The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis
Abstract
1. Introduction
2. Results and Discussion
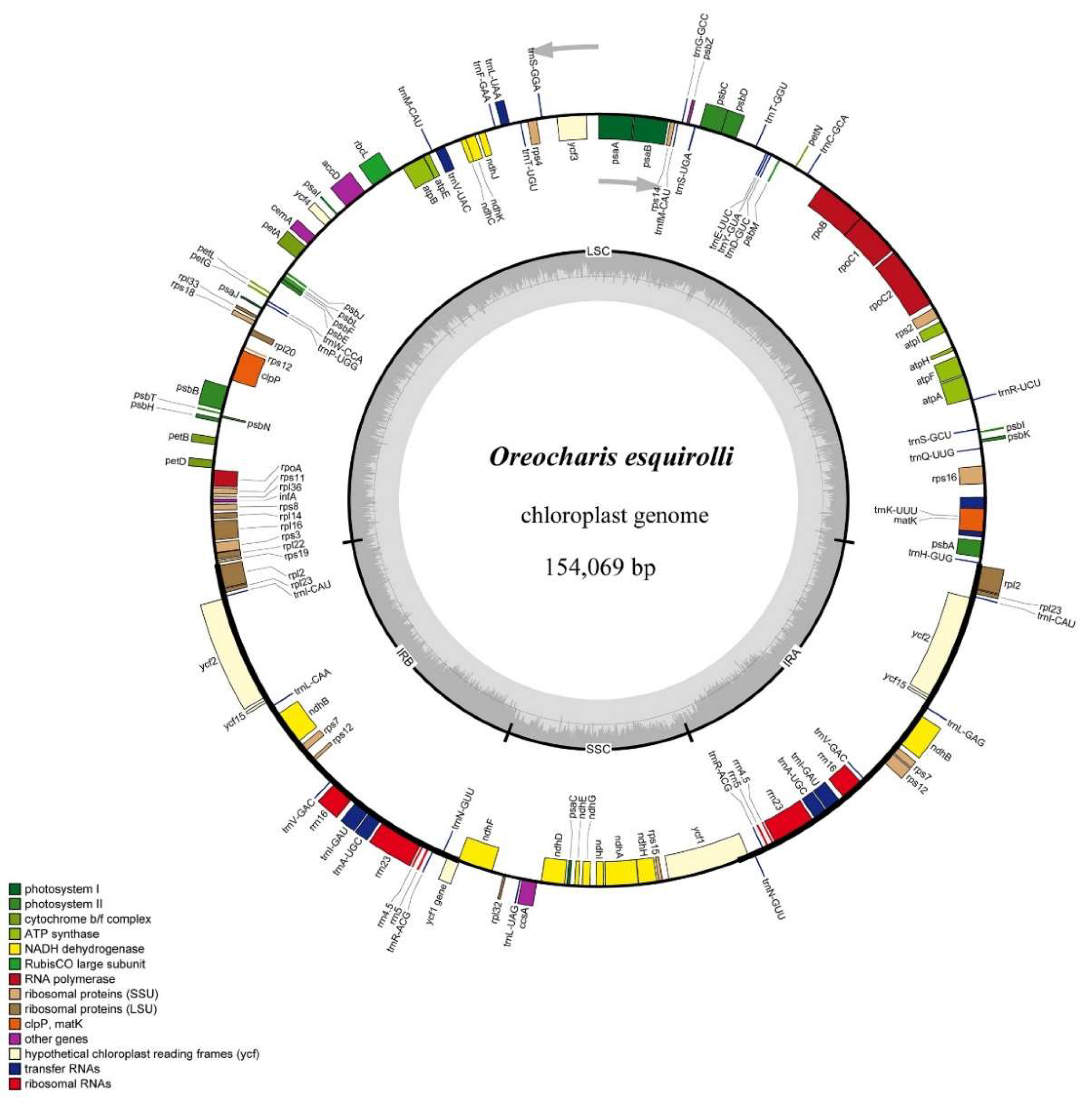
2.1. Gene Content and Structure of Chloroplast Genome of Oreocharis esquirolii
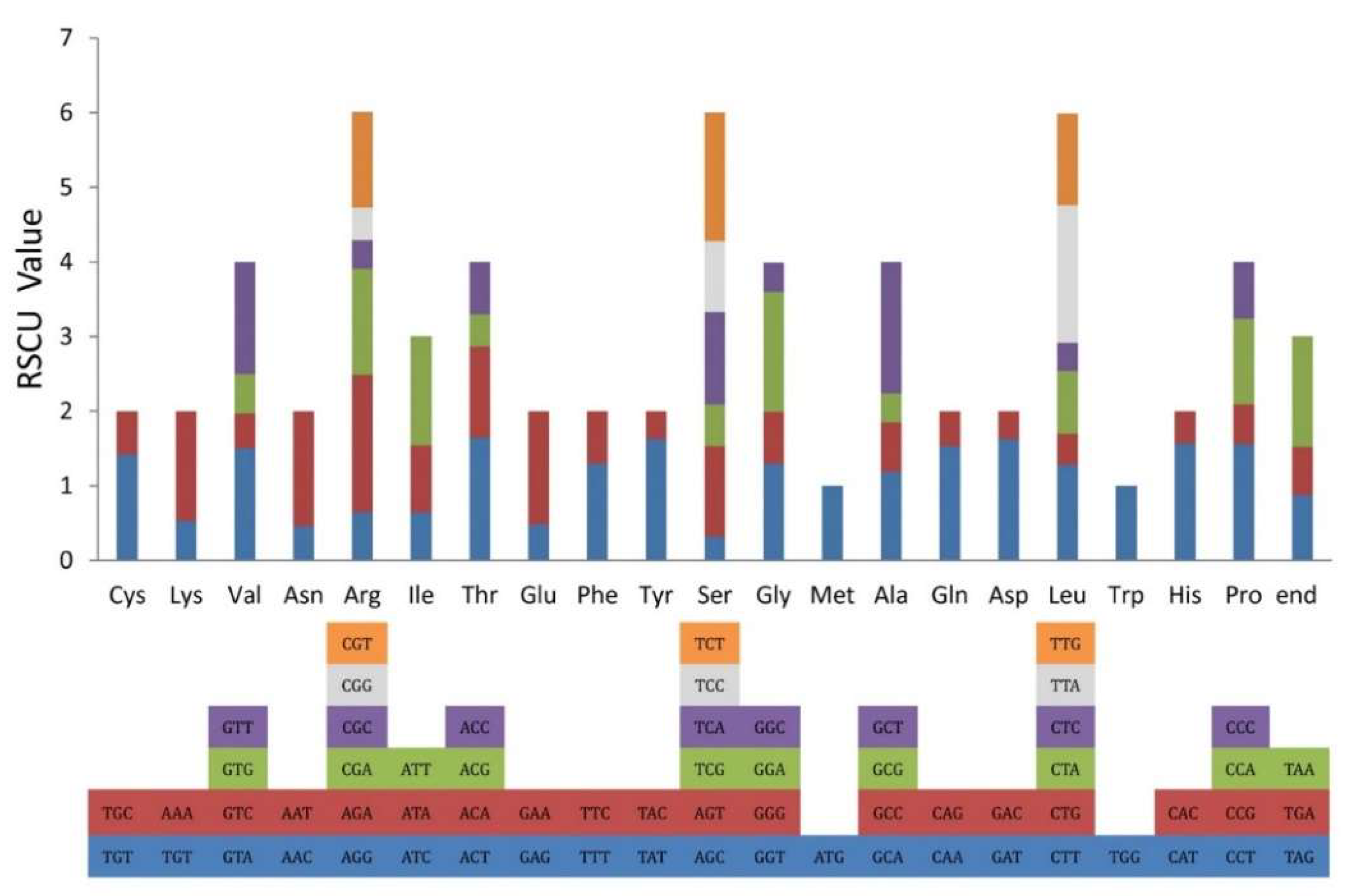
2.2. Codon Usage Bias Analysis
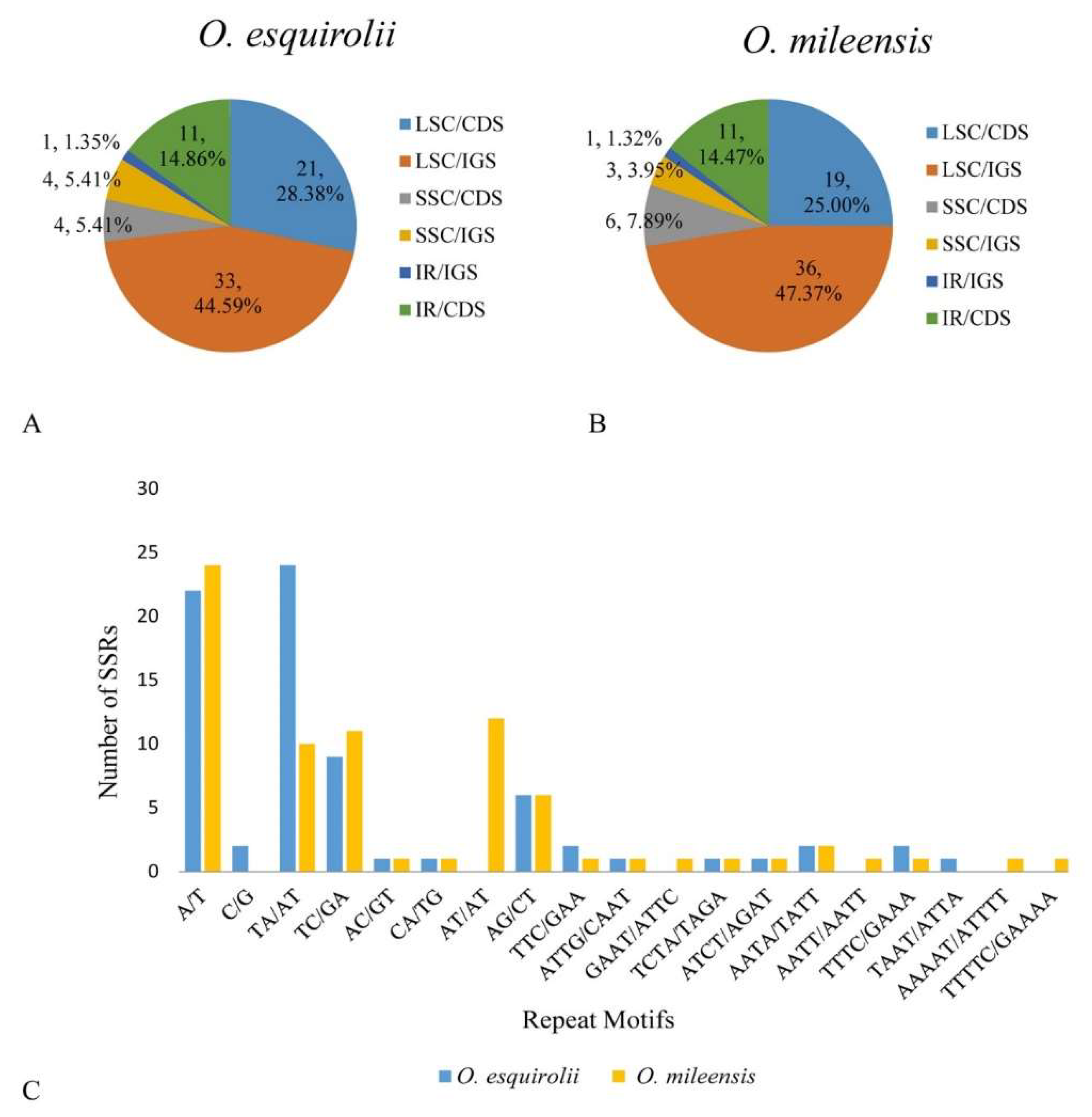
2.3. SSRs Analysis
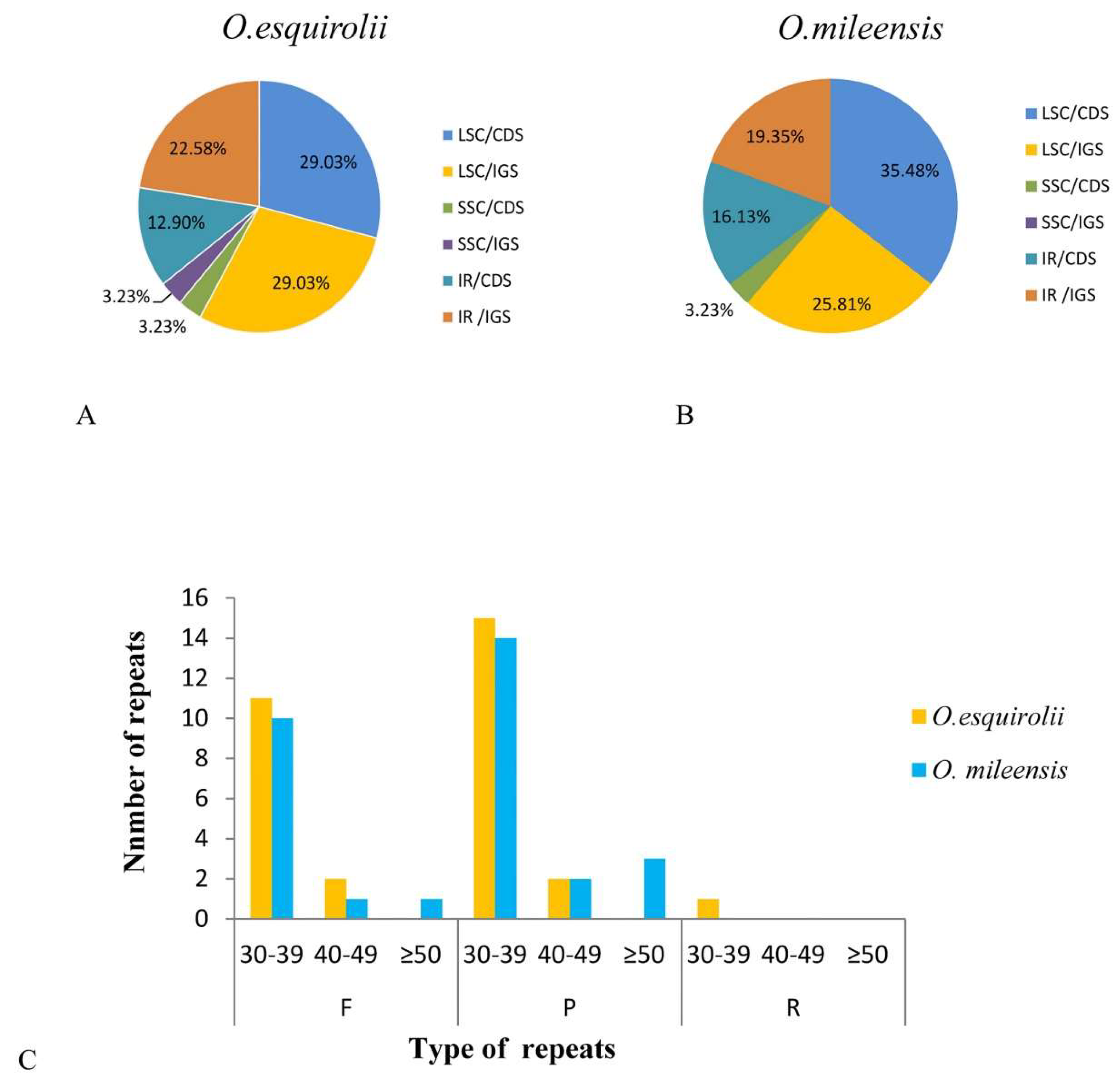
2.4. Analysis of Repeat Sequences
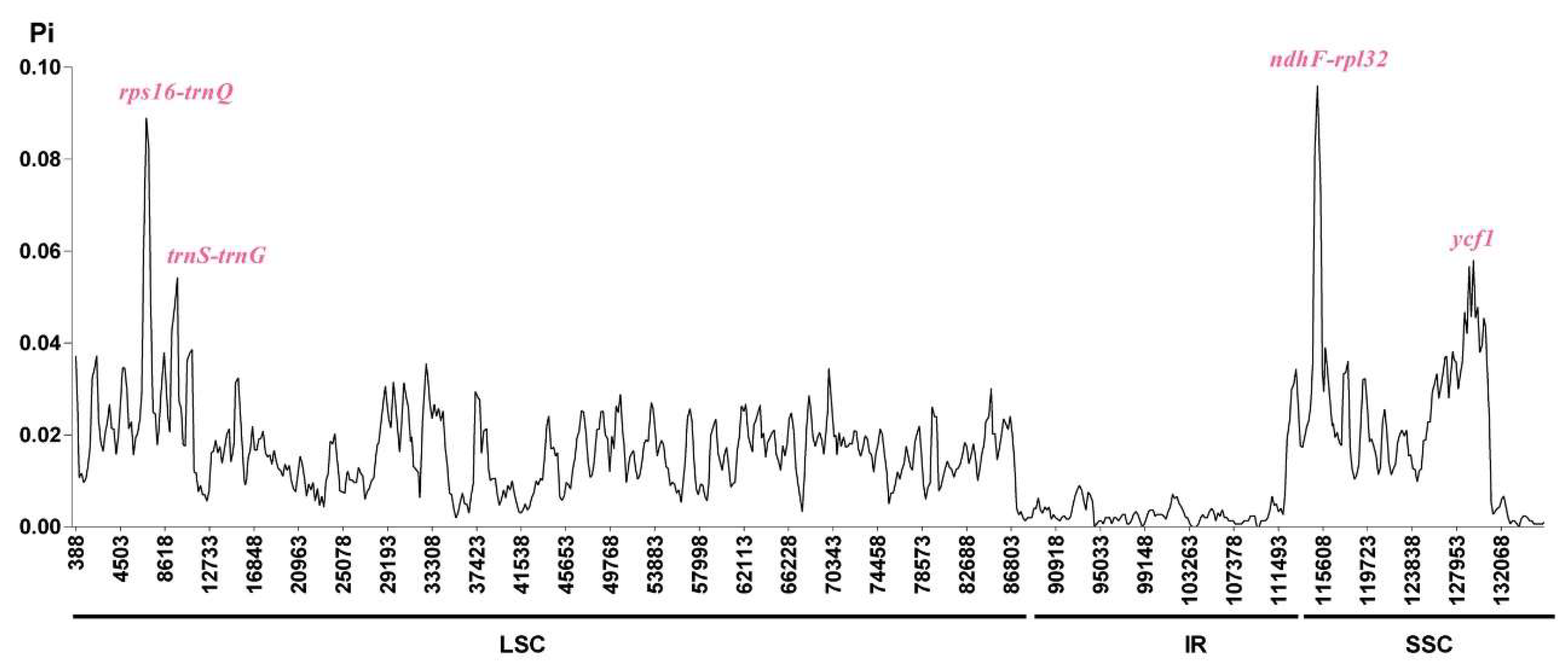
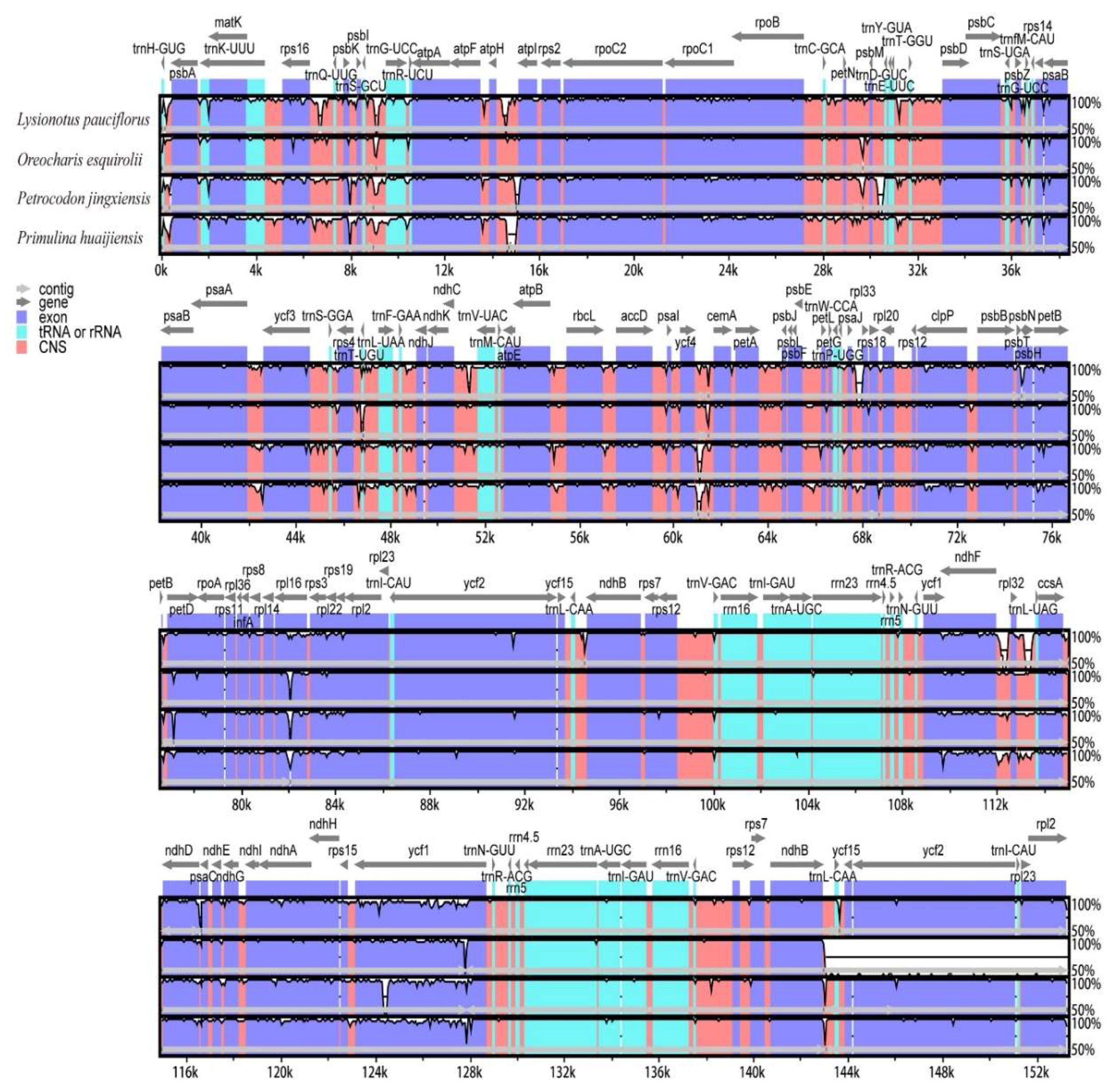
2.5. Comparisons of Chloroplast Genome among Oreocharis esquirolii and Closely Related Species
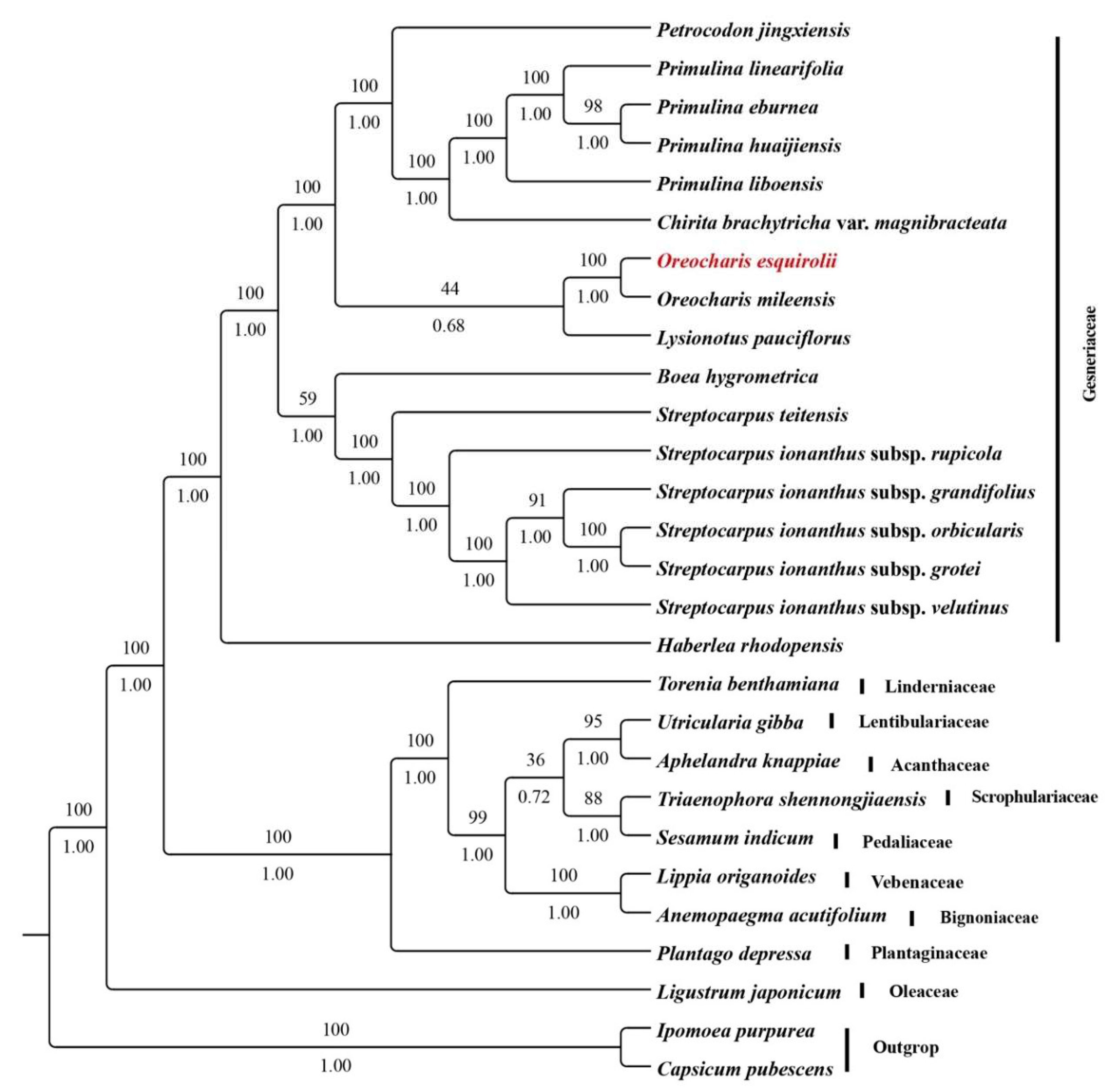
2.6. Phylogenetic Position of Oreocharis esquirolii
3. Materials and Methods
3.1. Plant Material, DNA Extraction, Sequencing, and Assembly
3.2. Genome Annotation and Sequence Submission
3.3. Codon Usage, Repetitive Sequence, and SSR Analysis
3.4. Genome Comparison
3.5. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bentham, G. Gesneriaceae. In Genera Plantarum; Bentham, G., Hooker, J.D., Eds.; Lovell Reeve & Co.: London, UK, 1876; pp. 990–1025. [Google Scholar]
- Wang, W.T.; Pan, K.Y.; Li, Z.Y.; Weitzman, A.L.; Skog, L.E. Gesneriaceae. In Flora of China; Wu, C.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 1998; Volume 18, pp. 244–401. [Google Scholar]
- Wang, Y.Z.; Liang, R.H.; Wang, B.H.; Li, J.M.; Qiu, Z.J.; Li, Z.Y.; Weber, A. Origin and phylogenetic relationships of the Old World Gesneriaceae with actinomorphic flowers inferred from ITS and trnL-trnF sequences. Taxon 2010, 59, 1044–1052. [Google Scholar] [CrossRef]
- Möller, M.; Middleton, D.; Nishii, K.; Wei, Y.G.; Sontag, S.; Weber, A. A new delineation for Oreocharis incorporating an additional ten genera of Chinese Gesneriaceae. Phytotaxa 2011, 23, 1–36. [Google Scholar] [CrossRef]
- Möller, M.; Forrest, A.; Wei, Y.G.; Weber, A. A molecular phylogenetic assessment of the advanced Asiatic and Malesian didymocarpoid Gesneriaceae with focus on non-monophyletic and monotypic genera. Plant Syst. Evol. 2011, 292, 223–248. [Google Scholar] [CrossRef]
- Möller, M. Species discovery in time: An example from Gesneriaceae in China. Guihaia 2019, 26, 1–16. [Google Scholar] [CrossRef]
- Wen, F.; Li, S.; Xin, Z.B.; Fu, L.F.; Hong, X.; Cai, L.; Qin, J.Q.; Pan, B.; Pan, F.G.; Wei, Y.G. The updated plant list of Gesneriaceae in China under the new Chinese naming rules. Guihaia 2019, 26, 37–63. [Google Scholar] [CrossRef]
- Middleton, D.J.; Möller, M. Tribounia, a new genus of Gesneriaceae from Thailand. Taxon 2012, 61, 1286–1295. [Google Scholar] [CrossRef]
- Do, T.V.; Wei, Y.G.; Wen, F. Oreocharis caobangensis (Gesneriaceae), a new species from Cao Bang Province, northern Vietnam. Phytotaxa 2017, 302, 65–70. [Google Scholar] [CrossRef]
- Chen, W.H.; Middleton, D.J.; Nguyen, H.Q.; Nguyen, H.T.; Averyanov, L.V.; Chen, R.Z.; Nguyen, K.S.; Möller, M.; Shui, Y.M. Two new species of Oreocharis (Gesneriaceae) from Northwest Vietnam. Gard. Bull. 2017, 69, 295–305. [Google Scholar] [CrossRef]
- Chen, W.H.; Nguyen, Q.H.; Chen, R.Z.; Nguyen, T.H.; Nguyen, S.K.; Nguyen, V.T.; Möller, M.; Middleton, D.J.; Shui, Y.M. Two new species of Oreocharis (Gesneriaceae) from Fan Si Pan, the highest mountain in Vietnam. Phytokeys 2018, 94, 95–106. [Google Scholar] [CrossRef]
- Léveillé, H. Decades plantarum novarum. LIV–LVII. Repert. Spec. Nov. Regni Veg. 1911, 9, 321–330. [Google Scholar]
- Wang, W.T. Genus novum primitivum Gesneriacearum e Sina. Acta Phytotax. Sin. 1981, 19, 485–489. [Google Scholar]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Mueller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cui, L.; Feng, K.; Deng, P.; Du, X.; Wan, F.; Song, W.; Nie, X. Comparative analysis of Asteraceae chloroplast genomes: Structural organization, RNA editing and evolution. Plant Mol. Biol. Rep. 2015, 33, 1526–1538. [Google Scholar] [CrossRef]
- Palmer, J.D. Plastid chromosomes: Structure and evolution. In The Molecular Biology of Plastids; Bogorad, L., Vasil, I.K., Eds.; Academic Press: Cambridge, MA, USA, 1991; pp. 5–53. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, X.; Zhang, D. Phylogenetic analysis of Fritillaria cirrhosa D. Don and its closely related species based on complete chloroplast genomes. PeerJ 2019, 7, e7480. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Givnish, T.J.; Zuluaga, A.; Spalink, D.; Soto Gomez, M.; Lam, V.K.Y.; Saarela, J.M.; Sass, C.; Iles, W.J.D.; de Sousa, D.J.L.; Leebens-Mack, J.; et al. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 2018, 11, 1888–1910. [Google Scholar] [CrossRef]
- He, Y.; Xiao, H.; Deng, C.; Xiong, L.; Yang, J.; Peng, C. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016, 17, 820. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Yang, J.; Lv, G. Complete chloroplast genome of seven Fritillaria species, variable DNA markers identification and phylogenetic relationships within the genus. PLoS ONE 2018, 13, e0194613. [Google Scholar] [CrossRef]
- Zong, D.; Zhou, A.; Zhang, Y.; Zou, X.; Li, D.; Duan, A.; He, C. Characterization of the complete chloroplast genomes of five Populus species from the western Sichuan plateau, southwest China: Comparative and phylogenetic analyses. PeerJ 2019, 7, e6386. [Google Scholar] [CrossRef]
- Rivas, D.L.; Lozano, J.; Ortiz, J.J.; Angel, R. Comparative analysis of chloroplast genomes: Functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 2002, 12, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huo, N.X.; Dong, L.L.; Wang, Y.; Zhang, S.X.; Young, H.A.; Feng, X.X.; Gu, Y.Q. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS ONE 2013, 8, e57533. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Wang, G.; Li, C.R.; Luo, Y. Characterization of the complete chloroplast genomes of three Chirita species (C. brachytricha, C. eburnea & C. liboensis) endemic to China. Conserv. Genet. Resour. 2018, 10, 597–600. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, L.; He, J. Complete plastid genome of the endangered species Paraisometrum mileense (Gesneriaceae) endemic to China. Mitochondrial DNA B Resour. 2019, 4, 3585–3586. [Google Scholar] [CrossRef]
- Xin, Z.B.; Fu, L.F.; Fu, Z.X.; Li, S.; Wei, Y.G.; Wen, F. Complete chloroplast genome sequence of Petrocodon jingxiensis (Gesneriaceae). Mitochondrial DNA B Resour. 2019, 4, 2771–2772. [Google Scholar] [CrossRef]
- Kyalo, C.M.; Gichira, A.W.; Li, Z.Z.; Saina, J.K.; Malombe, I.; Hu, G.W.; Wang, Q.F. Characterization and comparative analysis of the complete chloroplast genome of the critically endangered species Streptocarpus teitensis (Gesneriaceae). Biomed. Res. Int. 2018, 2018, 1507847. [Google Scholar] [CrossRef]
- Kyalo, C.M.; Li, Z.Z.; Mkala, E.M.; Malombe, I.; Hu, G.W.; Wang, Q.F. The first glimpse of Streptocarpus ionanthus (Gesneriaceae) phylogenomics: Analysis of five subspecies’ chloroplast genomes. Plants 2020, 9, 456. [Google Scholar] [CrossRef]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.E.; Li, C.; et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef]
- Liu, Q.; Dou, S.; Ji, Z.; Xue, Q. Synonymous codon usage and gene function are strongly related in Oryza sativa. Biosystems 2005, 80, 123–131. [Google Scholar] [CrossRef]
- Srivastava, D.; Shanker, A. Identification of Simple Sequence Repeats in chloroplast genomes of Magnoliids through bioinformatics approach. Interdiscip. Sci. 2015, 8, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kuang, X.J.; Zhu, X.X.; Zhu, Y.J.; Sun, C. Codon usage bias of Catharanthus roseus. Zhongguo Zhong Yao Za Zhi China J. Chin. Mater. Med. 2016, 41, 4165–4168. [Google Scholar] [CrossRef]
- Marino, S.M.; Gladyshev, V.N. Analysis and functional prediction of reactive Cysteine residues. J. Biol. Chem. 2012, 287, 4419–4425. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. Suppl. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Raza, A.; Mehmood, S.S.; Ashraf, F.; Khan, R.S.A. Genetic diversity analysis of Brassica species using PCR-based SSR markers. Gesunde Pflanz. 2018, 71, 1–7. [Google Scholar] [CrossRef]
- Torokeldiev, N.; Ziehe, M.; Gailing, O.; Finkeldey, R. Genetic diversity and structure of natural Juglans regia L. populations in the southern Kyrgyz Republic revealed by nuclear SSR and EST–SSR markers. Tree Genet. Genomes 2019, 15, 5. [Google Scholar] [CrossRef]
- Kuang, D.Y.; Wu, H.; Wang, Y.L.; Gao, L.M.; Zhang, S.Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef]
- Du, Y.P.; Bi, Y.; Yang, F.P.; Zhang, M.F.; Chen, X.Q.; Xue, J.; Zhang, X.H. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 5751. [Google Scholar] [CrossRef]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Zhao, F.; Drew, B.T.; Chen, Y.P.; Hu, G.X.; Li, B.; Xiang, C.L. The chloroplast genome of Salvia: Genomic characterization and phylogenetic analysis. Int. J. Plant Sci. 2020, 181, 812–830. [Google Scholar] [CrossRef]
- Neubig, K.M.; Whitten, W.M.; Carlsward, B.S.; Blanco, M.A.; Endara, L.; Williams, N.H.; Moore, M. Phylogenetic utility of ycf1 in orchids: Aplastid gene more variable than matK. Plant Syst. Evol. 2009, 277, 75–84. [Google Scholar] [CrossRef]
- Drew, B.T.; Sytsma, K.J. Testing the monophyly and placement of Lepechinia in the tribe Mentheae (Lamiaceae). Syst. Bot. 2011, 36, 1038–1049. [Google Scholar] [CrossRef]
- Möller, M.; Pfosser, M.; Jang, C.G.; Mayer, V.; Clark, A.; Hollingsworth, M.L.; Barfuss, M.H.J.; Wang, Y.Z.; Kiehn, M.; Weber, A. A preliminary phylogeny of the ‘didymocarpoid Gesneriaceae’ based on three molecular data sets: Incongruence with available tribal classifications. Am. J. Bot. 2009, 96, 989–1010. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemistry 1987, 19, 11–15. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Son, P.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. Suppl. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Hélène, C.; Frédérique, L.; Michel, C.; Alain, H. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 1998, 209, GC1–GC38. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. Suppl. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Munyao, J.N.; Dong, X.; Yang, J.X.; Mbandi, E.M.; Wanga, V.O.; Oulo, M.A.; Saina, J.K.; Musili, R.M.; Hu, G.W. Complete chloroplast genomes of Chlorophytum comosum and Chlorophytum gallabatense: Genome structures, comparative and phylogenetic analysis. Plants 2020, 9, 296. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Rozas, J.; Albert, F.M.; Juan, C.S.; Sara, G.R.; Pablo, L.; Sebastian, E.R.O.; Alejandro, S.G. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Liu, B.; Tan, Y.H.; Liu, S.; Olmstead, R.G.; Min, D.Z.; Chen, Z.D.; Joshee, N.; Vaidya, B.N.; Chung, R.C.K.; Li, B. Phylogenetic relationships of Cyrtandromoea and Wightia revisited: A new tribe in Phrymaceae and a new family in Lamiales. J. Syst. Evol. 2020, 1, 1–17. [Google Scholar] [CrossRef]
- Li, H.T.; Yi, T.S.; Gao, L.M.; Ma, P.F.; Zhang, T.; Yang, J.B.; Gitzendanner, M.A.; Fritsch, P.W.; Cai, J.; Luo, Y.; et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 2019, 5, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.T.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the SC10 Workshop on Gateway Computing Environments (GCE10), New Orleans, LA, USA, 14 November 2010. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Laget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.; Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]


| Category | Gene Group | Gene Names |
|---|---|---|
| Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpI, atpH |
| Subunits of NADH dehydrogenase | ndhA *, ndhB * (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK, | |
| Subunits of cytochrome | petA, petB, petD, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaJ, psaI | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbH, psbK, psbN, psbJ, psbF, psbL, psbI, psbM, psbT, psbZ | |
| Subunit of rubisco | rbcL | |
| Other genes | Subunit of Acetyl-CoA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsA | |
| Envelop membrane protein | cemA | |
| Protease | clpP ** | |
| Translational initiation | infA | |
| Maturase | matK | |
| Self-replication | Large subunit of ribosome | rpl2 * (×2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 |
| DNA dependent RNA polymerase | rpoA, rpoC2, rpoB, rpoC1 | |
| Small subunit of ribosome | rps12 ** (×2), rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps14, rps15, rps16 *, rps18, rps19 | |
| rRNA Genes | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) | |
| tRNA Genes | trnK-UUU *, trnI-GAU * (×2), trnA-UGC * (×2), trnV-UAC *, trnL-UAA *, trnS-UGA, trnS-GCU, trnS-GGA, trnY-GUA, trnL-CAA (×2), trnL-UAG, trnL-GAG, trnM-CAU, trnR-ACG (×2), trnP-UGG, trnW-CCA, trnD-GUC, trnH-GUG, trnF-GAA, trnT-UGU, trnE-UUC, trnN-GUU (×2), trnV-GAC (×2), trnT-GGU, trnQ-UUG, trnR-UCU, trnG-GCC, trnC-GCA, trnI-CAU (×2), trnfM-CAU | |
| Unknown function | Conserved open reading frames | ycf1 (×2, ψ), ycf2 (×2), ycf3 **, ycf4, ycf15 (×2) |
| Gene | Region | Exon1 (bp) | Intron1 (bp) | Exon2 (bp) | Intron2 (bp) | Exon3 (bp) |
|---|---|---|---|---|---|---|
| atpF | LSC | 144 | 707 | 411 | ||
| ndhA | SSC | 552 | 1062 | 540 | ||
| ndhB | IR | 777 | 679 | 756 | ||
| rpl2 | IR | 390 | 673 | 435 | ||
| rpl16 | LSC | 9 | 824 | 399 | ||
| rps16 | LSC | 42 | 921 | 210 | ||
| clpP | LSC | 69 | 814 | 291 | 644 | 228 |
| rpoC1 | LSC | 453 | 812 | 1611 | ||
| trnA-UGC | IR | 38 | 807 | 35 | ||
| trnI-GAU | IR | 37 | 941 | 35 | ||
| trnK-UUU | LSC | 37 | 2497 | 36 | ||
| trnL-UAA | LSC | 37 | 476 | 48 | ||
| trnV-UAC | LSC | 38 | 586 | 35 | ||
| ycf3 | LSC | 126 | 692 | 228 | 714 | 153 |
| Region | Length (bp) | A (%) | T (%) | G (%) | C (%) | GC (%) |
|---|---|---|---|---|---|---|
| LSC | 85,156 | 31.54 | 33.03 | 17.30 | 18.13 | 35.43 |
| SSC | 18,129 | 34.34 | 34.50 | 15.00 | 16.16 | 31.16 |
| IRA | 25,392 | 28.38 | 28.41 | 22.42 | 20.79 | 43.21 |
| IRB | 25,392 | 28.38 | 28.41 | 22.42 | 20.79 | 43.21 |
| CDS | 79,650 | 30.70 | 31.59 | 20.10 | 17.61 | 37.71 |
| Total genome | 154,069 | 30.83 | 31.67 | 18.45 | 19.04 | 37.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, L.; Su, T.; An, M.-T.; Hu, G.-X. The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis. Plants 2020, 9, 1692. https://doi.org/10.3390/plants9121692
Gu L, Su T, An M-T, Hu G-X. The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis. Plants. 2020; 9(12):1692. https://doi.org/10.3390/plants9121692
Chicago/Turabian StyleGu, Li, Ting Su, Ming-Tai An, and Guo-Xiong Hu. 2020. "The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis" Plants 9, no. 12: 1692. https://doi.org/10.3390/plants9121692
APA StyleGu, L., Su, T., An, M.-T., & Hu, G.-X. (2020). The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis. Plants, 9(12), 1692. https://doi.org/10.3390/plants9121692




