Abstract
Vacuolar H+-ATPases (V-ATPase) are multi-subunit complexes that function as ATP hydrolysis-driven proton pumps. They play pivotal roles in physiological processes, such as development, metabolism, stress, and growth. However, there have been very few studies on the characterisation of V-ATPase (VHA) genes in Rosaceae species. Therefore, in the present study, we performed a genome-wide analysis and identified VHA gene family members in five Rosaceae species (Pyrus bretschneideri, Malus domestica, Prunus persica, Fragaria vesca, and Prunus mume). A total of 159 VHA genes were identified, and were classified into 13 subfamilies according to the phylogenetic analysis. The structure of VHA proteins revealed high similarity among different VHA genes within the same subgroup. Gene duplication event analysis revealed that whole-genome duplications represented the major pathway for expansion of the Pyrus bretschneideri VHA genes (PbrVHA genes). The tissue-specific expression analysis of the pear showed that 36 PbrVHA genes were expressed in major tissues. Seven PbrVHA genes were significantly downregulated when the pollen tube growth stopped. Moreover, many PbrVHA genes were differentially expressed during fruit development and storage, suggesting that VHA genes play specific roles in development and senescence. The present study provides fundamental information for further elucidating the potential roles of VHA genes during development and senescence.
1. Introduction
Vacuoles are the largest compartments within plant cells, occupying up to 90% of the cellular volume [1]. In plants, vacuoles play important roles in growth and development and serve various functions, including storage and transport, cell homoeostasis, and stress response [2]. All vacuolar functions require enormous fluxes of metabolites and ions, which are transported by a series of vacuolar transport proteins [3]. Transport across the tonoplast is primarily energized by the ATP hydrolysis-driven proton pumps vacuolar H+-ATPases (V-ATPases) [3,4,5]. V-ATPases are highly conserved protein complexes, which are present in the vacuole as well as other intracellular compartments [5]. V-ATPases are large multi-subunit complexes comprising the cytosolic V1 domain and the membrane-integral V0 domain [6,7]. The two domains are joined by a stalk domain, which makes them a complex with integrated structure and function. The V1 domain harbors eight subunits (VHA-A, B, C, D, E, F, G, and H) and is responsible for ATP hydrolysis. The V0 domain consists of up to five subunits (VHA-a, c, c”, d, and e) and is involved in proton transport [6]. V-ATPases are the key regulators of functions of the vacuole and other endomembrane compartments in plants [8,9].
Research on plant V-ATPases began nearly 40 years ago, when the enzyme was discovered [10,11]. V-ATPase is a proton pump present in the vacuole and other intracellular compartments, including the endoplasmic reticulum (ER), Golgi, vesicles, and endosomes [6,12]. Moreover, V-ATPase activity in the trans-Golgi network/early endosome is essential for functional secretion and recycling [13], whereas V-ATPase activity in the tonoplast is required for ion homoeostasis and nutrient transport [3,14]. VHA genes, such as AtVHA-c5, AtVHA-d2, and JrVHAG1, have been speculated to be involved in stress response [11,15,16]. Likewise, other members of this gene family, such as PutVHA-c, AtVHA-a2, AtVHA-a3, and AtVHA-B2, are associated with plant growth or senescence [14,17]. To summarize, V-ATPases are involved in various physiological processes, including cell expansion, membrane trafficking, nutrient accumulation, leaf senescence, and environmental stress tolerance [17,18,19].
In the past decades, genes encoding various VHA subunits in plants have been identified, including 28 VHA genes in Arabidopsis thaliana [6], 54 VHA genes in Glycine max [12], 23 VHA genes in tomato [20], and 51 VHA genes in citrus fruit [18]. Moreover, VHA genes are conserved among green plants, although their number varies across different species [9,12]. V-ATPase has a highly complex subunit composition, and genes encoding at least 13 distinct subunits have been identified thus far [12]. Unlike P-type proton pumps encoded by multiple genes [21], most V1 subunits (A, C, D, F, and H) of V-ATPase are encoded by a single gene, whereas V0 subunits are encoded by multiple genes in Arabidopsis and some higher plant species [6,12]. Recently, genome databases have been used to identify VHA genes in numerous plant species, and genes encoding the V-ATPase subunit H in 11 major crops have been identified [22]. However, genome-wide identification and bioinformatic analysis of VHA genes have not been performed in Rosaceae species.
The Rosaceae family is an important family with an extraordinary spectrum of fruit types, including pear (Pyrus bretschneideri), apple (Malus x domestica), peach (Prunus persica), plum (Prunus mume), strawberry (Fragaria vesca) and so on [23]. Remarkably, a few biological functions of V-ATPases have been discovered in several Rosaceae species. Proteomic analysis revealed that abundance of the V-ATPase protein decreased during fruit senescence in apple and strawberry [24,25]. Moreover, the activity of V-ATPases and transcript expression patterns of the corresponding genes were upregulated during storage following 1-methylcyclopropene treatment of apple fruit [26]. Recently, the genome sequences of five main Rosaceae species (Pyrus bretschneideri, Malus x domestica, Prunus persica, Prunus mume and Fragaria vesca) had been published [27,28,29]. However, limited information is available regarding the roles of VHA genes in Rosaceae species. Therefore, identification and analyses of VHA genes in Rosaceae species are of great significance. Pear is one of the most important commercial fruits in the Rosaceae family and multiple transcriptome data could be gathered for this species. Pear is an excellent model for studying fruit development and senescence. In the present study, the V-ATPase (VHA) family genes in five Rosaceae species were identified, and the gene structure, conserved motifs, chromosomal distribution, evolution, gene ontology and expression patterns were simultaneously analysed in pear. The results of this study will help elucidate the biological functions and molecular mechanisms of action of the VHA gene family in several Rosaceae species.
2. Results
2.1. Identification of V-ATPase (VHA) Genes in Five Rosaceae Species
In Arabidopsis, 28 VHA genes encoding different V-ATPase subunits have been identified [6]. To identify and comprehensively analyze VHA genes in five Rosaceae species, sequences of A. thaliana were used to query the corresponding genome. Pfam and SMART databases were used to search for the conserved domain of each V-ATPase subunit. After filtering for domain integrity and redundancy of the candidate genes, 159 VHA genes were identified, including 48 in apple (Malus x domestica), 43 in Chinese white pear (Pyrus bretschneideri), 25 in Chinese plum ( Prunus mume), 23 in peach (Prunus persica), and 20 in wild strawberry (Fragaria vesca) (Table 1). Based on their similarity to the Arabidopsis VHA genes, the identified genes were preliminarily named VHA-x, where x represents the letter code for each subunit. Evidently, the Rosaceae genome database comprised at least one gene encoding one V-ATPase subunit. Detailed information on VHA genes, including name, identifier (ID), chromosomal distribution, exon number, and length, is provided in Table S1.

Table 1.
Numbers of VHA genes in Arabidopsis thaliana and five Rosaceae species.
2.2. Phylogenetic Analysis and haracterisation of VHA Genes
To further classify the VHA proteins, a neighbor-joining phylogenetic tree of VHA-like proteins in Arabidopsis and the five Rosaceae species was constructed. Phylogenetic analysis of the 159 genes in Rosaceae species, along with 28 genes in Arabidopsis, was performed across 13 subfamilies, namely VHA-A, VHA-B, VHA-C, VHA-D, VHA-E, VHA-F, VHA-G, VHA-H, VHA-a, VHA-c, VHA-c”, VHA-d, and VHA-e (Figure 1). The number of VHA protein family were divided in 13 well-conserved groups. The clades A, B, C, D, E, F, G, H, a, c, c”, d, and e contained 7, 7, 8, 8, 14, 9, 22, 6, 24, 28, 5, 7, and 14 members, respectively (Figure 1). VHA-c” contained the lowest number of VHA genes (n = 5), while VHA-c contained the highest number of VHA genes (n = 28). All VHA proteins belong to two V-ATPase subunits (V0 and V1) (Table 1), indicating the diverse functions of VHA gene in Rosaceae species. Detailed characteristics of VHA genes are provided in Table S1.
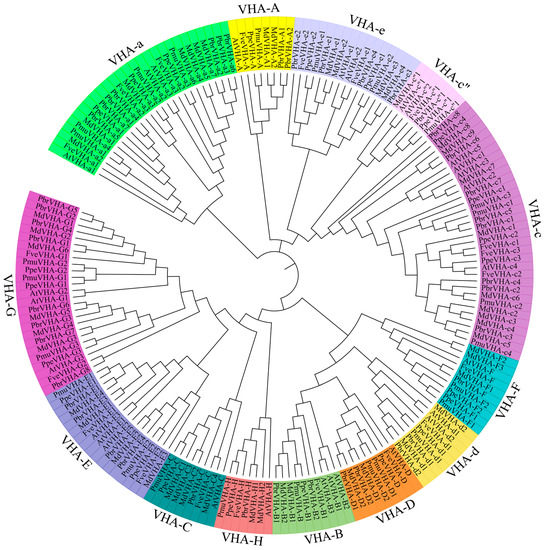
Figure 1.
Phylogenetic analysis of V-ATPase proteins from Arabidopsis and five Rosaceae species (Fragaria vesca, Prunus persica, Prunus mume, Pyrus bretschneideri and Malus x domestica). The phylogenetic tree was constructed using MEGA6.0 software (http://www.megasoftware.net/) by neighbor-joining method and diagram was drawn using online tool (iTOL, https://itol.embl.de/). The different colour indicate different subfamily respectively.
Pear is a typical Rosaceae species, which represents an excellent model for studying fruit development. The number of VHA genes in pear and apple were almost double the number in other analyzed species (Figure 1), suggested that it is necessary to make a systematic study in the complexity of PbrVHA genes. Furthermore, multiple transcriptome data are available for pear in the NCBI databases. Therefore, the VHA genes in five Rosaceae species were identified, and the gene structure, conserved motifs, chromosomal distribution, evolution, and expression patterns were emphatically analyzed in pear. Average length of the PbrVHA proteins was 70 (PbrVHA-e) to 868 (PbrVHA-a5) amino acids. Molecular weight of the PbrVHA proteins was 7584.44 (PbrVHA-e1) to 99,317 (PbrVHA-a5) Da, and their isoelectric points (pI) were 4.9 (PbrVHA-d2) to 9.61 (PbrVHA-c5) (Table S1).
2.3. Gene Structure, Conserved Motif, and Phylogenetic Analysis of the PbrVHA Genes
An unrooted tree was constructed to further explore the phylogeny of PbrVHA genes (Figure 2A). Consistent with the phylogenetic tree of Arabidopsis, our analysis supported the classification of PbrVHA genes into 13 subfamilies. Moreover, the exon–intron structure was analyzed based on genomic and cDNA sequences of PbrVHA genes. The structure of PbrVHA genes was relatively conserved within the same subfamily but varied across different subfamilies (Figure 2B). For instance, exon number in VHA genes ranged from 1 to 20, although the same subfamily generally had the same or similar exon number. Next, structures of conserved motifs of the 43 PbrVHA proteins were predicted using Multiple Em for Motif Elicitation (MEME) (Figure 2C). The block diagram and logos of 30 conserved motifs are shown in Figure S1. Motifs of the VHA proteins were mostly conserved within the same subfamily but varied across different subfamilies (Figure 2C). Motifs 1, 2, 3, 5, 7, 9, 14, 15, 16, 17, and 29 were found in all PbrVHA-a proteins, while motif 12 was present in the PbrVHA-G protein alone.
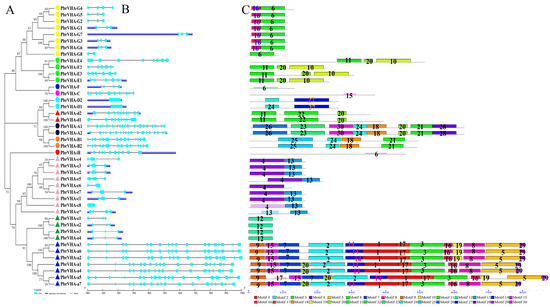
Figure 2.
Structural and phylogenetic analyses of PbrVHA genes. (A) A phylogenetic tree based on sequences of the PbrVHA proteins was constructed using MEGA 6.0 (B) The intron–exon structures were depicted by Gene Structure Display Server 2.0. UTRs and exons are represented by blue and light blue boxes, respectively, and introns are represented by grey lines. (C) Distribution of conserved motifs of PbrVHA proteins was described by Motif-based sequence analysis tools. Different motifs are indicated by different colours and numbered from 1 to 30; black lines represent non-conserved sequences.
The spatial structure of proteins plays a role in the biological function of proteins. Further, the secondary structures of these proteins were predicted using the online tool NPS@SOPMA (Figure S2A). The results showed that most VHA proteins were predicted to contain alpha helix, except for PbrVHA-G2, PbrVHA-G5, and PbrVHA-G8. Previous investigations suggested that the ATP-binding region on the A subunit may play an important role, so that one P-loop (GXXXXGKT/S) was identified in Figure S2B. It was predicted that proteins of VHA-a, VHA-c, VHA-c”, and VHA-e groups contained 2–8 cross-membrane domains (Figure S2). We also found that there were conserved proton-binding sites located in the fourth transmembrane domain of VHA-c and VHA-c” protein (Figure S2C,D). Similar to the motif analysis, the 3D structure analysis of the VHA proteins also showed that they shared high 3D structure similarity within the same subfamily (Figure S3). Thus, these results provided important information of their potential cellular roles.
2.4. Chromosomal Distribution and Gene Duplication of PbrVHA Genes
Chromosomal distribution and synteny of PbrVHA genes were analyzed to better understand the locations and duplication events in PbrVHA genes. A total of 38 PbrVHA genes were distributed on 14 of the 17 chromosomes (Figure 3), and 6 genes were located on scaffolds. The distribution of PbrVHA genes was random and uneven, and most PbrVHA genes were found on chromosomes 15 (n = 5), 10 (n = 4), and 12 (n = 4). Similarly, VHA genes in the other four species were randomly distributed (Figure S4). A total of 26 syntenic gene pairs were identified among the five Rosaceae species, including 10 in pear (Figure 3), 8 in apple (Figure S4A), 4 in peach (Figure S4B), 3 in Chinese plum (Figure S4C), and 1 in strawberry (Figure S4D). To further clarify their evolutionary history, we analyzed the orthologous VHA genes among five Rosaceae species. A total of 109 orthologous VHA gene pairs were identified, including 24, 37, 19, and 29 collinear gene pairs between pear and strawberry, apple, Chinese plum, and peach, respectively (Figure S5).
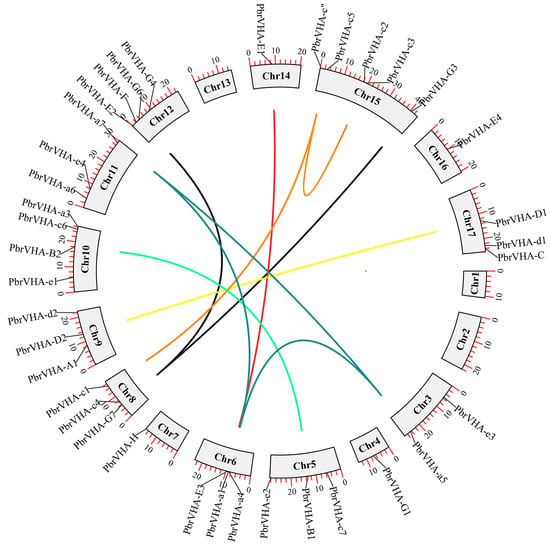
Figure 3.
Chromosomal distribution and synteny analysis of the PbrVHA genes. Gene pairs with a syntenic relationship were joined by a line. Chromosome number was indicated on the inner side.
To clarify the driving forces of gene duplication, Ka, Ks, and Ka/Ks values were calculated for paralogous gene pairs in pear (Table 2). The Ka/Ks value indicates selection pressure on duplicated PbrVHA genes during the course of evolution. Ka/Ks values of all syntenic PbrVHA gene pairs were less than 0.3, indicating that these genes are under the influence of purifying selection. Given their importance in the expansion of gene families, duplication events in paralogous gene pairs were analyzed. All 10 gene pairs were whole-genome duplication (WGD)-derived, suggesting that WGD represents the principal pathways for gene expansion of the PbrVHA gene family.

Table 2.
Ka, Ks, and Ka/Ks values of paralogous PbrVHA gene pairs.
2.5. Gene Ontology (GO) Analysis of PbrVHA Proteins
To further understand the functions of the PbrVHA proteins, GO annotation and GO enrichment analyses were performed. Our results showed that PbrVHA proteins were most enriched for 20 GO terms (Figure 4), which were divided into three categories: biological process (BP), cellular component (CC), and molecular function (MF). GO terms related to ATP hydrolysis coupled proton transport (GO:0015991), proton-transporting ATPase activity (GO:0046961), plant-type vacuole (GO:0000325), vacuolar proton-transporting V-type ATPase, V0 domain (GO:0000220), hydrogen ion transmembrane transporter activity (GO:0015078), and vacuolar acidification (GO:0007035), were obviously enriched GO categories from three categories. The GO enrichment suggested that PbrVHA genes play important roles in vacuole and trans-Golgi network. Furthermore, these results provide useful information for future gene characterization studies in pear.
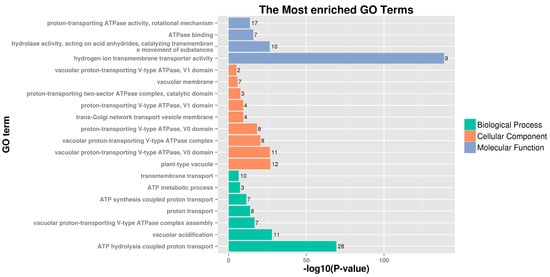
Figure 4.
Gene ontology analysis of PbrVHA proteins in biological processes, molecular functions, and cellular component categories.
2.6. Gene Expression Patterns of PbrVHA Genes in Different Tissues and During Fruit Development
To explore the putative functions of PbrVHA genes, their expression patterns in six major tissues (fruit, petal, shoot, leaf, ovary, and stigma) were analyzed based on previously reported RNA-Seq data [30]. Seven genes were not expressed or were weakly expressed in all tissues, which is probably explained by the presence of uncharacterized pseudogenes or specific gene expression at a particular stage. Moreover, 36 PbrVHA genes were expressed in major tissues and clustered into six subsets based on their expression patterns (Figure 5A). Among these genes, PbrVHA-c3, PbrVHA-c1, PbrVHA-c4, PbrVHA-c5, and PbrVHA-E1 exhibited the highest transcript abundances in different tissues, whereas PbrVHA-E2, PbrVHA-G8, and PbrVHA-a7 exhibited lower transcript abundances in six tissues (Figure 5A). Several genes were highly expressed in specific tissues, such as PbrVHA-F and PbrVHA-G1 in petal tissues. To further explore the possible roles of PbrVHA genes in fruit development, expression patterns of 26 genes at six different fruit developmental stages were evaluated based on publicly available RNA-Seq data [31]. The expression of PbrVHA-B2 showed an increasing trend throughout fruit maturity, while the expression of PbrVHA-G1, PbrVHA-E3, PbrVHA-E1, and PbrVHA-d1 was upregulated at the S3 stage (Figure 5B). Interestingly, PbrVHA-D1, PbrVHA-D2, PbrVHA-E1, PbrVHA-F, and PbrVHA-G1 showed relatively high expression levels at the S6 stage (Figure 5B), indicating their crucial role in fruit ripening. Moreover, the expression profiles of PbrVHAs during fruit development were cultivar dependent (Figure S6).
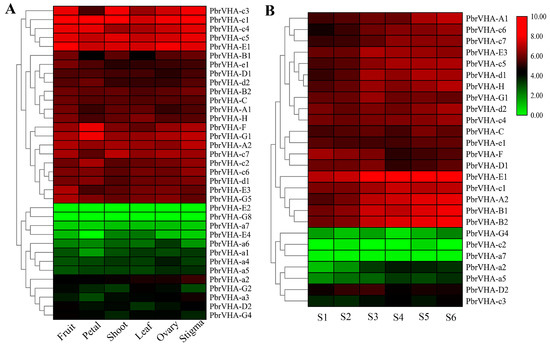
Figure 5.
Expression patterns of PbrVHA genes in different tissues and during fruit development of “Yali” pear. (A) Expression patterns of PbrVHA genes in six different tissues, including fruit, petal, shoot, leaf, ovary, and stigma. (B) Expression profiles of PbrVHA genes during fruit development, including the fruit-setting stage (S1), physiological fruit dropping stage (S2), fruit rapid enlargement stage (S3), a month after fruit enlargement stage (S4), pre-mature stage (S5), and mature stage (S6). The expression levels were calculated as log2-transformed (RPKM + 1) values. Green, black, and red indicate low, moderate, and high levels, respectively.
2.7. Expression Profiles of PbrVHA Genes during Pollen Tube Development and Fruit Senescence
To explore the possible functions of PbrVHA genes, their expression patterns during pollen tube development [32] and fruit senescence [31] were also analyzed. A total of 27 PbrVHA genes were expressed during pollen tube growth and senescence, with PbrVHA-G8 specifically expressed in the growing pollen tube (Figure 6A). Moreover, seven PbrVHA genes (PbrVHA-c1, PbrVHA-c3, PbrVHA-c4, PbrVHA-D2, PbrVHA-E3, PbrVHA-e1, and PbrVHA-G7) were significantly downregulated when the pollen tube growth stopped, suggesting their important roles in pollen tube growth. PbrVHA-c3 was highly expressed during the hydrous and growth stages (Figure 6A), suggesting that it positively regulates pollen tube growth. To analyze whether PbrVHA is involved in core browning, we compared the transcript levels of PbrVHA genes during browning in “Whangkeumbae” pear [33]. Two PbrVHAs (PbrVHA-c1 and PbrVHA-D2) were differentially expressed during browning (Figure 6B), suggesting that these genes function in core browning. To further explore the potential roles of PbrVHA genes during fruit senescence, their expression profiles during postharvest storage were analyzed [31]. A total of 21 PbrVHA genes were transcribed during the storage of “Housui” pear, with diverse expression patterns (Figure S7). The expression levels of 10 genes (PbrVHA-G4, PbrVHA-G1, PbrVHA-E3, PbrVHA-E1, PbrVHA-F, PbrVHA-C, PbrVHA-D2, PbrVHA-A1, PbrVHA-H, and PbrVHA-a7) were significantly reduced in “Housui” pear (Figure S7). Moreover, the expression profiles of PbrVHA genes during fruit senescence were cultivar dependent (Figure S7).
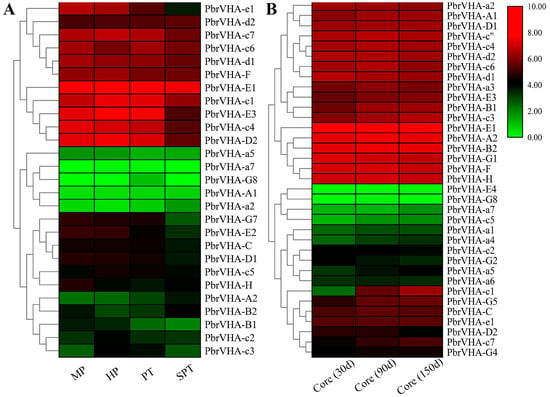
Figure 6.
Expression profiles of PbrVHA genes during pollen tube development and fruit senescence. (A) The expression of PbrVHA genes during four development stages of pear pollen, including mature pollen grains (MP), hydrated pollen grains (HP), growing pollen tubes (PT) and stopped-growth pollen tubes (SPT). (B) Expression profiles of PbrVHA genes in the pear core during fruit storage at three stage (30d, 90d and 150d after harvesting). Color scale represents log2-transformed FPKM + 1 values. Green, black, and red indicate low, moderate, and high levels, respectively.
3. Discussion
V-ATPases are found in a wide variety of endomembrane system of all eukaryotes, which function as ATP hydrolysis-driven proton pumps [5]. With extensive research, an increasing number of biological functions of VHA genes in plants are gradually being discovered [14,19]. V-ATPases energize secondary active transport and serve as important regulators of membrane trafficking [5]. Moreover, V-ATPases play vital roles in plant development and stress response, functioning as a house-keeping and stress-responsive enzyme [22]. Whole-genome sequencing has enabled the investigation of VHA genes and their potential functions. The VHA gene families in A. thaliana, rice, and tomato, among other plants, have been described [6,12,20]. However, there has been no comprehensive study on genome-wide identification of VHA genes in Rosaceae species.
The present study is the first to reveal detailed characteristics of the VHA gene family in Rosaceae species. A total of 159 VHA genes were identified in apple (n = 48), pear (n = 43), Chinese plum (n = 25), peach (n = 23), and strawberry (n = 20). Our results indicate that the number of genes encoding V-ATPase proteins may vary across plant species. The similar number of VHA proteins between pear and apple may indicate the parallel evolutionary relationship of this gene family between these two species [27,34]. Phylogenetic analysis showed that the VHA proteins of the Rosaceae species were clustered with the 13 previously described, corresponding subfamilies in Arabidopsis [6], confirming the suitability of our methods of gene identification and phylogenetic tree construction.
To explore the functions of PbrVHA genes, phylogeny, exon–intron structures, conserved motif structures, and spatial structure were analyzed in this research. PbrVHA proteins were classified into 13 subfamilies, consistent with the previously reported classification of A. thaliana [12]. The number and distribution of introns and exons were mostly conserved within the same subfamily but varied across different subfamily (Figure 2B). MEME analysis showed that VHA proteins within the same subfamily contained the same basal motifs (Figure 2C). Therefore, VHA proteins in the same subfamily may show similar structures and functions. Accordingly, the functions of VHA genes in Rosaceae species can be predicted based on the known functions of homologous genes in other species. For example, overexpression of VHA-B from the halophyte Halostachys caspica enhanced salt tolerance of transgenic Arabidopsis [35], indicating that its homologous gene may serve similar functions. Apple V-ATPase subunit B1 is involved in drought and salt tolerance [36], which further supports our prediction. The localization of the V-ATPase protein was previously shown to be determined by the isoforms of subunit a (VHA-a) in Arabidopsis [3,11]. It was predicted that PbrVHA-a subunit that consists of a C-terminal hydrophobic domain with six transmembrane domains (Figure S2), suggested that PbrVHA-a subunit may be the determinant that targets the V-ATPase to the TGN/EE or to the tonoplast as previously described [11]. The assembly of V-ATPases requires the presence of all V1 and V0 subunits [7], so that it is very important to identify the functional domain of every subunit. Conserved proton-binding sites were identified in the fourth transmembrane domain of VHA-c and VHA-c” protein, suggested that subunit c and its isoforms c” may form a H+-binding rotor ring in H+-transporting V-ATPases [7,8]. The V0 subunits are integral membrane proteins except for subunit d, which is consistent with the number of transmembrane domains in this research.
Gene duplication is an important evolutionary event for the expansion of gene families. Gene duplication can be classified into five types, including WGD, tandem duplication, proximal duplication, dispersed duplication, and transposed duplication [37]. Our results indicated that VHA genes have expanded mainly through WGD, which contributes to expansion of PbrVHA genes (Table 2). WGD may greatly contribute to the evolution of physiological diversity in pear [37]. The number of VHA genes in pears and apples was much higher than that in peach, strawberry, and Chinese plum (Table 1). These findings corroborate the previous report that pear and apple may have undergone a recent lineage-specific WGD, while strawberry does not share a recent WGD event [27]. In previous studies, the Ka/Ks values of various gene pairs were variable, while WGD genes are usually conserved with smaller Ka/Ks ratios [37]. Ka/Ks values of all paralogous VHA gene pairs were less than 0.3 (Table 2), suggesting that these gene pairs have undergone strong purifying selection pressure [38]. Together, these results indicate that PbrVHA proteins are highly conserved and that purifying selection plays a key role in the expansion of the PbrVHA gene family.
The roles of VHA genes have been studied previously. These genes are involved in numerous biological processes, including development, metabolism, and stress response [14,35]. VHA-c5 (Arabidopsis), VHA-C (Arabidopsis), VHA-E (Wheat), VHA-B1 (Apple), and VHA-G1 (Juglans regia) are involved in plant responses to abiotic stress [15,39,40]. Based on GO analysis, PbrVHA genes were involved in proton transport, vacuolar acidification, transmembrane transport, and so on, which is consistent with previous research about the function of VHA genes [12,41]. Gene expression patterns are usually consistent with gene functions. In this study, transcriptomic analysis was conducted to determine the possible biological functions of PbrVHA genes. Expression patterns of VHA genes varied across six tissues, which may be consistent with their diverse physiological roles. VHA-c genes in Arabidopsis are differentially expressed in expanding cells to support growth. For instance, AtVHA-c1 and AtVHA-c2 are highly expressed in roots and shoots, whereas AtVHA-c3 is mainly expressed in root caps and pollen [42]. PbrVHA-c1, PbrVHA-c3, and PbrVHA-c4 were highly expressed in different tissues (Figure 5A), but their expression varied at different pollen tube developmental stages (Figure 6A), indicating the crucial roles of V-ATPases during cell expansion.
The previous studies suggested that VHA genes play critical roles in abiotic stress tolerance as well as plant growth and senescence [22,39,43]. The conserved VHA-c subunit plays an important role in plant growth [43]. Transcript analysis of VHA genes revealed their possible involvement in citrate accumulation in fruit [18,44]. The functional roles of PbrVHA genes at different developmental stages were also investigated in this study. The expression of some genes (PbrVHA-B2, PbrVHA-G1, PbrVHA-E3, PbrVHA-E1, and PbrVHA-d1) was significantly upregulated at the S3 stage (Figure 5B), suggesting their pivotal roles in pear fruit growth and ripening. There is some evidence that VHA genes perform important functions during senescence. Molecular genetic studies have shown that OsPSL1/VHA-A1 and AtVHA-B1 are implicated in leaf senescence [19,45]. In the present study, transcript levels of seven VHA genes were downregulated when the pollen tube stopped growing, which was consistent with changes in V-ATPase activity [46]. Proteomic analysis revealed that V-ATPase protein abundance decreased during fruit storage, suggesting that V-ATPase activity is associated with fruit senescence. To further confirm this speculation, we analyzed the expression patterns of PbrVHA genes during the storage of “Housui” pear and found that the expression levels of 10 PbrVHA genes were significantly downregulated following storage, indicating their role in pear senescence and fruit quality maintenance.
Vacuolar H+-ATPases are multi-subunit protein complexes, which were divided into two structurally distinct subcomplexes (V1 and V0 subunits) [6,12,41]. The water-soluble V1 domain was located in the cytosol, which catalyzes ATP hydrolyzing and binding; the membrane-embedded V0 domain contained the proton pore and transported protons [41]. The core of the V1 domain consists of A and B subunits, whereas the V0 domain is composed of subunit VHA-c, VHA-c”, VHA-a and VHA-e. V 0 and V 1 complexes are joined by a central stalk (subunits D, F, and d) and three peripheral stators (subunits C, E, G, H, and a) [41]. The assembly of the plant V-ATPase is dependent on all the 13 subunits. The diversity of functions of the V-ATPase may owe to roles of its various subunits and numerous functional regulation sites and motifs [41]. We also found that the expression patterns of some subunits were uniformly expressed, while some were differently expressed (Figure 5 and Figure 6), which suggested that the complexity of the cells coordinates the expression of certain subunits during development and senescence. Genome-wide identification of the VHA genes in five Rosaceae species was an important first step in the system research for VHA genes, this study laid a foundation for clarify the particular roles of each subunit.
4. Materials and Methods
4.1. Identification and Characterization of VHA Genes
Protein sequences of previously identified VHA gene family members in Arabidopsis were downloaded from the Arabidopsis Information Resource (http://www.Arabidopsis.org/) [6]. These amino acid sequences of A. thaliana VHA proteins were used as queries to perform BLASTP searches against the National Center for Biotechnology Information Support Center (NCBI) database and the Rosaceae genome, with an e-value cut-off of 1.0 × 10−10. In this study, we investigated five Rosaceae species genomes. Pear and Chinese plum genome sequences were downloaded from the Pear Genome Project (http://peargenome.njau.edu.cn/) [27] and Prunus mume Genome Project (http://prunusmumegenome.bjfu.edu.cn) [28] respectively. Genomic sequences of peach, apple, and strawberry were downloaded from Phytozome [29]. To further confirm the validity of the identified VHA genes, SMART (http://smart.embl-heidelberg.de/) and Pfam (https://pfam.xfam.org) databases were used to search for the functional domains of potential VHA proteins [47]. Molecular weights and pI values of VHA proteins were predicted using ProtParam (http://web.expasy.org/protparam/) [48].
4.2. Phylogenetic Analysis
Multiple sequence alignment was performed using ClustalW program based on the amino acid sequences of A. thaliana, pear, peach, apple, Chinese plum, and strawberry. A neighbour-joining phylogenetic tree was constructed with 1000 bootstrap replicates using MEGA 6.0 [49]. The diagram was drawn using online tool (iTOL, https://itol.embl.de/), as previously described [50].
4.3. Gene Structure and Conserved Motif Analysis
A schematic diagram of the VHA gene structure was constructed using Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/index.php) [51] by aligning the coding sequence with the genomic sequence. Conserved motifs of the VHA proteins were analyzed using MEME Version 5.1.0 (http://meme-suite.org/tools/meme) [52]. The optimized parameters were as follows: zero or one site per sequence; optimum width of each motif, 6–100 residues; maximum number of motifs, 30. The secondary structures of VHA proteins were predicted by the online tool NPS@SOPMA (https://npsa-prabi.ibcp.fr/). The 3D models of tertiary structures were constructed using the Swiss-Model (https://www.swissmodel.expasy.org/) [53]. Multiple alignments of V-ATPase proteins from pear and Arabidopsis were performed using DNAman soft.
4.4. Chromosomal Distribution and Synteny Analysis
Chromosomal distribution of VHA genes was analyzed based on the Rosaceae genome annotation data. MCScanX was used for the detection and evolutionary analysis of gene synteny and collinearity among the five Rosaceae species [34]. The Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/downloads) was used to identify segmentally duplicated genes among the five Rosaceae genomes [37]. Chromosomal distribution and syntenic relationships of VHA genes were plotted using TBtools and Circos. Ka and Ks values of the duplicated pairs were calculated using TBtools [54].
4.5. Gene ontology (GO) Items and Expression Pattern Analysis
Global gene expression profiles in various tissues and at different developmental stages were investigated based on publicly available datasets in the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database. RNA-Seq datasets (SRA number: PRJNA590622) were used to determine tissue-specific expression profiles [30]. Raw reads data were used to quantify the expression levels of PbrVHA genes in pear fruit at different developmental (SRA numbers: PRJNA309745) or storage stages (SRA numbers: PRJNA309745 and PRJNA597402) [31,33]. Moreover, PbrVHA genes involved in pollen tube growth were identified using RNA-Seq datasets (SRA number: PRJNA299117) [32]. Heatmaps were constructed using TBtools [54]. GO annotation analysis of the PbrVHA genes was conducted using the Blast2GO program [55].
5. Conclusions
In the present study, genome-wide identification and bioinformatic analyses of VHA genes in pear and four other Rosaceae species were performed. A total of 159 members of the VHA gene family were identified and further classified into 13 subfamilies based on a phylogenetic tree. Most PbrVHA genes were derived through WGD under purifying selection. In addition, the expression patterns of PbrVHA genes in various tissues and at different fruit developmental or storage stages were investigated using RNA-Seq data. In comparative transcriptomic analyses, some PbrVHA genes were differentially expressed during fruit growth and storage, suggesting that VHA genes play specific roles in development and senescence. Expression profiles of PbrVHA genes were tissue- and cultivar-specific. The present study can serve as a foundation for further investigation of biological functions of VHA genes in Rosaceae species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/12/1661/s1, Figure S1: Logos of PbrVHA protein conserved motifs. A total of 30 motifs were identified by the MEME tool. Figure S2. Secondary structure and multiple alignments of domain analysis of the V-ATPase proteins. A: The secondary structures of VHA proteins were predicted using the online tool NPS@SOPMA (https://npsa-prabi.ibcp.fr/); B: Multiple alignments of the P-loop motif in VHA-A; C: The cross membrane domain and proton binding sites of VHA-c and VHA-c”; D: The cross membrane domain and amino acids identified as essential for H+-pumping of VHA-a; E: The cross membrane domain of VHA-e. Alpha helix was colored in red, extend strand was colored in blue, and random coli in purple. The cross membrane domain was marked with a blue box. The cross-membrane domain was analyzed using online tool TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). The P-loop is highlighted with a green box. Amino acids identified as essential for H+ -pumping are marked with purple box. Figure S3. The three-dimensional (3D) structural models of V-ATPase proteins in pear. The structure of PbrVHA proteins was constructed using SWISS MODEL. Figure S4: Chromosome location and synteny analysis of the VHA genes in the other four Rosaceae species. Gene pairs with a syntenic relationship were joined by a line. A: apple; B: peach; C: Chinese plum; D: strawberry. Figure S5. Chromosomal distribution and orthologous VHA gene pairs of the VHA genes between pear and strawberry, apple, Chinese plum, peach, respectively. A: Between pear and strawberry; B: Between pear and apple; C: Between pear and Chinese plum; D: Between pear and peach. Figure S6. Expression patterns of VHA genes at six key fruit developmental stages of four pear cultivars. Figure S7. Expression profiles of PbrVHA genes of five pear cultivars during fruit senescence after harvesting. Table S1: Detailed information of all VHA gene family genes identified in the five Rosaceae species
Author Contributions
H.Z. and P.L. designed the experiments of the study. W.H. and H.H. analyzed the data; S.L. and L.Z. performed the experiments; H.Z. and Y.Z. wrote and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 31901746), the Natural Science Foundation of Jiangsu Province (Grant No. BK20160589), Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF) [CX(19)3113].
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Shimada, T.; Takagi, J.; Ichino, T.; Shirakawa, M.; Hara-Nishimura, I. Plant Vacuoles. Annu. Rev. Plant Biol. 2018, 69, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Li, K.; Wang, Z.; Zhu, K.; Tan, X.; Cao, J. A Review of Plant Vacuoles: Formation, Located Proteins, and Functions. Plants 2019, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.; Beyhl, D.; Gorlich, E.; Al-Rasheid, K.A.; Marten, I.; Stierhof, Y.D.; Hedrich, R.; Schumacher, K. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 2010, 107, 3251–3256. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.T.; Tang, R.J.; Zhang, Y.J.; Xue, H.W.; Ferjani, A.; Luan, S.; Lin, W.H. Two tonoplast proton pumps function in Arabidopsis embryo development. New Phytol. 2020, 225, 1606–1617. [Google Scholar] [CrossRef]
- Vasanthakumar, T.; Rubinstein, J.L. Structure and roles of V-type ATPases. Trends Biochem. Sci. 2020, 45, 295–307. [Google Scholar] [CrossRef]
- Sze, H.; Schumacher, K.; Müller, M.L.; Padmanaban, S.; Taiz, L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 2002, 7, 157–161. [Google Scholar] [CrossRef]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef]
- Saroussi, S.; Nelson, N. The little we know on the structure and machinery of V-ATPase. J. Exp. Biol. 2009, 212, 1604–1610. [Google Scholar] [CrossRef]
- Schumacher, K.; Krebs, M. V-ATPases: Rotary engines for transport and traffic. In Transporters and Pumps in Plant Signaling. Signaling and Communication in Plants; Springer: Berlin, Germany, 2011; Volume 7, pp. 293–312. [Google Scholar] [CrossRef]
- Hager, A.; Helmle, M. Properties of an ATP-fueled, Cl−1-dependent proton pump localized in membranes of microsomal vesicles from maize coleoptiles. Z. Nat. C 1981, 36, 997–1008. [Google Scholar] [CrossRef]
- Feng, S.; Peng, Y.; Liu, E.; Ma, H.; Qiao, K.; Zhou, A.; Liu, S.; Bu, Y. Arabidopsis V-ATPase d2 Subunit Plays a Role in Plant Responses to Oxidative Stress. Genes 2020, 11, 701. [Google Scholar] [CrossRef]
- Schumacher, K.; Krebs, M. The V-ATPase: Small cargo, large effects. Curr. Opin. Plant Biol. 2010, 13, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Scholl, S.; Doering, A.; Zhang, Y.; Irani, N.G.; Rubbo, S.D.; Neumetzler, L.; Krishnamoorthy, P.; Van Houtte, I.; Mylle, E.; et al. V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nat. Plants 2015, 1, 15094. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Takano, T.; Liu, S. The role of endomembrane-localized VHA-c in plant growth. Plant Signal. Behav. 2018, 13, e1382796. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Liu, E.; Ma, H.; Feng, S.; Gong, S.; Wang, J. NaCl-induced expression of AtVHA-c5 gene in the roots plays a role in response of Arabidopsis to salt stress. Plant Cell Rep. 2018, 37, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ge, Y.; Zhang, W.; Zhao, Y.; Yang, G. The walnut JrVHAG1 gene is involved in cadmium stress response through ABA-signal pathway and MYB transcription regulation. BMC Plant Biol. 2018, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, A.; Andres, Z.; Medzihradszky, A.; Kruger, F.; Scholl, S.; Delang, S.; Patir-Nebioglu, M.G.; Gute, G.; Yang, H.; Murphy, A.S.; et al. Job sharing in the endomembrane system: Vacuolar acidification requires the combined activity of V-ATPase and V-PPase. Plant Cell 2015, 27, 3383–3396. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.Y.; Hussain, S.B.; Guo, L.X.; Yang, H.; Ning, D.Y.; Liu, Y.Z. Genome-wide identification and transcript analysis of vacuolar-ATPase genes in citrus reveal their possible involvement in citrate accumulation. Phytochemistry 2018, 155, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ji, Y.; Zhou, J.; Xing, D. Phosphatidylinositol 3-kinase promotes V-ATPase activation and vacuolar acidification and delays methyl jasmonate-induced leaf senescence. Plant Physiol. 2016. [Google Scholar] [CrossRef]
- Coker, J.S.; Jones, D.; Davies, E. Identification, conservation, and relative expression of V-ATPase cDNAs in tomato plants. Plant Mol. Biol. Rep. 2003, 21, 145–158. [Google Scholar] [CrossRef]
- Axelsen, K.B.; Palmgren, M.G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. Biochem. 2001, 126, 696–706. [Google Scholar] [CrossRef]
- Kang, C.; Sun, F.; Yan, L.; Li, R.; Bai, J.; Caetano-Anolles, G. Genome-wide identification and characterization of the vacuolar H+-ATPase subunit H gene family in crop plants. Int. J. Mol. Sci. 2019, 20, 5125. [Google Scholar] [CrossRef] [PubMed]
- Farinati, S.; Rasori, A.; Varotto, S.; Bonghi, C. Rosaceae Fruit Development, Ripening and Post-harvest: An Epigenetic Perspective. Front. Plant Sci. 2017, 8, 1247. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, Z.; Huang, X.; Zhang, L.; Zhao, P.; Ma, H.; Li, X.; Ban, Z.; Liu, X. Label-free quantitative proteomics to investigate strawberry fruit proteome changes under controlled atmosphere and low temperature storage. J. Proteom. 2015, 120, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Y.; Qin, G.; Tian, S. iTRAQ-based quantitative proteomic analysis reveals the role of the tonoplast in fruit senescence. J. Proteom. 2016, 146, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Y.; Qin, G.; Tian, S. Molecular basis of 1-methylcyclopropene regulating organic acid metabolism in apple fruit during storage. Postharvest Biol. Technol. 2016, 117, 57–63. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, M.; Zhang, H.; Zhang, S.; Qian, M.; Zhang, Z.; Luo, W.; Fan, J.; Liu, Z.; Wang, L. Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 2019, 19, 587. [Google Scholar] [CrossRef]
- Zhang, M.; Xue, C.; Xu, L.; Sun, H.; Qin, M.; Zhang, S.; Wu, J. Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (Pyrus L.). Sci. Rep. 2016, 6, 28130. [Google Scholar] [CrossRef]
- Zhou, H.; Yin, H.; Chen, J.; Liu, X.; Zhang, S. Gene-expression profile of developing pollen tube of Pyrus bretschneideri. Gene Expr. Patterns 2016, 20, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tian, M.; Huang, W.; Luo, S.; Hu, H.; Zhang, Y.; Zhang, L.; Li, P. Physiological and transcriptomic analysis of ‘Whangkeumbae’ pear core browning during low-temperature storage. Gene Expr. Patterns 2020, 36, 119113. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Qi, K.; Liu, X.; Yin, H.; Wang, P.; Chen, J.; Wu, J.; Zhang, S. Genome-wide identification and comparative analysis of the cation proton antiporters family in pear and four other Rosaceae species. Mol. Genet. Genom. 2016, 291, 1727–1742. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zeng, Y.; Guan, B.; Zhang, F. Overexpression of a vacuolar H+-pyrophosphatase and a B subunit of H+-ATPase cloned from the halophyte Halostachys caspica improves salt tolerance in Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2011, 108, 63–71. [Google Scholar] [CrossRef]
- Hu, D.; Sun, M.; Sun, C.; Liu, X.; Zhang, Q.; Zhao, J.; Hao, Y. Conserved vacuolar H+-ATPase subunit B1 improves salt stress tolerance in apple calli and tomato plants. Sci. Hortic. 2015, 197, 107–116. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Li, X.; Gao, F.; Feng, J.; Zhou, Y. Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef]
- Xu, Z.; Zhao, Y.; Ge, Y.; Peng, J.; Dong, M.; Yang, G. Characterization of a vacuolar H+-ATPase G subunit gene from Juglans regia (JrVHAG1) involved in mannitol-induced osmotic stress tolerance. Plant Cell Rep. 2017, 36, 407–418. [Google Scholar] [CrossRef]
- Adem, G.D.; Roy, S.J.; Huang, Y.; Chen, Z.-H.; Wang, F.; Zhou, M.; Bowman, J.P.; Holford, P.; Shabala, S. Expressing Arabidopsis thaliana V-ATPase subunit C in barley (Hordeum vulgare) improves plant performance under saline condition by enabling better osmotic adjustment. Funct. Plant Biol. 2017, 44. [Google Scholar] [CrossRef]
- Ma, B.; Xiang, Y.; An, L. Structural bases of physiological functions and roles of the vacuolar H+-ATPase. Cell. Signal. 2011, 23, 1244–1256. [Google Scholar] [CrossRef]
- Padmanaban, S.; Lin, X.; Perera, I.; Kawamura, Y.; Sze, H. Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 2004, 134, 1514–1526. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Bu, Y.; Takano, T.; Zhang, X.; Liu, S. Conserved V-ATPase c subunit plays a role in plant growth by influencing V-ATPase-dependent endosomal trafficking. Plant Biotechnol. J. 2016, 14, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.X.; Shi, C.Y.; Liu, X.; Ning, D.Y.; Jing, L.F.; Yang, H.; Liu, Y.Z. Citrate accumulation-related gene expression and/or enzyme activity analysis combined with metabolomics provide a novel insight for an orange mutant. Sci. Rep. 2016, 6, 29343. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gong, P.; Li, K.; Huang, F.; Cheng, F.; Pan, G. A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J. Exp. Bot. 2016, 67, 2761–2776. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, H.; Chen, J.; Jiang, X.; Tao, S.; Wu, J.; Zhang, S. Mitochondrial dysfunction mediated by cytoplasmic acidification results in pollen tube growth cessation in Pyrus pyrifolia. Physiol. Plant. 2015, 153, 603–615. [Google Scholar] [CrossRef]
- Wang, W.; Sun, Y.-Q.; Li, G.-L.; Zhang, S.-Y. Genome-wide identification, characterization, and expression patterns of the BZR transcription factor family in sugar beet (Beta vulgaris L.). BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Totowa, N.J., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Ali, M.; Luo, D.X.; Khan, A.; Haq, S.U.; Gai, W.X.; Zhang, H.X.; Cheng, G.X.; Muhammad, I.; Gong, Z.H. Classification and Genome-Wide Analysis of Chitin-Binding Proteins Gene Family in Pepper (Capsicum annuum L.) and Transcriptional Regulation to Phytophthora capsici, Abiotic Stresses and Hormonal Applications. Int. J. Mol. Sci. 2018, 19, 2216. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. Tbtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).