ER-Phagy and Its Role in ER Homeostasis in Plants
Abstract
1. Introduction
2. Autophagy and the ER
3. The ER as an Autophagosome Membrane Source
4. Function of Plant ER Stress Response Pathways in ER Homeostasis
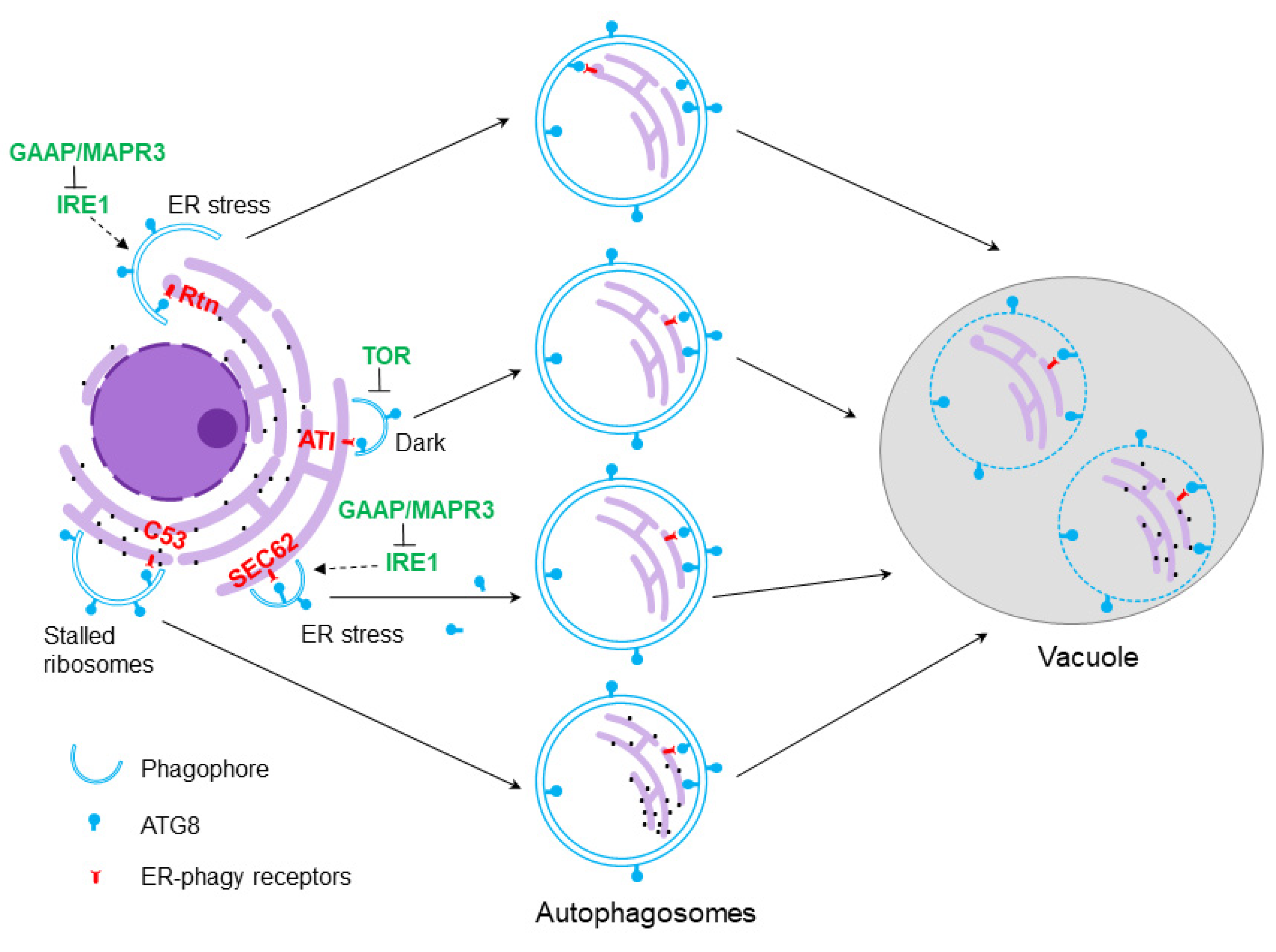
5. Selective Autophagy of the ER
6. ER-Phagy Receptors in Plants and Their Functions
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, J. Communications Between the Endoplasmic Reticulum and Other Organelles During Abiotic Stress Response in Plants. Front. Plant Sci. 2019, 10, 749. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, B.; Zhang, W.; Jiang, L. ER-Phagy and ER Stress Response (ERSR) in Plants. Front. Plant Sci. 2019, 10, 1192. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, F.; Xie, Q. Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 2020, 226, 345–350. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy in plants. Autophagy 2013, 9, 622–623. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, X.; Marshall, R.S.; Paez-Valencia, J.; Lacey, P.; Vierstra, R.D.; Otegui, M.S. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. eLife 2020, 9. [Google Scholar] [CrossRef]
- Srivastava, R.; Li, Z.; Russo, G.; Tang, J.; Bi, R.; Muppirala, U.; Chudalayandi, S.; Severin, A.; He, M.; Vaitkevicius, S.I.; et al. Response to Persistent ER Stress in Plants: A Multiphasic Process That Transitions Cells from Prosurvival Activities to Cell Death. Plant Cell 2018, 30, 1220–1242. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Honig, A.; Galili, G. Variations on a theme: Plant autophagy in comparison to yeast and mammals. Protoplasma 2012, 249, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for Self-Eating in Plant Cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781.e24. [Google Scholar] [CrossRef] [PubMed]
- Le Bars, R.; Marion, J.; Le Borgne, R.; Satiat-Jeunemaitre, B.; Bianchi, M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 2014, 5, 4121. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.T.F.; Yu, C.; Wong, J.S.K.; Lo, H.S.; Benlekbir, S.; Jiang, L.; Lau, W.C.Y. Subnanometer resolution cryo-EM structure of. Autophagy 2020, 16, 575–583. [Google Scholar] [CrossRef]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E426–E435. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Pleskot, R.; Zang, J.; Winkler, J.; Wang, J.; Yperman, K.; Zhang, T.; Wang, K.; Gong, J.; Guan, Y.; et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 2019, 10, 5132. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Richardson, C.; Hawes, C.; Hussey, P.J. Arabidopsis NAP1 Regulates the Formation of Autophagosomes. Curr. Biol. 2016, 26, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Park, J.M. Endoplasmic Reticulum Plays a Critical Role in Integrating Signals Generated by Both Biotic and Abiotic Stress in Plants. Front. Plant Sci. 2019, 10, 399. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein Quality Control in the Endoplasmic Reticulum of Plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Tsai, B.; Arvan, P. New Insights into the Physiological Role of Endoplasmic Reticulum-Associated Degradation. Trends Cell Biol. 2017, 27, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Bassham, D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013, 8, e24297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bao, Y.; Howell, S. The Unfolded Protein Response Supports Plant Development and Defense as well as Responses to Abiotic Stress. Front. Plant Sci. 2017, 8, 344. [Google Scholar] [CrossRef]
- Deng, Y.; Srivastava, R.; Quilichini, T.D.; Dong, H.; Bao, Y.; Horner, H.T.; Howell, S.H. IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J. 2016, 88. [Google Scholar] [CrossRef]
- Zhang, S.S.; Yang, H.; Ding, L.; Song, Z.T.; Ma, H.; Chang, F.; Liu, J.X. Tissue-Specific Transcriptomics Reveals an Important Role of the Unfolded Protein Response in Maintaining Fertility upon Heat Stress in Arabidopsis. Plant Cell 2017, 29, 1007–1023. [Google Scholar] [CrossRef]
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651. [Google Scholar] [CrossRef]
- Yang, X.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016, 85, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Pu, Y.; Yu, X.; Gregory, B.D.; Srivastava, R.; Howell, S.H.; Bassham, D.C. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Wang, W.; Fan, W.; Wang, Z.; Zhu, M.; Tang, X.; Wu, W.; Yang, X.; Shao, X.; Sun, Y.; et al. Arabidopsis GAAP1 and GAAP3 Modulate the Unfolded Protein Response and the Onset of Cell Death in Response to ER Stress. Front. Plant Sci. 2018, 9, 348. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Tang, X.; Wang, Z.; Xu, W.; Zhou, Y.; Wang, W.; Li, X.; Li, R.; Guo, K.; Sun, Y.; et al. Arabidopsis GAAPs interacting with MAPR3 modulate the IRE1-dependent pathway upon endoplasmic reticulum stress. J. Exp. Bot. 2019, 70, 6113–6125. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232.e11. [Google Scholar] [CrossRef]
- Chino, H.; Hatta, T.; Natsume, T.; Mizushima, N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol. Cell 2019, 74, 909–921.e6. [Google Scholar] [CrossRef]
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell 2019, 74, 891–908.e10. [Google Scholar] [CrossRef]
- Chen, J.; Stefano, G.; Brandizzi, F.; Zheng, H. Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 2011, 124, 2241–2252. [Google Scholar] [CrossRef]
- Fumagalli, F.; Noack, J.; Bergmann, T.J.; Cebollero, E.; Pisoni, G.B.; Fasana, E.; Fregno, I.; Galli, C.; Loi, M.; Soldà, T.; et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016, 18, 1173–1184. [Google Scholar] [CrossRef]
- Grumati, P.; Morozzi, G.; Hölper, S.; Mari, M.; Harwardt, M.I.; Yan, R.; Müller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, R.M.; Grumati, P.; Garcia-Pardo, J.; Kalayil, S.; Covarrubias-Pinto, A.; Chen, W.; Kudryashev, M.; Dikic, I.; Hummer, G. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun. 2019, 10, 2370. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Zou, C.X.; Liu, X.M.; Jiang, Z.D.; Yu, Z.Q.; Suo, F.; Du, T.Y.; Dong, M.Q.; He, W.; Du, L.L. A UPR-Induced Soluble ER-Phagy Receptor Acts with VAPs to Confer ER Stress Resistance. Mol. Cell 2020, 79, 963–977.e3. [Google Scholar] [CrossRef] [PubMed]
- Nthiga, T.M.; Kumar Shrestha, B.; Sjøttem, E.; Bruun, J.A.; Bowitz Larsen, K.; Bhujabal, Z.; Lamark, T.; Johansen, T. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J. 2020, 39, e103649. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ye, H.; Cui, Y.; Jiang, L. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant Biol. 2020, 62, 181–200. [Google Scholar] [CrossRef]
- Stephani, M.; Picchianti, L.; Gajic, A.; Beveridge, R.; Skarwan, E.; Sanchez de Medina Hernandez, V.; Mohseni, A.; Clavel, M.; Zeng, Y.; Naumann, C.; et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 2020, 9. [Google Scholar] [CrossRef]
- Tolley, N.; Sparkes, I.A.; Hunter, P.R.; Craddock, C.P.; Nuttall, J.; Roberts, L.M.; Hawes, C.; Pedrazzini, E.; Frigerio, L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 2008, 9, 94–102. [Google Scholar] [CrossRef]
- Michaeli, S.; Honig, A.; Levanony, H.; Peled-Zehavi, H.; Galili, G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014, 26, 4084–4101. [Google Scholar] [CrossRef]
- Honig, A.; Avin-Wittenberg, T.; Ufaz, S.; Galili, G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012, 24, 288–303. [Google Scholar] [CrossRef]
- Michaeli, S.; Clavel, M.; Lechner, E.; Viotti, C.; Wu, J.; Dubois, M.; Hacquard, T.; Derrien, B.; Izquierdo, E.; Lecorbeiller, M.; et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc. Natl. Acad. Sci. USA 2019, 116, 22872–22883. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Michaeli, S.; Galili, G.; Peled-Zehavi, H. ATG8-interacting (ATI) 1 and 2 define a plant starvation-induced ER-phagy pathway and serve as MSBP1 (MAPR5) cargo-receptors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Naumann, C.; Müller, J.; Sakhonwasee, S.; Wieghaus, A.; Hause, G.; Heisters, M.; Bürstenbinder, K.; Abel, S. The Local Phosphate Deficiency Response Activates Endoplasmic Reticulum Stress-Dependent Autophagy. Plant Physiol. 2019, 179, 460–476. [Google Scholar] [CrossRef]
- Kim, J.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ER-Anchored Transcription Factors bZIP17 and bZIP28 Regulate Root Elongation. Plant Physiol. 2018, 176, 2221–2230. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Bassham, D.C.; Howell, S.H. A Functional Unfolded Protein Response Is Required for Normal Vegetative Development. Plant Physiol. 2019, 179, 1834–1843. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Srivastava, R.; Howell, S. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 19633–19638. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Hidema, J.; Makino, A.; Ishida, H. Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 2013, 161, 1682–1693. [Google Scholar] [CrossRef]
- Hua, Z.; Vierstra, R.; Merchant, S.; Briggs, W.; Ort, D. The Cullin-RING Ubiquitin-Protein Ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Hafrén, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Song, W.M.; Wang, P.; Yu, X.; Li, B.; Jiang, C.; Shiu, S.H.; Zhang, H.; Bassham, D.C. COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 7482–7493. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y. Links between drought stress and autophagy in plants. Plant Signal. Behav. 2020, 15, 1779487. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, Y.; Bassham, D.C. ER-Phagy and Its Role in ER Homeostasis in Plants. Plants 2020, 9, 1771. https://doi.org/10.3390/plants9121771
Bao Y, Bassham DC. ER-Phagy and Its Role in ER Homeostasis in Plants. Plants. 2020; 9(12):1771. https://doi.org/10.3390/plants9121771
Chicago/Turabian StyleBao, Yan, and Diane C. Bassham. 2020. "ER-Phagy and Its Role in ER Homeostasis in Plants" Plants 9, no. 12: 1771. https://doi.org/10.3390/plants9121771
APA StyleBao, Y., & Bassham, D. C. (2020). ER-Phagy and Its Role in ER Homeostasis in Plants. Plants, 9(12), 1771. https://doi.org/10.3390/plants9121771





