Sci. 2004, 167, 1343–1349. [Google Scholar] [CrossRef]
- Skiada, F.; Grigoriadou, K.; Maliogka, V.; Katis, N.; Eleftheriou, E. Elimination of grapevine leafroll-associated virus 1 and grapevine rupestris stem pitting-associated virus from grapevine cv. Agiorgitiko, and a micropropagation protocol for mass production of virus-free plantlets. J. Plant Pathol. 2009, 91, 177–184. [Google Scholar]
- Skiada, F.G.; Maliogka, V.I.; Katis, N.I.; Eleftheriou, E.P. Elimination of grapevine rupestris stem pitting-associated virus (GRSPaV) from two vitis vinifera cultivars by in vitro chemotherapy. Eur. J. Plant Pathol. 2013, 135, 407–414. [Google Scholar] [CrossRef]
- Wang, Q.; Valkonen, J.P. Cryotherapy of shoot tips: Novel pathogen eradication method. Trends Plant Sci. 2009, 14, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Panattoni, A.; Luvisi, A.; Triolo, E. Elimination of viruses in plants: Twenty years of progress. Span. J. Agric. Res. 2013, 11, 173–188. [Google Scholar] [CrossRef]
- Ben Mahmoud, K.; Najar, A.; Jemai, N.; Jemmali, A. Advances in sanitation methods for fruit tree species through in vitro technologies: Possibilities and limits. J. New Sci. 2017, 45, 2483–2495. [Google Scholar]
- Damba, Y.; Quainoo, A.; Sowley, E. Effectiveness of Somatic Embryogenesis in Eliminating the Cassava Mosaic Virus from Infected Cassava (Manihot Esculenta Crantz) Plant Materials. Int. J. Sci. Technol. Res. 2013, 2, 282–287. [Google Scholar]
- Quainoo, A. Age of callus tissues and cotyledonary materials on the selection of cocoa swollen shoot virus-free somatic embryos. Res. Biotechnol. 2011, 2, 75–81. [Google Scholar]
- Meziane, M.; Frasheri, D.; Carra, A.; Boudjeniba, M.; D’Onghia, A.M.; Mercati, F.; Djelouah, K.; Carimi, F. Attempts to eradicate graft-transmissible infections through somatic embryogenesis in citrus ssp. And analysis of genetic stability of regenerated plants. Eur. J. Plant Pathol. 2017, 148, 85–95. [Google Scholar] [CrossRef]
- D’onghia, A.; Carimi, F.; De Pasquale, F.; Djelouah, K.; Martelli, G. Elimination of citrus psorosis virus by somatic embryogenesis from stigma and style cultures. Plant Pathol. 2001, 50, 266–269. [Google Scholar] [CrossRef]
- Parmessur, Y.; Aljanabi, S.; Saumtally, S.; Dookun-Saumtally, A. Sugarcane yellow leaf virus and sugarcane yellows phytoplasma: Elimination by tissue culture. Plant Pathol. 2002, 51, 561–566. [Google Scholar] [CrossRef]
- Goussard, P.; Wiid, J.; Kasdorf, G. The effectiveness of in vitro somatic embryogenesis in eliminating fanleaf virus and leafroll associated viruses from grapevines. S. Afr. J. Enol. Viticult. 1991, 12, 77–81. [Google Scholar] [CrossRef]
- Borroto-Fernandez, E.G.; Sommerbauer, T.; Popowich, E.; Schartl, A.; Laimer, M. Somatic embryogenesis from anthers of the autochthonous vitis vinifera cv. Domina leads to arabis mosaic virus-free plants. Eur. J. Plant Pathol. 2009, 124, 171–174. [Google Scholar] [CrossRef]
- Peiro, R.; Gammoudi, N.; Yuste, A.; Olmos, A.; Gisbert, C. Mature seeds for in vitro sanitation of the grapevine leafroll associated virus (glrav-1 and glrav-3) from grape (vitis vinifera l.). Span. J. Agric. Res. 2015, 13, 1005. [Google Scholar] [CrossRef]
- Popescu, C.; Buciumeanu, E.-C.; Visoiu, E. Somatic embryogenesis a reliable method for grapevine fleck virus-free regeneration. In Proceedings of the 14th Meeting of International Council for the Study of Virus and Virus-Like Diseases of the Grapevine, Bari, Italy, 12–17 September 2003. [Google Scholar]
- Gambino, G.; Bondaz, J.; Gribaudo, I. Detection and elimination of viruses in callus, somatic embryos and regenerated plantlets of grapevine. Eur. J. Plant Pathol. 2006, 114, 397–404. [Google Scholar] [CrossRef]
- Gambino, G.; Matteo, D.; Gribaudo, I. Elimination of grapevine fanleaf virus from three vitis vinifera cultivars by somatic embryogenesis. Eur. J. Plant Pathol. 2009, 123, 57–60. [Google Scholar] [CrossRef]
- Gambino, G.; Navarro, B.; Vallania, R.; Gribaudo, I.; Di Serio, F. Somatic embryogenesis efficiently eliminates viroid infections from grapevines. Eur. J. Plant Pathol. 2011, 130, 511–519. [Google Scholar] [CrossRef]
- Gribaudo, I.; Gambino, G.; Cuozzo, D.; Mannini, F. Attempts to eliminate grapevine rupestris stem pitting-associated virus from grapevine clones. J. Plant Pathol. 2006, 88, 293–298. [Google Scholar]
- Malenica, N.; Jagić, M.; Pavletić, B.; Bauer, N.; Vončina, D.; Zdunić, G.; Leljak Levanić, D. Somatic embryogenesis as a tool for virus elimination in croatian indigenous grapevine cultivars. Acta Bot. Croat. 2020, 79, 26–34. [Google Scholar] [CrossRef]
- San Pedro, T.; Gammoudi, N.; Peiro, R.; Olmos, A.; Gisbert, C. Somatic embryogenesis from seeds in a broad range of vitis vinifera l. Varieties: Rescue of true-to-type virus-free plants. BMC Plant Biol. 2017, 17, 216. [Google Scholar] [CrossRef] [PubMed]
- Bouamama-Gzara, B.; Selmi, I.; Chebil, S.; Melki, I.; Mliki, A.; Ghorbel, A.; Carra, A.; Carimi, F.; Mahfoudhi, N. Elimination of grapevine leafroll associated virus-3, grapevine rupestris stem pitting associated virus and grapevine virus a from a tunisian cultivar by somatic embryogenesis and characterization of the somaclones using ampelographic descriptors. Plant Pathol. J. 2017, 33, 561–571. [Google Scholar] [PubMed]
- Eichmeier, A.; Kominkova, M.; Pecenka, J.; Kominek, P. High-throughput small rna sequencing for evaluation of grapevine sanitation efficacy. J. Virol. Methods 2019, 267, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Czotter, N.; Molnar, J.; Szabó, E.; Demian, E.; Kontra, L.; Baksa, I.; Szittya, G.; Kocsis, L.; Deak, T.; Bisztray, G. NGS of virus-derived small RNAs as a diagnostic method used to determine viromes of hungarian vineyards. Front. Microbiol. 2018, 9, 122. [Google Scholar] [CrossRef]
- Demian, E.; Jaksa-Czotter, N.; Molnar, J.; Tusnady, G.E.; Kocsis, L.; Varallyay, E. Grapevine rootstocks can be a source of infection with non-regulated viruses. Eur. J. Plant Pathol. 2020, 156, 897–912. [Google Scholar] [CrossRef]
- Gualandri, V.; Bianchedi, P.; Morelli, M.; Giampetruzzi, A.; Valenzano, P.; Bottalico, G.; Campanale, A.; Saldarelli, P. Pp 51-production of grapevine pinot gris virus-free germplasm: Techniques and tools. In Proceedings of the 18th Congress of ICVG, Ankara, Turkey, 7–11 September 2015. [Google Scholar]
- Komínek, P.; Komínková, M.; Jandová, B. Effect of repeated ribavirin treatment on grapevine viruses. Acta Virol. 2016, 60, 400. [Google Scholar] [CrossRef]
- Maliogka, V.; Skiada, F.; Eleftheriou, E.; Katis, N. Elimination of a new ampelovirus (glrav-pr) and grapevine rupestris stem pitting associated virus (GRSPaV) from two vitis vinifera cultivars combining in vitro thermotherapy with shoot tip culture. Sci. Hortic. 2009, 123, 280–282. [Google Scholar] [CrossRef]
- Wang, M.-R.; Cui, Z.-H.; Li, J.-W.; Hao, X.-Y.; Zhao, L.; Wang, Q.-C. In vitro thermotherapy-based methods for plant virus eradication. Plant Methods 2018, 14, 87. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for rna extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Czotter, N.; Molnár, J.; Pesti, R.; Demián, E.; Baráth, D.; Varga, T.; Várallyay, É. Use of sirnas for diagnosis of viruses associated to woody plants in nurseries and stock collections. Methods Mol. Biol. 2018, 1746, 115–130. [Google Scholar]
- Suzuki, R.M.; Kerbauy, G.B. Effects of light and ethylene on endogenous hormones and development of catasetum fimbriatum (orchidaceae). Braz. J. Plant Physiol. 2006, 18, 359–365. [Google Scholar] [CrossRef][Green Version]
- Olah, R. The use of activated charcoal in grapevine tissue culture. Grapevine Res. 2017, 56, 161–171. [Google Scholar]
- Oláh, R.; Zok, A.; Pedryc, A.; Howard, S.; Kovacs, L. Somatic embryogenesis in a broad spectrum of grape genotypes. Sci. Hortic. 2009, 120, 134–137. [Google Scholar] [CrossRef]
- Gribaudo, I.; Gambino, G.; Vallania, R. Somatic embryogenesis from grapevine anthers: The optimal developmental stage for collecting explants. Am. J. Enol. Vitic. 2004, 55, 427–430. [Google Scholar]
| Cultivar/Clone | Status of the Plant | Library Code |
|---|---|---|
| Muscat Ottonel 7 | Mother Plant | 1_MO7 |
| Muscat Ottonel 14 | 2_MO14 | |
| Trilla | 3_T | |
| Sziren | 4_SZ | |
| Muscat Ottonel 7 | Lines Prepared with Somatic Embryogenesis | 5_MO7_SE |
| Muscat Ottonel 14 | 6_MO14_SE | |
| Trilla | 7_T_SE | |
| Sziren | 8_SZ_SE | |
| Trilla | Lines Prepared with Meristem Culture | 9_T_ME |
| Sziren | 10_SZ_ME |
| Library Code | Virus Diagnostics | Viruses | Viroids | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GFkV | GRVFV | GSyV-1 | GRSPaV | GVT | GPGV | HSVd | GYSVd-1 | ||
| MO7 | sRNS HTS | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 |
| RT-PCR | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| MO14 | sRNS HTS | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| RT-PCR | 1 | (1) | 1 | 1 | 1 | 1 | 1 | 1 | |
| T | sRNS HTS | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 |
| RT-PCR | 0 | 0 | 1 | 1 | (1) | 1 | 1 | 1 | |
| SZ | sRNS HTS | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 |
| RT-PCR | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Cultivars/Clones | Numbers of Anthers | Numbers of Calli | Numbers of Embryogenic Calli | Numbers of Regenerated Lines | Numbers of in Vitro Lines Tested via sRNA HTS * |
|---|---|---|---|---|---|
| Muscat ottonel H-7-3 | 243 | 17 | 5 | 18 | 7 |
| Muscat ottonel H-14-1 | 273 | 15 | 2 | 19 | 8 |
| ‘Trilla’ | 280 | 15 | 9 | 55 | 12 |
| ’Sziren’ | 251 | 34 | 9 | 32 | 11 |
| Cultivar/Clone | Numbers of Meristems | Numbers of Growing Shoots | Numbers of Small Plantlets with no Roots | Numbers of Regenerated Independent Lines Plants with Roots Tested via sRNA HTS * |
|---|---|---|---|---|
| Muscat Ottonel H-7-3 | 5 | 0 | 0 | 0 |
| Muscat Ottonel H-14-1 | 7 | 0 | 0 | 0 |
| ’Trilla’ | 42 | 15 | 11 | 5 |
| ’Sziren’ | 25 | 10 | 9 | 4 |
| Library Code | Virus Diagnostics | Viruses | Viroids | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GFkV | GRVFV | GSyV-1 | GRSPaV | GVT | GPGV | HSVd | GYSVd-1 | |||
| MO7_SE | sRNS HTS | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| RT-PCR | pool | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| lines | n | N | n | 0/7 | 0 | n | 0/7 | 1/7 | ||
| MO14_SE | sRNS HTS | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| RT-PCR | pool | 0 | 0 | 0 | 0 | (1) | 0 | 0 | 1 | |
| lines | n | n | n | 0/8 | 0/8 | n | 0/8 | 2/8 | ||
| T_SE | sRNS HTS | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| RT-PCR | pool | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| lines | n | n | n | 0/12 | 0/12 | 0/12 | 2/12 | 11/12 | ||
| T_ME | sRNS HTS | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| RT-PCR | pool | 0 | 0 | 0 | (1) | 0 | 0 | 1 | 1 | |
| lines | n | n | n | 3/5 | (1)/5 | (2)/5 | 5/5 | 5/5 | ||
| SZ_SE | sRNS HTS | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| RT-PCR | pool | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | |
| lines | n | n | n | 6/11 | 0/11 | 0/11 | 2/11 | 2/11 | ||
| SZ_ME | sRNS HTS | 0 | 0 | 0 | 1 | 1 | 1 | 1 | (1) | |
| RT-PCR | pool | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | |
| lines | n | n | n | 2/4 | 2/4 | 2/4 | 2/4 | 0/4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turcsan, M.; Demian, E.; Varga, T.; Jaksa-Czotter, N.; Szegedi, E.; Olah, R.; Varallyay, E. HTS-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine. Plants 2020, 9, 1782. https://doi.org/10.3390/plants9121782
Turcsan M, Demian E, Varga T, Jaksa-Czotter N, Szegedi E, Olah R, Varallyay E. HTS-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine. Plants. 2020; 9(12):1782. https://doi.org/10.3390/plants9121782
Chicago/Turabian StyleTurcsan, Mihaly, Emese Demian, Tunde Varga, Nikoletta Jaksa-Czotter, Erno Szegedi, Robert Olah, and Eva Varallyay. 2020. "HTS-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine" Plants 9, no. 12: 1782. https://doi.org/10.3390/plants9121782
APA StyleTurcsan, M., Demian, E., Varga, T., Jaksa-Czotter, N., Szegedi, E., Olah, R., & Varallyay, E. (2020). HTS-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine. Plants, 9(12), 1782. https://doi.org/10.3390/plants9121782