Molecular Phylogeny and Phylogeography of Potentilla multifida L. agg. (Rosaceae) in Northern Eurasia with Special Focus on Two Rare and Critically Endangered Endemic Species, P. volgarica and P. eversmanniana
Abstract
1. Introduction
- To assess the genetic variability of P. volgarica and P. eversmanniana and to test whether they represent two separate species.
- To assess the genetic distinctions of both species from P. multifida agg. sensu Soják [19] and other related species of the section Multifidae.
- To assess the phylogenetic relationships of P. multifida agg. species and pinpoint the origin of disjunct isolated populations of P. volgarica and P. eversmanniana in the Russian Plain and the foothills of the Southern Urals, respectively.
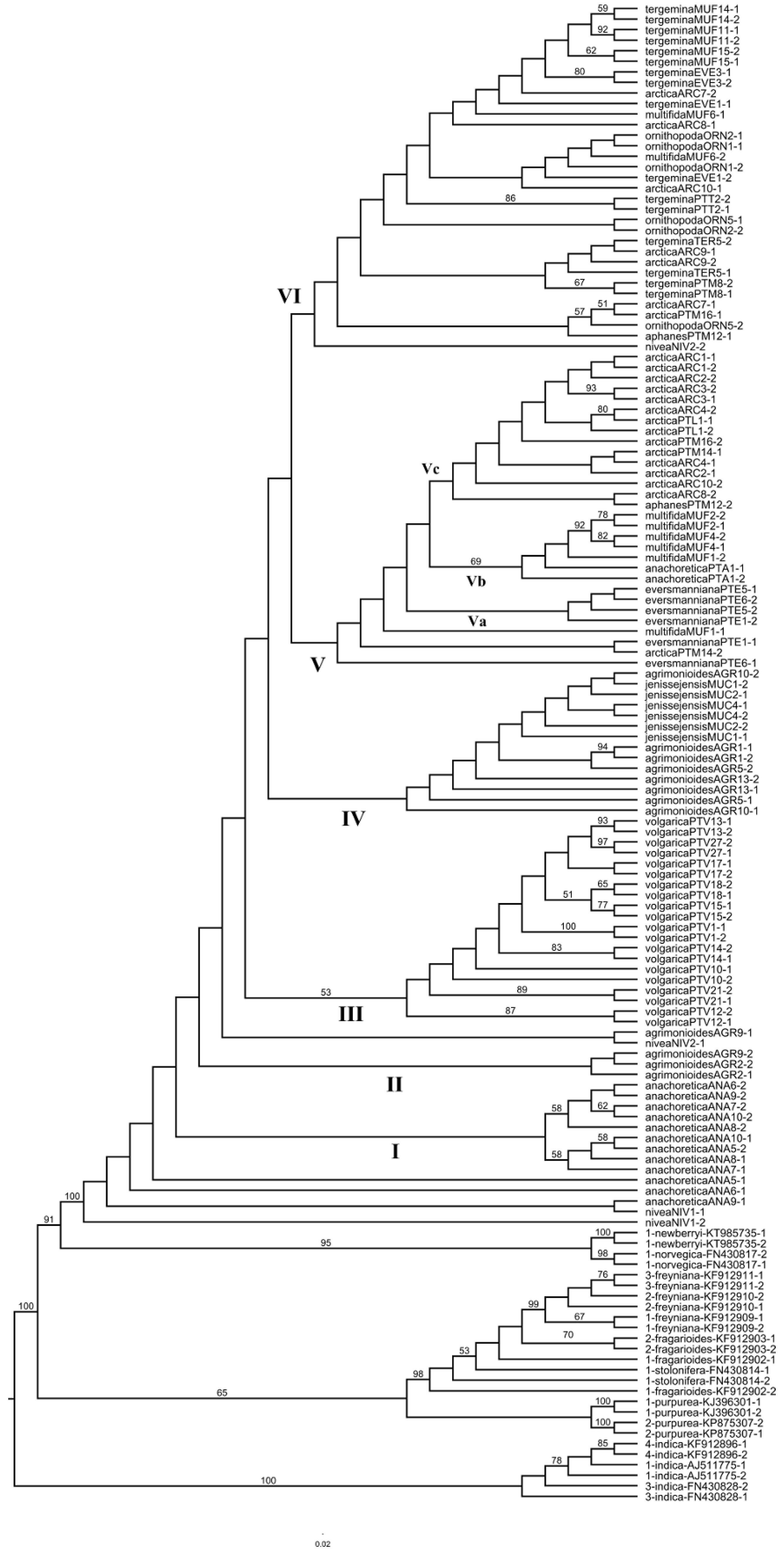
2. Results
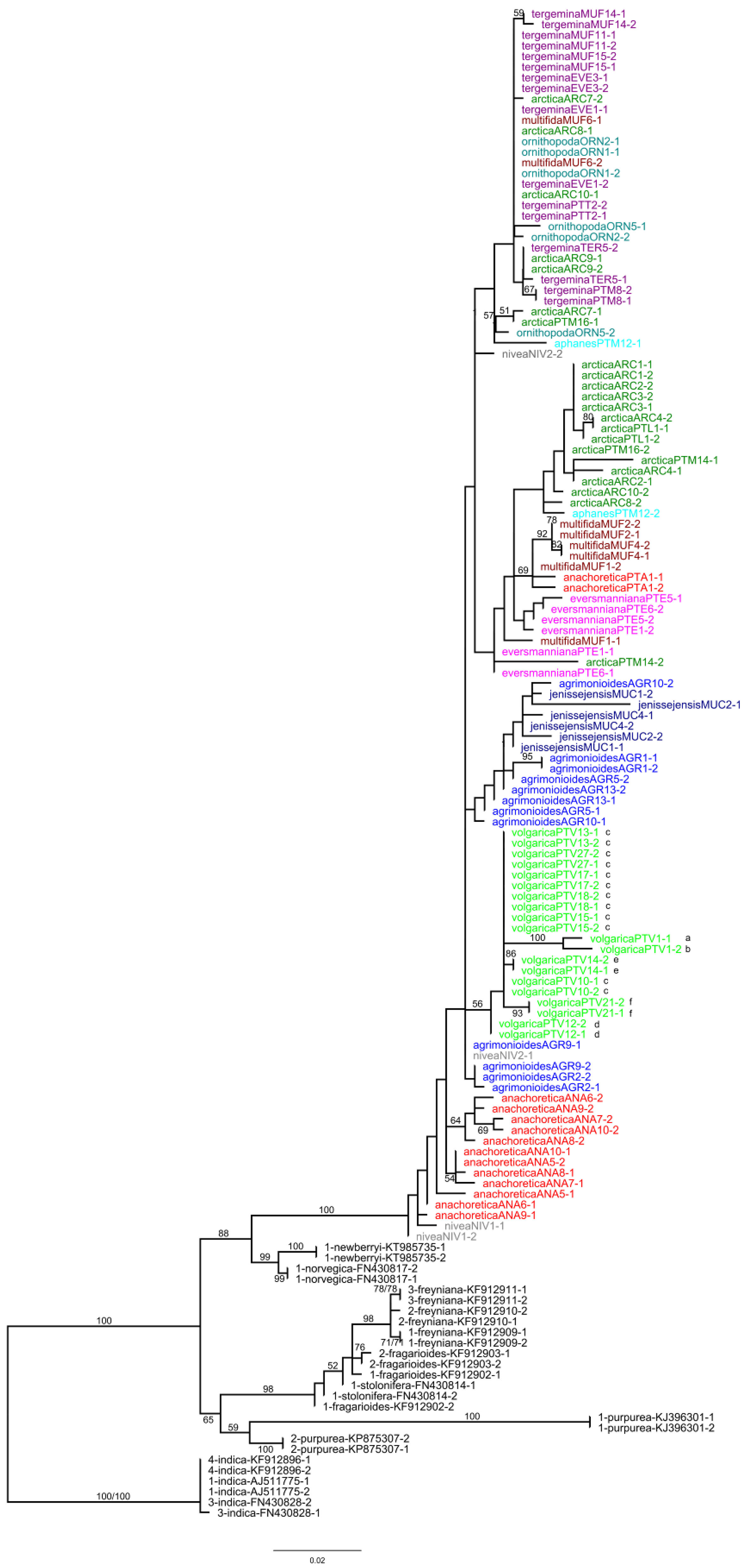
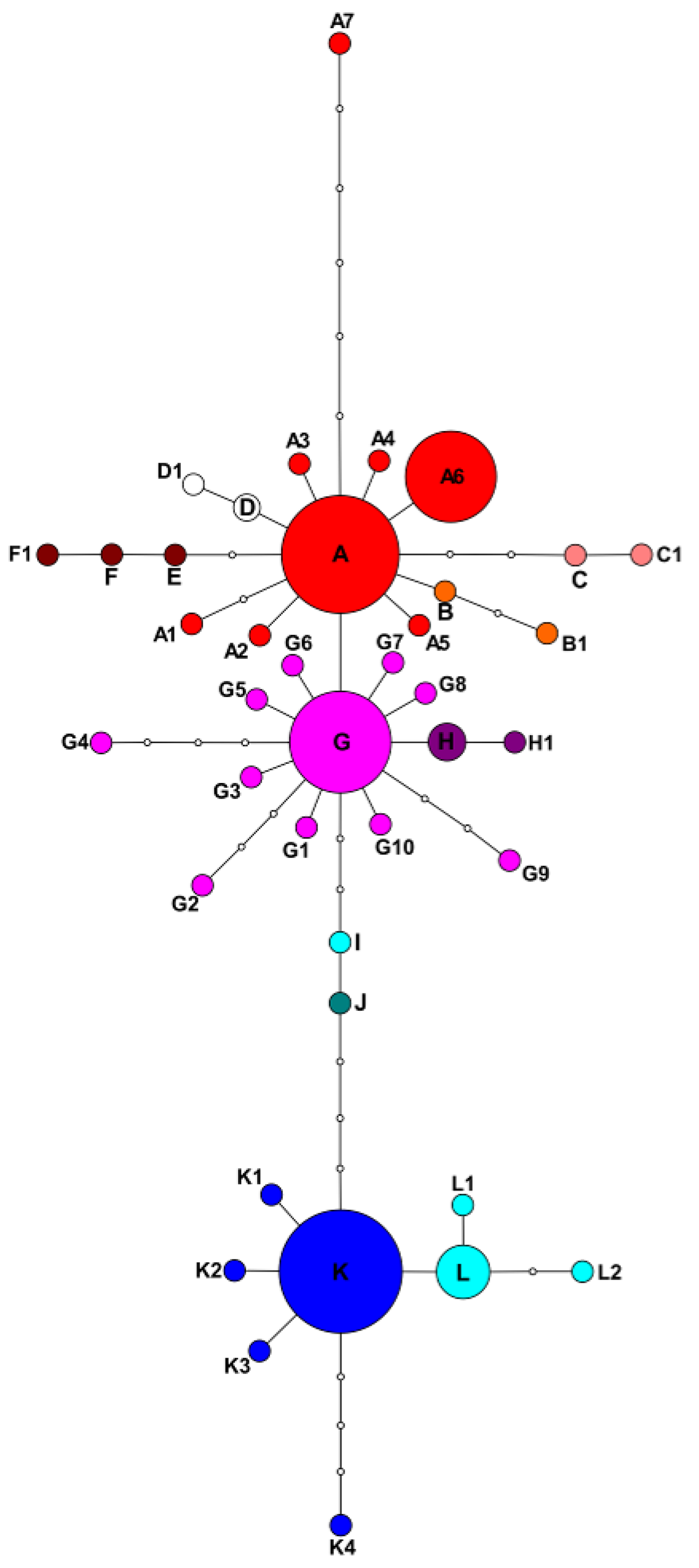
2.1. Plastid Data Analyses
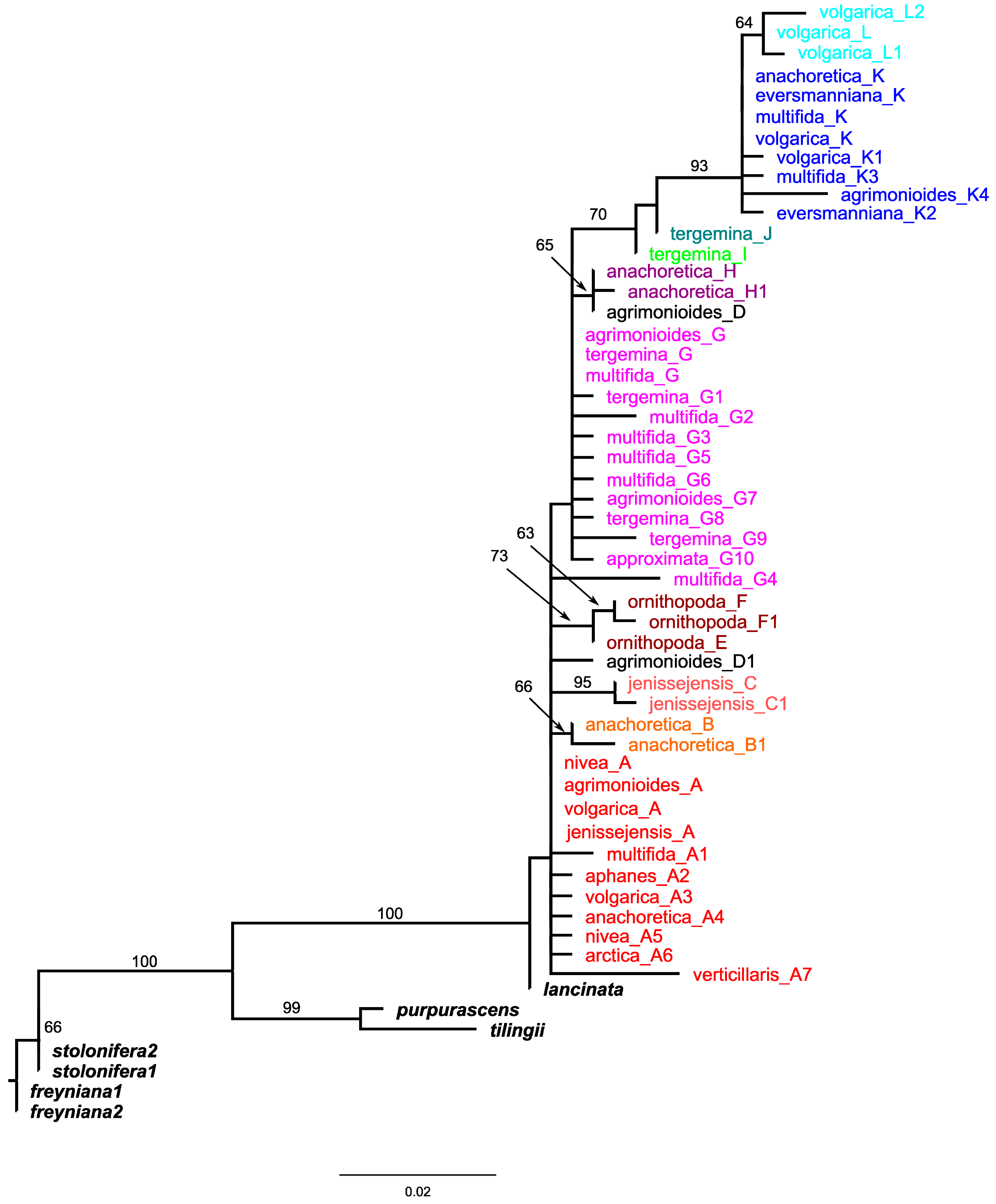
2.2. Nuclear ITS Data Analyses
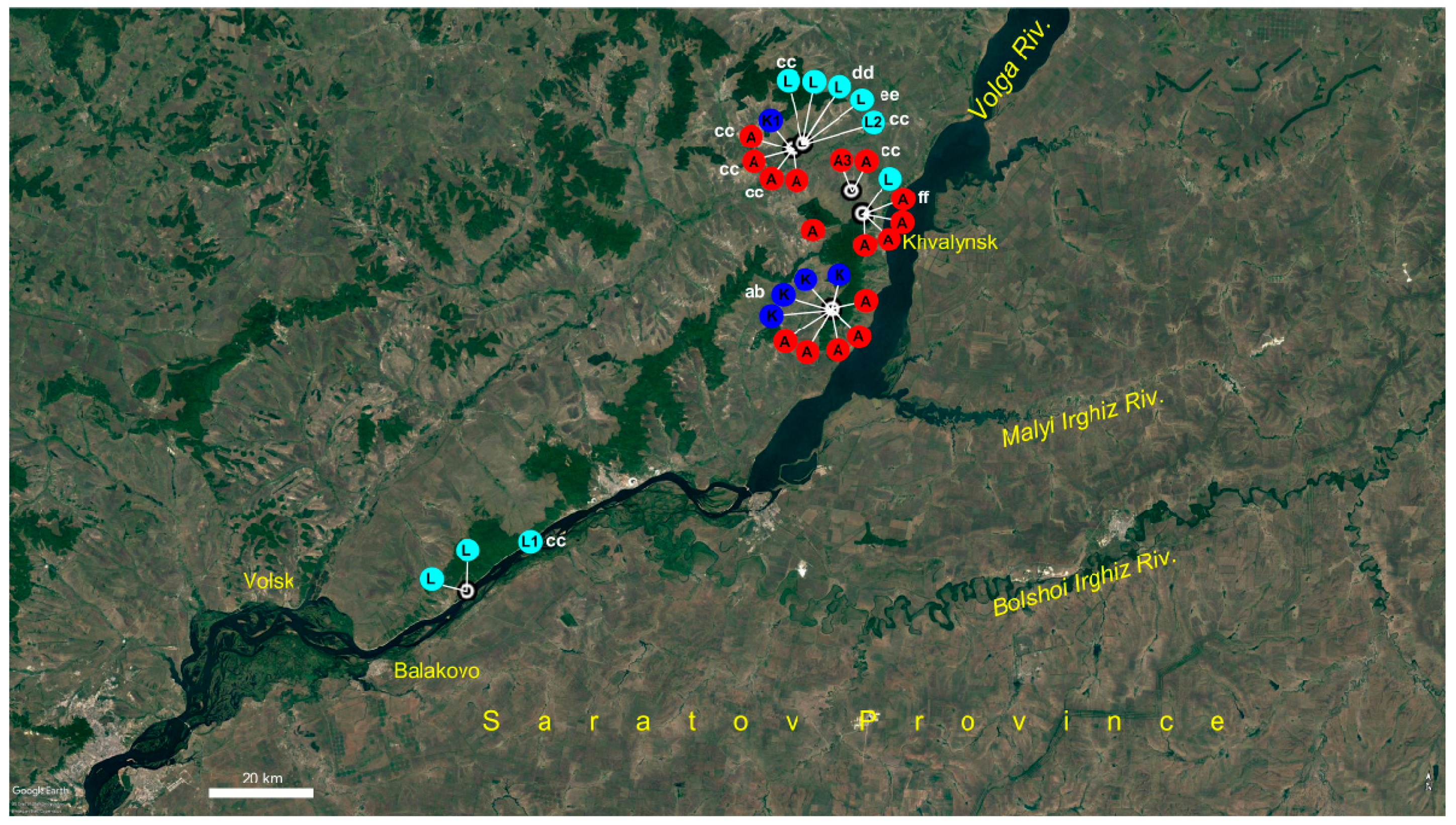
2.3. Potentilla volgarica and P. eversmanniana Population Structure Analyses
3. Discussion
4. Materials and Methods
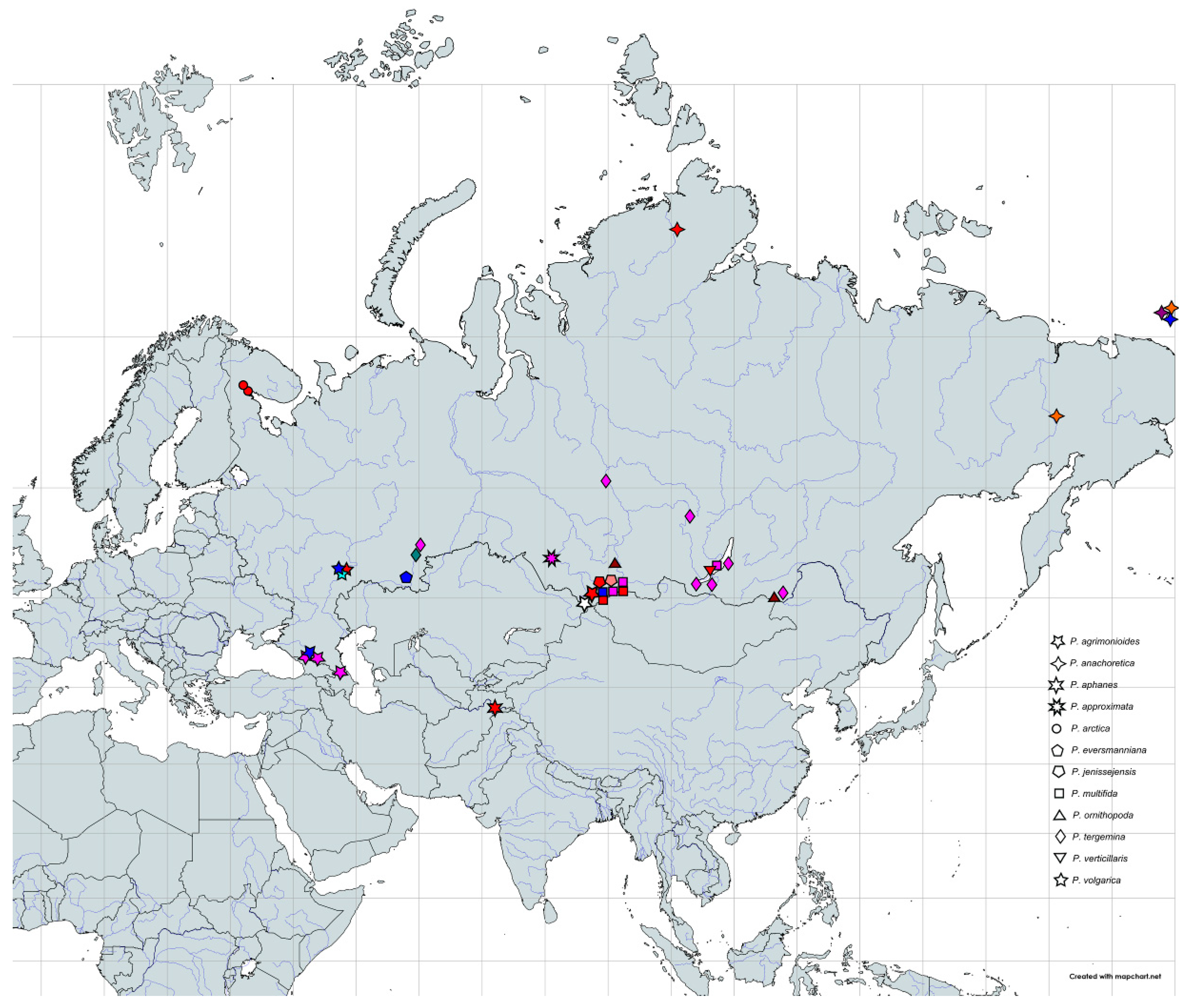
4.1. Taxon Sampling
4.2. Molecular Methods
4.3. Sequence Editing and Alignment
4.4. Data Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Samples and Accessions | Specimen Voucher | Plastid Haplotype | ndhC-trnV | psbA- trnH | ITS |
|---|---|---|---|---|---|
| P. agrimonioides (AGR1) | [Russia] Karachay-Cherkessia, W spurs of Mt. Elbrus, right slope of the Biytik-Tyubyu Riv. basin, ca. 5 km upstream the Kyukyurtlyu Riv. mouth, meadow-steppe slope, 4.08.2009, A.S. Zernov No. 7269, 43.411297° N, 42.344777° E (MW) | G | MW046910 | MW046965 | MW042130 |
| P. agrimonioides (AGR10) | [Russia] Altai Republic, Kosh-Agachsky distr., Saylyugem ridge, the Chagan-Burgazy Riv. basin, middle part of the left slope of the Bayan-Chagan Riv. valley ca. 5 km upstream its flow into the Karasu Riv. 2690 m alt., 11.08.1999, D.A. Petelin, N.V. Fedotkina No. 99-462 (specimen 2), 49.516667° N, 88.766667° E (MW) | D | MW046906 | MW046961 | MW042127 |
| P. agrimonioides (AGR11) | [Russia] Altai Republic, Kosh-Agachsky distr., Saylyugem ridge, the Chagan-Burgazy Riv. basin, middle part of the left slope of the Bayan-Chagan Riv. valley ca. 5 km upstream its flow into the Karasu Riv. 2690 m alt., 11.08.1999, D.A. Petelin, N.V. Fedotkina No. 99-462 (specimen 3), 49.516667° N, 88.766667° E (MW) | D1 | MW046907 | MW046962 | – |
| P. agrimonioides (AGR12) | [Russia] Altai Republic, Kosh-Agachsky distr., Chikhachev ridge, the Bar-Burgazy Riv. valley, left shingle bank of the river, alt. 2075 m, 05.07.1982, A. Maneev, A. Krasnikov, 49.916667° N, 89.416667° E (MW) | D | MW046908 | MW046963 | – |
| P. agrimonioides (AGR13) | [Russia] Altai Republic, Kosh-Agachsky distr., Chikhachev ridge, the Tekelyu Riv. valley (left tributary of the Bguzun Riv.), rocks of the left bank, alt. 2300 m, 27.07.1981, A. Maneyev, A. Krasnikov (left sample), 50.083333° N, 89.416667° E (MW) | A | MW046909 | MW046964 | MW042129 |
| P. agrimonioides (AGR2) | [Russia] Karachay-Cherkessia, Karachaevsky distr., vicinity of Verkhnyaya Mara vill., Mount Kyokle-Bashi, S spurs, alt. ca. 1800 m, meadow-steppe slope near summit, 13.07.2009, A.S. Zernov, S.A. Senator No. 7103, 43.775° N, 42.117778° E (MW) | K4 | MW046911 | MW046966 | MW042131 |
| P. agrimonioides (AGR3) | [Russia] Stavropol Territory, Elbrus distr., village Uchkulan, upstream of the Kuban Riv., mountain steppe, 17. 07.1967, T.A. Lezhankina, 43.449018° N, 42.088726° E (MW) | G | MW046912 | MW046967 | MW042132 |
| P. agrimonioides (AGR4) | [Russia] North Ossetian ASSR, the Ardon Riv. basin, left slope of Alagir gorge above Nuzal vill., dry stony slopes, alt. 1100 m, 18.05.1977, A.M. Amirkhanov 42.824758° N, 44.019145° E (MW) | G | MW046913 | MW046968 | MW042133 |
| P. agrimonioides (AGR5) | [Russia] North Ossetian ASSR, the Ardon Riv. basin, the Main Dividing Ridge system, W slope of Tseyskiy Ridge above Buron vill., left slope of Alagir gorge, scree SE slopes, alt. 1200 m, 23.05.1977, A.M. Amirkhanov 42.794852° N, 44.02317° E (MW) | G | MW046914 | MW046969 | MW042134 |
| P. agrimonioides (AGR6) | [Russia] North Ossetian ASSR, the Ardon Riv. basin, the Main Dividing Ridge system, W slope of Tseyskiy Ridge above Buron vill., left slope of Alagir gorge, scree SE slopes, alt. 1250 m, 23.05.1977, A.M. Amirkhanov 42.794852° N, 44.002317° E (MW) (left specimen) | G | MW046915 | MW046970 | MW042135 |
| P. agrimonioides (AGR7) | [Russia] North Ossetian ASSR, the Ardon Riv. basin, the Main Dividing Ridge system, W slope of Tseyskiy Ridge above Buron vill., left slope of Alagir gorge, scree SE slopes, alt. 1250 m, 23.05.1977, A.M. Amirkhanov 42.794852° N, 44.002317° E (MW) (right specimen) | G7 | MW046916 | MW046971 | – |
| P. agrimonioides (AGR9) | [Russia] Dagestan Republic, the Mularchay Riv., shoals of the left river bank, alt. 2740–2500 m, 05.08.1989, A.M. Amirkhanov No. 21, 41.28661° N, 47.741299° E (MW) | G | MW046917 | MW046972 | MW042136 |
| P. anachoretica (ANA10) | [Russia] Chukotka AR, Wrangel Island, middle course of the Mamontovaya Riv. (near inflow of Khrustalny and Vesely streams), meadow on Vesely stream valley slope, 12.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-112 (upper right specimen), 71.175511° N, 179.75655° E (MW) | H | MW046918 | MW046973 | MW042137 |
| P. anachoretica (ANA4) | [Russia] Chukotka AR, Wrangel Island, middle course of the Mamontovaya Riv. (near inflow of Khrustalny and Vesely streams), steppe meadow at Vesely stream mouth, 04.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-109 (upper specimen), 71.175511° N, 179.75655 W (MW) | H1 | MW046919 | MW046974 | – |
| P. anachoretica (ANA5) | [Russia] Chukotka AR, Wrangel Island, middle course of the Mamontovaya Riv. (near inflow of Khrustalny and Vesely streams), steppe meadow at Vesely stream mouth, 04.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-109 (lower specimen), 71.175511° N, 179.75655 W (MW) | H | MW046920 | MW046975 | MW042138 |
| P. anachoretica (ANA6) | [Russia] Chukotka AR, Wrangel Island, upstream of the Neizvestnaya Riv., the northern ‘Bobovaya gorka’ area, tundra, 12.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-049 (upper specimen) 71.169347° N, 179.434647 W (MW) | H | MW046921 | MW046976 | MW042139 |
| P. anachoretica (ANA7) | [Russia] Chukotka AR, Wrangel Island, upstream of the Neizvestnaya Riv., the northern ‘Bobovaya gorka’ area, steppe meadow at rocks, 12.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-111 (upper specimen) 71.178525° N, 179.408767W (MW) | – | – | – | MW042140 |
| P. anachoretica (ANA8) | [Russia] Chukotka AR, Wrangel Island, upstream of the Neizvestnaya Riv., the northern ‘Bobovaya gorka’ area, thinned meadow, 12.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-049 (left specimen) 71.169931° N, 179.435272 W (MW) | B | MW046922 | MW046977 | MW042141 |
| P. anachoretica (ANA9) | [Russia] Chukotka AR, Wrangel Island, middle course of the Mamontovaya Riv. (near inflow of Khrustalny and Vesely streams), steppe meadow at Vesely stream mouth, 06.07.2014, I.N. Pospelov, E.B. Pospelova No. 14-110 71.176561° N, 179.7578 W (MW) | H | MW046923 | MW046978 | MW042142 |
| P. anachoretica (PTA1) | [Russia] Central Taimyr Peninsula, Byrranga Mountains, middle course of the Bolshaya Bootankaga Riv. near Vetvisty stream inflow, lower part of matted schistose slope, tundra, 16.07.1990, E.B. Pospelova No. 90-472v, 74.31025° N, 98.07938° E (MHA) | A4 | MN849358 | MN871334 | MN872925 |
| P. anachoretica (PTA2) | [Russia] Chukotka AR, Wrangel Island, Somnitelnaya Bay, 19.07.1984, B.Yurtsev, 70.979137° N, 179.463733 W (MHA) | K | MN849359 | MN871335 | – |
| P. anachoretica (PTM11) | [Russia] Magadan Prov., Bilibinsky distr., right bank of the Omolon Riv., 18 km downstream of the Kedon Riv. mouth, steppe slope, 05.07.1976, G.L. Antropova, A.P. Khokhryakov, 65.717199N, 159.079131E (MHA) | B1 | MN849377 | MN871353 | – |
| P. aphanes (PTM12) | Tajikistan, Western Pamir, Ishkashim distr., vicinity of Vigost vill., scrub, alt. ca. 3500 m, 12.07.1983, G.M. Proskuryakova, Z.R. Alferova, 36.7° N, 71.6° E (MHA) | A2 | MN849378 | MN871354 | MN872943 |
| P. approximata (PTM6) | [Russia] Novosibirsk Prov., Ordynsky distr., vicinity of Kirza vill., steppe meadow, alt. 200 m, 01.07.1991, I.M. Krasnoborov No. NS56, 54.12° N, 81.40° E (MHA) | G10 | MN849385 | MN871361 | – |
| P. arctica (ARC1) | [Russia] Murmansk Prov., Kandalaksha distr., Tonnye Islands, Oleny archipelago, sea-side meadow on rocks, 22.06.2016, E.V. Kudr, K.A. Savina, 67.11089° N, 32.40611° E (MW) | A6 | MW046926 | MW046981 | MW042145 |
| P. arctica (ARC10) | [Russia] Murmansk Prov., Terskiy distr., White Sea, Kandalaksha Gulf, Porya Bay, Perunok island, southern rocky cape, rocky meadow, 10.07.2011, M.N. Kozhin No. M-1775, 66.77747° N, 33.62591° E (MW) | A6 | MW046924 | MW046979 | MW042143 |
| P. arctica (ARC11) | [Russia] Murmansk Prov., White Sea, Porya Bay, Karbonatnaya Luda island, 15.07.2008, M.N. Kozhin No. M-0623, 66.73993° N, 33.63885° E (MW) | A6 | MW046925 | MW046980 | – |
| P. arctica (ARC2) | [Russia] Murmansk Prov., Kandalaksha distr., Kandalaksha Gulf of the White Sea, Luven’gsky archipelago, Golomyanny Vlasov island, cracked sea-side rocks, 16.08.2017, E.I. Vuzman, 67.07184° N, 32.72459° E (MW) | A6 | MW046927 | MW046982 | MW042146 |
| P. arctica (ARC3) | [Russia] Murmansk Prov., Kandalaksha distr., Tonnye Islands, Oleny archipelago, sea-side meadow on rocks, 22.06.2016, T.S. Taskina, 67.11089° N, 32.40611° E (MW) | A6 | MW046928 | MW046983 | MW042147 |
| P. arctica (ARC4) | [Russia] Karelia, Loukhsky distr., Vysoky Island, S shore, on rocks, 4.07.2003, L.A. Abramova, P.A. Volkova, K.A. Astafjev, 66.574624° N, 32.90934° E (MW) | – | – | – | MW042148 |
| P. arctica (ARC6) | [Russia] Murmansk Prov., Terskiy distr., White Sea, Kandalaksha Gulf, Porya Bay, Chayachya Luda Island, SW part, rock crevices, 4.08.2011, M.N. Kozhin No. M-1858, 66.78037° N, 33.57952° E (MW) | A6 | MW046929 | MW046984 | – |
| P. arctica (ARC7) | [Russia] Murmansk Prov., Terskiy distr., White Sea, Kandalaksha Gulf, Porya Bay, Krachny Baklysh Island, seashore rocks, 14.07.2011, M.N. Kozhin No. M-2075, 66.78828° N, 33.63157° E (MW) | A6 | MW046930 | MW046985 | MW042149 |
| P. arctica (ARC8) | [Russia] Murmansk Prov., Terskiy distr., White Sea, Kandalaksha Gulf, Porya Bay, Medvezhy Island, SE cape, rocks, 21.07.2008, M.N. Kozhin No. M-0601, 66,7° N, 33,6° E (MW) | A6 | MW046931 | MW046986 | MW042150 |
| P. arctica (ARC9) | [Russia] Murmansk Prov., Terskiy distr., White Sea, Kandalaksha Gulf, Porya Bay, Chayachya Luda Island, SW part, rock crevices, 4.08.2011, M.N. Kozhin No. M-1858 (2nd sheet), 66.78037° N, 33.57952° E (MW) | A6 | MW046932 | MW046987 | MW042151 |
| P. arctica (PTL1) | [Russia] Murmansk Prov., Kandalaksha distr., 2.5 km NE of Kovda vill., Ovechy Island, seashore, in rock crevices, 30.07.2004, N.M. Reshetnikova, 66.7° N, 39.2° E (MHA) | A6 | MN849376 | MN871352 | MN872940 |
| P. arctica (PTM14) | [Russia] Murmansk Prov., Kandalaksha distr., vicinity of Kovda vill., Sedlovaty Island, seashore rocks, 27.07.2019, E.G. Petrash, 66.690052° N, 33.060953° E (1st sheet) (MHA) | A6 | MN849379 | MN871355 | MN872944 |
| P. arctica (PTM15) | [Russia] Murmansk Prov., Kandalaksha distr., vicinity of Kovda vill., Sedlovaty Island, seashore rocks, 27.07.2019, E.G. Petrash, 66.690052° N, 33.060953° E (2nd sheet) (MHA) | A6 | MN849380 | MN871356 | – |
| P. arctica (PTM16) | [Russia] Murmansk Prov., Kandalaksha distr., vicinity of Kovda vill., Sedlovaty Island, seashore rocks, 27.07.2019, E.G. Petrash, 66.690052° N, 33.060953° E (3rd sheet) (MHA) | A6 | MN849381 | MN871357 | MN872946 |
| P. arctica (PTM17) | [Russia] Murmansk Prov., Kandalaksha distr., vicinity of Kovda vill., Sedlovaty Island, seashore rocks, 27.07.2019, E.G. Petrash, 66.690052° N, 33.060953° E (4th sheet) (MHA) | A6 | MN849382 | MN871358 | – |
| P. eversmanniana (PTE1) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-13, 51.943102° N, 56.771153° E (MHA) | K | MN849367 | MN871343 | MN872926 |
| P. eversmanniana (PTE10) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-02, 51. 922852° N, 56. 767177° E (MHA) | K | MN849360 | MN871336 | – |
| P. eversmanniana (PTE11) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-03, 51. 922852° N, 56. 767177° E (MHA) | K | MN849361 | MN871337 | – |
| P. eversmanniana (PTE12) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-04, 51. 922852° N, 56. 767177° E (MHA) | K2 | MN849362 | MN871338 | – |
| P. eversmanniana (PTE13) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-05, 51. 922852° N, 56. 767177° E (MHA) | K | MN849363 | MN871339 | – |
| P. eversmanniana (PTE14) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-06, 51. 922852° N, 56. 767177° E (MHA) | K | MN849364 | MN871340 | – |
| P. eversmanniana (PTE15) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-07, 51. 922852° N, 56. 767177° E (MHA) | K | MN849365 | MN871341 | – |
| P. eversmanniana (PTE2) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-14, 51.943102° N, 56.771153° E (MHA) | K | MN849368 | MN871344 | – |
| P. eversmanniana (PTE3) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-15, 51.943102° N, 56.771153° E (MHA) | K | MN849369 | MN871345 | – |
| P. eversmanniana (PTE4) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-16, 51.943102° N, 56.771153° E (MHA) | K | MN849370 | MN871346 | – |
| P. eversmanniana (PTE5) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-17, 51.943102° N, 56.771153° E (MHA) | K | MN849371 | MN871347 | MN872930 |
| P. eversmanniana (PTE6) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-18, 51.943102° N, 56.771153° E (MHA) | K | MN849372 | MN871348 | MN872931 |
| P. eversmanniana (PTE7) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-26, 51.943102° N, 56.771153° E (MHA) | K | MN849373 | MN871349 | – |
| P. eversmanniana (PTE8) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., Bashkirskaya Chumaza vill., ca. 500 m downstream the Bolshaya Samaza Rivulet, flat place on a hill above the village, overgrazed feather-grass steppe, 26.05.2019, I.A. Schanzer, A.V. Fedorova No. SH260519-27, 51.943102° N, 56.771153° E (MHA) | K | MN849374 | MN871350 | – |
| P. eversmanniana (PTE9) | [Russia] Rep. of Bashkortostan, Zianchurinskii distr., ca. 2 km S of Bashkirskaya Chumaza vill., a small valley between hills, stony slopes at a hill summit, 27.05.2019, I.A. Schanzer, A.V. Fedorova No. SH270519-01, 51. 922852° N, 56. 767177° E (MHA) | K | MN849375 | MN871351 | – |
| P. jenissejensis (MUC1) | [Russia] Altai, Kosh-Agachsky distr., Chikhachev ridge, the Bar-Burgazy Riv. valley, thinned grassy slope in the flood plain, alt. 2100 m, 5.07.1982, A. Maneyev, A. Krasnikov, 49.916667° N, 89.416667° E (1st sheet) (MW) | C | MW046936 | MW046991 | MW042156 |
| P. jenissejensis (MUC2) | [Russia] Tuvinskaya ASSR, Mongun-Taiginsky distr., Chikhachev ridge, the Ustu-Gimateh Riv. valley, talus, 11.08.1980, A. Krasnikov, A. Maneyev, N. Sidorenko (right specimen), 50.453164° N, 89.991742° E (MW) | A | MW046937 | MW046992 | MW042157 |
| P. jenissejensis (MUC4) | [Russia] Altai, Kosh-Agachsky distr., Chikhachev ridge, the Bar-Burgazy Riv. valley, thinned grassy slope in the flood plain, alt. 2100 m, 5.07.1982, A. Maneyev, A. Krasnikov, 49.916667° N, 89.416667° E (2nd sheet) (MW) | C1 | MW046939 | MW046994 | MW042158 |
| P. multifida (MUF1) | [Russia] Tuvinskaya ASSR, Mongun-Taiginsky distr., Chikhachev ridge, the Alty-Gimateh Riv. valley 2 km upstream the mouth, meadow, 04.08.1980, A. Krasnikov, A. Maneyev, V. Shein, 50.453164° N, 89.991742° E (MW) | A1 | MW046944 | MW046999 | MW042162 |
| P. multifida (MUF13) | [Russia] Irkutsk Prov., [Lake Baikal], Olkhon Island, 2 km° E of Kharantsy vill., shore of the ‘Minor Sea’, 09.07.1979, students, 53.238731° N, 107.459352° E (MW) | G4 | MW046941 | MW046996 | – |
| P. multifida (MUF2) | [Russia] Krasnoyarsky Territory, W Sayan Mountains, Sayano-Shushensky Nature Reserve, left bank of the B. Ura Riv., meadow steppe, alt. 1200 m, 12.08.1988, V. Kuvaev No. 1214-18 (right specimen), 51.990188° N, 91.842219° E (MW) | G | MW046945 | MW047000 | MW042163 |
| P. multifida (MUF4) | [Russia] Tuvinskaya ASSR, Mongun-Taiginsky distr., Chikhachev ridge, the Alty-Gimateh Riv. valley 2 km upstream the mouth, pebble bank, 01.08.1980, A. Krasnikov, A. Maneyev, 50.453164° N, 89.991742° E (MW) | G | MW046946 | MW047001 | MW042164 |
| P. multifida (MUF5) | [Russia] Altai, Kosh-Agachsky distr., Chikhachev ridge, the Baylugem Riv. valley, alt. 2270 m, 21.06.1982, A. Maneyev, A. Krasnikov (upper specimen) 50.25° N, 89.33° E (MW) | G | MW046947 | MW047002 | – |
| P. multifida (MUF6) | [Russia] Altai, Kosh-Agachsky distr., Chikhachev ridge, the Baylugem Riv. valley, alt. 2270 m, 21.06.1982, A. Maneyev, A. Krasnikov (middle specimen) 50.25° N, 89.33° E (MW) | G3 | MW046948 | MW047003 | MW042165 |
| P. multifida (MUF7) | [Russia] Altai, Kosh-Agachsky distr., Chikhachev ridge, the Baylugem Riv. valley, alt. 2270 m, 21.06.1982, A. Maneyev, A. Krasnikov (lower specimen) 50.25° N, 89.33° E (MW) | G2 | MW046949 | MW047004 | – |
| P. multifida (PTM3) | [Russia] Altai, Kosh-Agachsky distr., the Tarhata Riv. valley, meadow on river bank, alt. 2150 m, 08.07.1982, M. Lomonosova, A. Vanyaev, 49.45° N, 88.30° E (MHA) | K | MN849383 | MN871359 | – |
| P. multifida (PTM4) | [Russia] Altai Mountains, vicinity of Kosh-Agach, a quarry in Leymus-steppe, alt. 1770 m, 20.07.1982, I. Krasnoborov, A. Krasnikov No. 1309, 50.00° N, 88.40° E (MHA) | K3 | MN849384 | MN871360 | – |
| P. nivea (NIV1) | [Russia] Karachay-Cherkessia, Teberdinsky State Reserve, N spurs of Malaya Khatipara Mt., stony slope, 12.08.2006, A.S. Zernov No. 5524, 43.438549° N, 41.681227° E (MW) | A | MW046952 | MW047007 | MW042167 |
| P. nivea (NIV2) | [Russia] Karachay-Cherkessia, Karachaevsky distr., S slope of Sadyrla range, alt. ca. 2900, talus, 9.08.2011, A.S. Zernov, A.N. Filin No. 7619, 43.434741° N, 42.266179° E (MW) | A5 | MW046951 | MW047006 | MW042166 |
| P. ornithopoda (ORN1) | [Russia] Khakassia Rep., Altaiskyi distr., less than 1 km S of Izykhskiye Kopi vill., alt. ca. 322 m., shallow gully, on wet salty loamy soil, I. Schanzer, N. Stepanova No. 11IK75, 53.539° N, 91.28675° E (MHA) | F | MW046953 | MW047008 | MW042168 |
| P. ornithopoda (ORN2) | [Russia] Khakassia Rep., Altaiskyi distr., less than 1 km S of Izykhskiye Kopi vill., alt. ca. 322 m., shallow gully, on wet salty loamy soil, I. Schanzer, N. Stepanova No. 11IK83, 53.539° N, 91.28675° E (MHA) | E | MW046954 | MW047009 | MW042169 |
| P. ornithopoda (ORN5) | [Russia] Chitinskaya Prov., Ononsky distr., near lake Zun-Torey, 24.07.1977, T. Sofeykova et al., 50,023955° N, 115,907498° E (MHA) | F1 | MW046956 | MW047011 | MW042170 |
| P. tergemina (EWE1) | [Russia] Sverdlovskaya Prov., Nevyansk distr., vicinity of Murzinka railway station, 4 km south of lake Tavatuy, 10.06.1991, N. Shvedchikova, 57.055421° N, 60.174235° E (1st sheet) (MW) | G | MW046933 | MW046988 | MW042153 |
| P. tergemina (EWE3) | [Russia] Sverdlovskaya Prov., Nevyansk distr., vicinity of Murzinka railway station, 4 km south of lake Tavatuy, 10.06.1991, N. Shvedchikova, 57.055421° N, 60.174235° E (3rd sheet) (MW) | G | MW046934 | MW046989 | MW042154 |
| P. tergemina (EWE4) | [Russia] Southern Urals, between sources of the Ui and Miass Rivers, field road ca. 1.5 km NW of Starobayramovo vill., Uchalinsky distr. of Bashkortostan, 16.07.1993, M.S. Knyazev 54,740913° N, 59,742309° E (MW) | J | MW046935 | MW046990 | MW042155 |
| P. tergemina (MUF11) | [Russia] Trans-Baikal Territory, Aleksandrovo-Zavodsky distr., near Mankovo vill., the Verkhniy Gazimur Riv. flood plain, steppe meadow, alt. 885 m, 22.07.2011, N.S. Gamova, S.V. Dudov No. 11_0010, 50.8184° N, 117.6888° E (MW) | G | MW046940 | MW046995 | MW042159 |
| P. tergemina (MUF14) | [Russia] Irkutsk Prov., Ust-Ilimsk distr., vicinity of Ust-Ilimsk, right bank of the Ust-Ilimsk reservoir, 4.5 km° E of the railway station, groundwork edge, 09.08.2007, A. Seregin, A. Khokhlov No. S-303, 57.916667° N, 102.852778° E (MW) | G5 | MW046942 | MW046997 | MW042160 |
| P. tergemina (MUF15) | [Russia] Irkutsk Prov., Slyudyansky distr., 23 km to° E from Baikalsk, S shore of Lake Baikal, at the Pankovka Riv. mouth, concrete plates at the railway basement, alt. 240 m, 29.07.2007, A. Seregin No. S-15 51.461111° N, 104.491667° E (MW) | G6 | MW046943 | MW046998 | MW042161 |
| P. tergemina (MUF9) | [Russia] Republic of Buryatia, Barguzinsky distr., near Ust-Barguzin vill., right slope of the Barguzin Riv. valley, abandoned field, 24.05.2011, S.V. Dudov No. 2011-Bur-022, 53.4431° N, 109.0754° E (MW) | G | MW046950 | MW047005 | – |
| P. tergemina (PTM8) | [Russia, Krasnoyarsky Territory, Turukhansky distr.] Left bank of the Yenisei Riv., Zotino vill., meadow, 15.07.2001, V. Kuvaev, M. Skrebtsova, A. Sidorov No. 2887-13, 60.901355° N 89.680666° E (MHA) | G | MN849386 | MN871362 | MN872942 |
| P. tergemina (PTM9) | [Russia] Udmurtskaya ASSR, Glazov-Tuktym section of the railroad, 19.06.1983, A.N. Puzyrev, 58.097 ° N, 52.863° E (MHA) | G | MN849387 | MN871363 | – |
| P. tergemina (PTT1) | [Russia] Buryatia, vicinity of Gusinoozersk, steppe, 04.08.1993, N. Shevyriova, T. Konovalova, 51.17 ° N, 106.32° E (MHA) | G1 | MN849388 | MN871364 | – |
| P. tergemina (PTT2) | [Ukraine] Kiev, Kiev-Tovarnaya railway station, on gravel along tracks, 28.04.1990, S.L. Mosyakin, 50.428762° N, 30.506669° E (MHA) | I | MN849389 | MN871365 | MN872948 |
| P. tergemina (TER1) | [Russia] Tverskaya Prov., Bologovsky distr., vicinity of Kuzhenkino railway station, siding tracks, 10.07.2004, A. Notov, N. Markelova, 57.727525° N, 33.975539° E (MW) | G8 | MW046957 | MW047012 | – |
| P. tergemina (TER3) | [Russia] Moscow, near Kanatchikovo railway station of the Moscow Circle Railway, on groundwork, 24.06.1997. S.R. Mayorov, D.D. Sokolov, 55.70139° N, 37.591641° E (MW) | G | MW046958 | MW047013 | – |
| P. tergemina (TER5) | [Russia] Moscow, sandy areas of the embankment of the Leningrad railway between stations NATI and Mosselmash, 14.06.1980, M.S. Ignatov, 55.855475° N, 37.543115° E (MW) | G9 | MW046959 | MW047014 | MW042171 |
| P. verticillaris (VER1) | [Russia] Irkutskaya Prov., W coast of Lake Baikal, S end of Olkhon Island, steppe, 24.08.1993, N. Shevyreva, T. Konovalova, 53,05° N, 106,966667° E (MHA) | A7 | MW046960 | MW047015 | – |
| P. volgarica (PTE16) | [Russia] Saratovskaya Prov., Khvalynsky distr., Sosnovaya Maza, 14.07.1993, A.K. Skvortsov, 52.505911° N, 47.903532° E (MHA) | A | MN849366 | MN871342 | – |
| P. volgarica (PTV1) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-18, 20.05.2019, 52,372381° N, 47,967634° E (MHA) | K | MN849400 | MN871376 | MN872949 |
| P. volgarica (PTV10) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda Piche-Panda upland, at the border of feather-grass lined steppe at the hill summit and an upper gentle part of the S slope, I. Schanzer et al. No. SH210519-17, 21.05.2019, 52.654234° N, 47.861662° E (MHA) | L2 | MN849390 | MN871366 | MN872965 |
| P. volgarica (PTV11) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda Piche-Panda upland, at the border of feather-grass lined steppe at the hill summit and an upper gentle part of the S slope, I. Schanzer et al. No. SH210519-18, 21.05.2019, 52.654234° N, 47.861662° E (MHA) | L | MN849391 | MN871367 | – |
| P. volgarica (PTV12) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda Piche-Panda upland, at the border of feather-grass lined steppe at the hill summit and an upper gentle part of the S slope, I. Schanzer et al. No. SH210519-19, 21.05.2019, 52.654234° N, 47.861662° E (MHA) | L | MN849392 | MN871368 | MN872951 |
| P. volgarica (PTV13) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda Piche-Panda upland, at the border of feather-grass lined steppe at the hill summit and an upper gentle part of the S slope, I. Schanzer et al. No. SH210519-20, 21.05.2019, 52.654234° N, 47.861662° E (MHA) | L | MN849393 | MN871369 | MN872952 |
| P. volgarica (PTV14) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda Piche-Panda upland, at the border of feather-grass lined steppe at the hill summit and an upper gentle part of the S slope, I. Schanzer et al. No. SH210519-21, 21.05.2019, 52.654234° N, 47.861662° E (MHA) | L | MN849394 | MN871370 | MN872953 |
| P. volgarica (PTV15) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda upland, hills near Yeriomkino vill., low deforestated chalky hill, 21.05.2019, I. Schanzer, et al. No. SH210519-28, 52.645109° N, 47.834526° E | K1 | MN849395 | MN871371 | MN872954 |
| P. volgarica (PTV16) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda upland, hills near Yeriomkino vill., low deforestated chalky hill, 21.05.2019, I. Schanzer, et al. No. SH210519-29, 52.645109° N, 47.834526° E | A | MN849396 | MN871372 | – |
| P. volgarica (PTV17) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda upland, hills near Yeriomkino vill., low deforestated chalky hill, 21.05.2019, I. Schanzer, et al. No. SH210519-29, 52.645109° N, 47.834526° E | A | MN849397 | MN871373 | MN872956 |
| P. volgarica (PTV18) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda upland, hills near Yeriomkino vill., low deforestated chalky hill, 21.05.2019, I. Schanzer, et al. No. SH210519-30, 52.645109° N, 47.834526° E | A | MN849398 | MN871374 | MN872957 |
| P. volgarica (PTV19) | [Russia] Saratovskaya Prov., Khvalynsky distr., Piche-Panda upland, hills near Yeriomkino vill., low deforestated chalky hill, 21.05.2019, I. Schanzer, et al. No. SH210519-31, 52.645109° N, 47.834526° E | A | MN849399 | MN871375 | – |
| P. volgarica (PTV2) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-19, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | A | MN849411 | MN871387 | – |
| P. volgarica (PTV20) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-18, 52.538878° N, 48.040852° E (MHA) | A | MN849401 | MN871377 | – |
| P. volgarica (PTV21) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-19, 52.538878° N, 48.040852° E (MHA) | A | MN849402 | MN871378 | MN872959 |
| P. volgarica (PTV22) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-20, 52.538878° N, 48.040852° E (MHA) | L | MN849403 | MN871379 | – |
| P. volgarica (PTV23) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-21, 52.538878° N, 48.040852° E (MHA) | A | MN849404 | MN871380 | – |
| P. volgarica (PTV24) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-22, 52.538878° N, 48.040852° E (MHA) | A | MN849405 | MN871381 | – |
| P. volgarica (PTV25) | [Russia] Saratovskaya Prov., Khvalynsky distr., 4.5 km NW of Khvalynsk, chalk hills near a quarry N of the Balakovo-Syzran highway. 22.05.2019 I. Schanzer et al. No. SH-220519-23, 52.538878° N, 48.040852° E (MHA) | A | MN849406 | MN871382 | – |
| P. volgarica (PTV26) | [Russia] Saratovskaya Prov., Khvalynsky distr., chalk hills near a road to Popovka, 16.05.1983, L.H., 52.576915° N, 48.006993° E (1st specimen) (SARBG 1498) | A3 | MN849407 | MN871383 | – |
| P. volgarica (PTV27) | [Russia] Saratovskaya Prov., Khvalynsky distr., chalk hills near a road to Popovka, 16.05.1983, L.H., 52.576915° N, 48.006993° E (2nd specimen) (SARBG 1498) | A | MN849408 | MN871384 | MN872962 |
| P. volgarica (PTV28) | [Russia] Saratovskaya Prov., Khvalynsky distr., 3–4 km N of Voskresensk, chalk outcrops of the bank of the Volga Riv., 25.06.1988, E.A. Kireev, 51.852827° N, 46.991139° E (1st specimen) (SARBG 1499) | L | MN849409 | MN871385 | – |
| P. volgarica (PTV29) | [Russia] Saratovskaya Prov., Khvalynsky distr., 3–4 km N of Voskresensk, chalk outcrops of the bank of the Volga Riv., 25.06.1988, E.A. Kireev, 51.852827° N, 46.991139° E (2nd specimen) (SARBG 1499) | L | MN849410 | MN871386 | – |
| P. volgarica (PTV3) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-20, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | K | MN849413 | MN871389 | – |
| P. volgarica (PTV30) | [Russia] Saratovskaya Prov., Volsky distr., 5 km S of Rybnoye vill., 27.06.1988, E.A. Kireev, 51.944471° N, 47.158729° E (SARBG 1500) | L1 | MN849412 | MN871388 | MN872964 |
| P. volgarica (PTV4) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-21, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | K | MN849414 | MN871390 | – |
| P. volgarica (PTV5) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-22, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | K | MN849415 | MN871391 | – |
| P. volgarica (PTV6) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-23, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | A | MN849416 | MN871392 | – |
| P. volgarica (PTV7) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-29, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | A | MN849417 | MN871393 | – |
| P. volgarica (PTV8) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-30, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | A | MN849418 | MN871394 | – |
| P. volgarica (PTV9) | [Russia] Saratovskaya Prov., Khvalynsky distr., near Novaya Yablonka vill., chalk hill slope over the Syzran-Saratov highway, I. Schanzer et al. No. SH200519-31, 20.05.2019, 52.372381° N, 47.967634° E (MHA) | A | MN849419 | MN871395 | – |
References
- Wolf, T. Monographie der Gattung. Potentilla. Bibl. Bot. 1908, 71, 1–715. [Google Scholar]
- Dobeš, C.; Paule, J. A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): Implications for its geographic origin, phylogeography and generic circumscription Mol. Phylogenet. Evol. 2010, 56, 156–175. [Google Scholar] [CrossRef] [PubMed]
- Persson, N.L.; Toresen, I.; Andersen, H.L.; Smedmark, J.E.E.; Eriksson, T. Detecting destabilizing species in the phylogenetic backbone of Potentilla (Rosaceae) using low-copy nuclear markers. AoB Plants 2020, 12, plaa017. [Google Scholar] [CrossRef] [PubMed]
- Müntzing, A.; Müntzing, G. Some new results concerning apomixis, sexuality and polymorphism in Potentilla Bot. Not. 1941, 94, 237–278. [Google Scholar]
- Asker, S. Apomixis and sexuality in the Potentilla argentea complex. Hereditas 1970, 66, 127–144. [Google Scholar] [CrossRef]
- Goswami, A.D.; Matfield, B. Pseudogamy in the genus Potentilla L. New Phytol. 1974, 73, 1243–1247. [Google Scholar] [CrossRef]
- Kamelin, R.V. Potentilla L. Flora Europae Orientalis; Tzvelev, N.N., Ed.; Mir i Semia: Petropoli, Russia, 2001; Volume 10, pp. 394–452. [Google Scholar]
- Soják, J. Copies of seven species and twenty hybrids of Potentilla (Rosaceae) obtained through experimental hybridization. Notes on Potentilla XXVI. Thaiszia–J. Bot. 2012, 22, 33–48. [Google Scholar]
- Töpel, M.; Lundberg, M.; Eriksson, T.; Eriksen, B. Molecular data and ploidal levels indicate several putative allopolyploidization events in the genus Potentilla (Rosaceae). PLoS Curr. Tree Life 2011, 18, 1. [Google Scholar] [CrossRef]
- Nardi, F.D.; Dobeš, C.; Müller, D.; Grasegger, T.; Myllynen, T.; Alonso-Marcos, H.; Tribsch, A. Sexual intraspecific recombination but not de novo origin governs the genesis of new apomictic genotypes in Potentilla puberula (Rosaceae). Taxon 2018, 67, 1108–1131. [Google Scholar] [CrossRef]
- Eriksson, T.; Donoghue, M.J.; Hibbs, M.S. Phylogenetic analysis of Potentilla using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS), and implications for the classification of Rosoideae (Rosaceae). Plant Syst. Evol. 1998, 211, 155–179. [Google Scholar] [CrossRef]
- Eriksson, T.; Hibbs, M.S.; Yoder, A.D.; Delwiche, C.F.; Donoghue, M.J. The phylogeny of Rosoideae (Rosaceae) based on sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA and the trnL/F region of chloroplast DNA. Int. J. Plant Sci. 2003, 164, 197–211. [Google Scholar] [CrossRef]
- Paule, J. Evolutionary Patterns and Processes in the Genus Potentilla L. (Rosaceae). Doctoral Dissertation, University of Heidelberg, Heidelberg, Germany, 23 July 2010. [Google Scholar]
- Faghir, M.B.; Attar, F.; Farazmand, A.; Kazempour Osaloo, S. Phylogeny of the genus Potentilla (Rosaceae) in Iran based on nrDNA ITS and cpDNA trnL-F sequences with a focus on leaf and style characters’ evolution. Turk. J. Bot. 2014, 38, 417–429. [Google Scholar] [CrossRef]
- Feng, T.; Moore, M.J.; Yan, M.-H.; Sun, Y.-X.; Zhang, H.-J.; Meng, A.-P.; Li, X.-D.; Jian, S.-G.; Li, J.-Q.; Wang, H.-C. Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J. Sytematics Evol. 2017, 55, 177–191. [Google Scholar] [CrossRef]
- Kurbatsky, V. Notes about genus Potentilla L. in the flora of Siberia. Turczaninowia 1999, 2, 10–18. [Google Scholar]
- Li, C.-L.; Ikeda, H.; Ohba, H. Potentilla Linnaeus. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003; Volume 9, pp. 291–327. [Google Scholar]
- Soják, J. Potentilla L. (Rosaceae) and related genera in the former USSR (identification key, checklist and figures). Notes on Potentilla XVI. Bot. Jahrb. Syst. 2004, 125, 253–340. [Google Scholar] [CrossRef]
- Soják, J. Potentilla L. s.l. (Rosaceae) in Flora Europae Orientalis (Notes on Potentilla XVIII). Candollea 2005, 60, 59–78. [Google Scholar]
- Soják, J. Potentilla L. (Rosaceae) in the former USSR; second part: Comments. Notes on Potentilla XXIV. Feddes Reper. 2009, 120, 185–217. [Google Scholar] [CrossRef]
- Kechaykin, A.A.; Shmakov, A.I. A system of subtribe Potentillinae J. Presl (Rosaceae Juss.) Turczaninowia 2016, 19, 114–128. [Google Scholar] [CrossRef]
- Walter, K.S.; Gillett, H.J. (Eds.) 1997 IUCN Red List of Threatened Plants; Compiled by the World Conservation Monitoring Centre; IUCN–The World Conservation Union: Gland, Switzerland; Cambridge, UK, 1998; pp. 494–495. [Google Scholar]
- Kamelin, R.V. Potentilla eversmanniana Fisch. ex Claus; Potentilla volgarica Juz. In Red Data Book of Russian Federation (Plants and, Fungi); Trutnev, Y.P., Gizatulin, R.R., Mitvol, O.L., Amirkhanov, A.M., Kamelin, R.V., Bardunov, L.V., Novikov, V.S., Orlov, V.A., Stepanitsky, V.B., Belanovich, D.M., et al., Eds.; KMK Press: Moscow, Russia, 2008; pp. 495–498. [Google Scholar]
- Paule, J.; Scherbantin, A.; Dobeš, C. Implications of hybridisation and cytotypic differentiation in speciation assessed by AFLP and plastid haplotypes—A case study of Potentilla alpicola La Soie. BMC Evol. Biol. 2012, 12, 132. [Google Scholar] [CrossRef]
- Dobeš, C.; Rossa, J.; Paule, J.; Hülber, K. Morphology, DNA-molecular variation, karyology, ecogeography, and phytosociology suggest allopatric differentiation and species rank for Potentilla rigoana (Rosaceae). Taxon 2013, 62, 733–745. [Google Scholar] [CrossRef]
- Paule, J.; Kolář, F.; Dobeš, C. Arctic-alpine and serpentine differentiation in polyploid Potentilla crantzii. Preslia 2015, 87, 195–215. [Google Scholar]
- Urgamal, M.; Oyuntsetseg, B.; Gundegmaa, V.; Munkh-Erdene, T.; Solongo, K. Additions to the vascular flora of Mongolia–III (Since the “Conspectus of the vascular plants of Mongolia 2014”). Proc. Mong. Acad. Sci. 2016, 56, 32–38. [Google Scholar] [CrossRef]
- IPCN. Index to Plant Chromosome Numbers. In IPCN Database; Goldblatt, P., Johnson, D.E., Eds.; Missouri Botanical Garden: St. Louis, MO, USA, 1979; Available online: www.tropicos.org/project/ipcn (accessed on 30 September 2020).
- Kechaykin, A.A.; Skaptsov, M.V.; Smirnov, S.V.; Kutsev, M.G.; Shmakov, A.I. Study of genome size representatives of the genus Potentilla L. (Rosaceae Juss.). Biol. Bull. Bogdan Chmelnitskiy Melitopol State Pedagog. Univ. 2016, 6, 229–233. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (block mapping and gathering with entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.S.; Blattner, F.R. A plastid genealogy of Hordeum (Poaceae): Long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 2006, 23, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Gurushidze, M.; Fritsch, R.M.; Blattner, F.R. Species-level phylogeny of Allium subgenus Melanocrommyum: Incomplete lineage sorting, hybridization and trnF gene duplication. Taxon 2010, 59, 829–840. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 1993, 134, 959–969. [Google Scholar]
- Stephens, M.; Smith, N.; Donelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Stephens, M.; Donelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003, 73, 1162–1169. [Google Scholar] [CrossRef]
- Zhang, H.J.; Feng, T.; Landis, J.B.; Deng, T.; Zhang, X.; Meng, A.P.; Sun, H.; Wang, H.C.; Sun, Y.X. Molecular phylogeography and ecological niche modeling of Sibbaldia procumbens s.l. (Rosaceae). Front. Genet. 2019, 10, 201. [Google Scholar] [CrossRef]
- Gao, Y.D.; Zhang, Y.; Gao, X.F.; Zhu, Z.M. Pleistocene glaciations, demographic expansion and subsequent isolation promoted morphological heterogeneity: A phylogeographic study of the alpine Rosa sericea complex (Rosaceae). Sci. Rep. 2015, 5, 11698. [Google Scholar] [CrossRef] [PubMed]
- Kurbatsky, V.I. Opredelitel Vidov Roda Potentilla L. (Lapchatka) Aziatskoi Rossii; Izdatelstvo TGU: Tomsk, Russia, 2016; pp. 6–34. [Google Scholar]
- Wen, J.; Zimmer, E. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA. Mol. Phylogenet. Evol. 1996, 6, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2020. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models, Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v.1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 September 2020).
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
| Source of Variation | d.f. | Variance Components | Percentage of Variation | Fixation Indices |
|---|---|---|---|---|
| Potentilla volgarica vs. P. eversmanniana | ||||
| Among groups | 1 | 3.23474 Va | 16.18 | FSC = 0.63879 |
| Among populations within groups | 7 | 10.70696 Vb | 53.55 | FST = 0.69722 |
| Within populations | 37 | 6.05440 Vc | 30.28 | FCT = 0.16177 |
| Potentilla volgarica | ||||
| Among populations | 6 | 13.34836 Va | 58.94 | FST = 0.58942 |
| Within populations | 24 | 9.29815 Vb | 41.06 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schanzer, I.A.; Fedorova, A.V.; Shelepova, O.V.; Suleymanova, G.F. Molecular Phylogeny and Phylogeography of Potentilla multifida L. agg. (Rosaceae) in Northern Eurasia with Special Focus on Two Rare and Critically Endangered Endemic Species, P. volgarica and P. eversmanniana. Plants 2020, 9, 1798. https://doi.org/10.3390/plants9121798
Schanzer IA, Fedorova AV, Shelepova OV, Suleymanova GF. Molecular Phylogeny and Phylogeography of Potentilla multifida L. agg. (Rosaceae) in Northern Eurasia with Special Focus on Two Rare and Critically Endangered Endemic Species, P. volgarica and P. eversmanniana. Plants. 2020; 9(12):1798. https://doi.org/10.3390/plants9121798
Chicago/Turabian StyleSchanzer, Ivan A., Alina V. Fedorova, Olga V. Shelepova, and Guzyaliya F. Suleymanova. 2020. "Molecular Phylogeny and Phylogeography of Potentilla multifida L. agg. (Rosaceae) in Northern Eurasia with Special Focus on Two Rare and Critically Endangered Endemic Species, P. volgarica and P. eversmanniana" Plants 9, no. 12: 1798. https://doi.org/10.3390/plants9121798
APA StyleSchanzer, I. A., Fedorova, A. V., Shelepova, O. V., & Suleymanova, G. F. (2020). Molecular Phylogeny and Phylogeography of Potentilla multifida L. agg. (Rosaceae) in Northern Eurasia with Special Focus on Two Rare and Critically Endangered Endemic Species, P. volgarica and P. eversmanniana. Plants, 9(12), 1798. https://doi.org/10.3390/plants9121798






