Pseudomonas putida Represses JA- and SA-Mediated Defense Pathways in Rice and Promotes an Alternative Defense Mechanism Possibly through ABA Signaling
Abstract
1. Introduction
2. Results
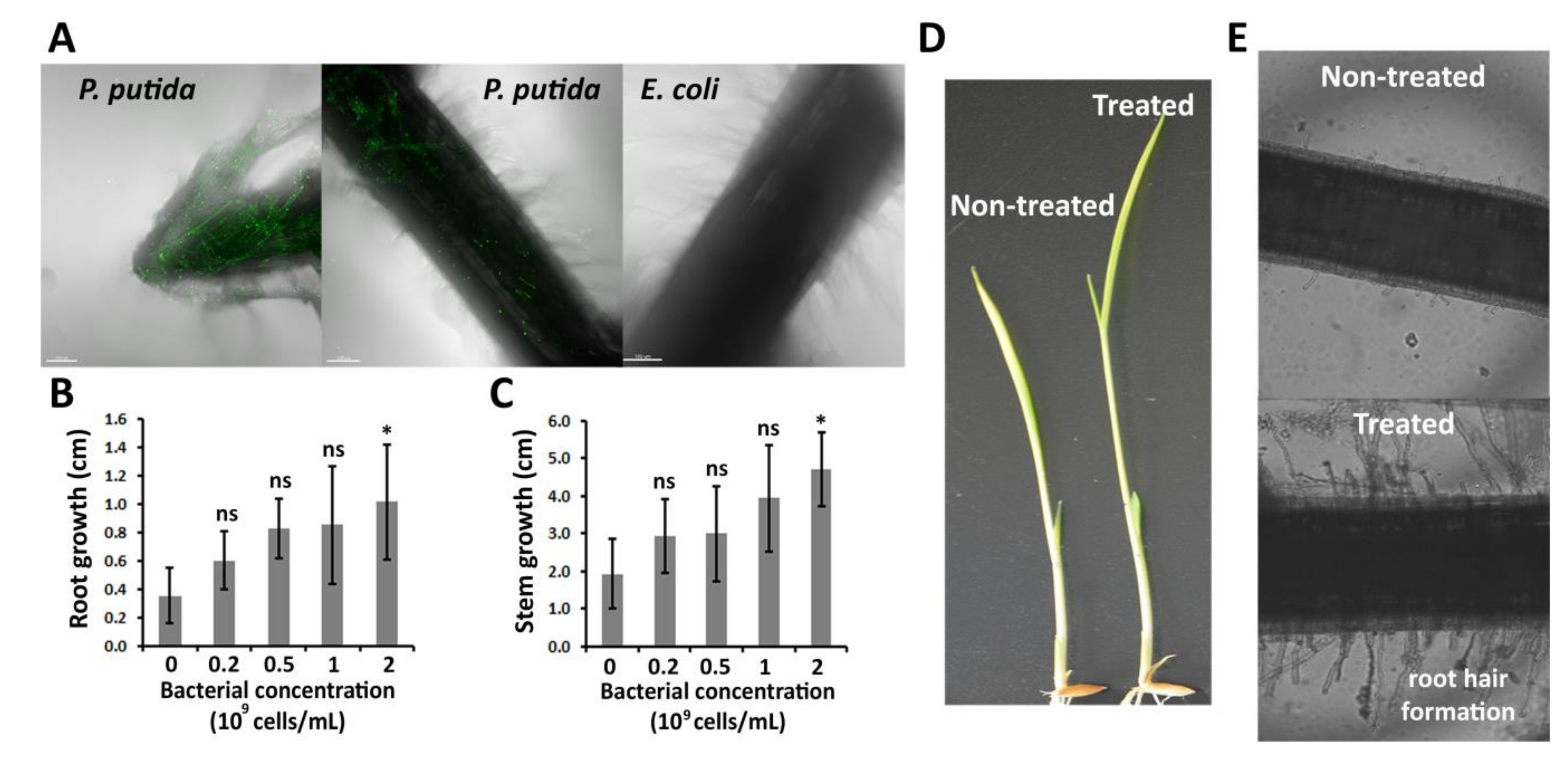
2.1. P. putida Can Colonize Rice Roots, and Promotes Root and Stem Elongation
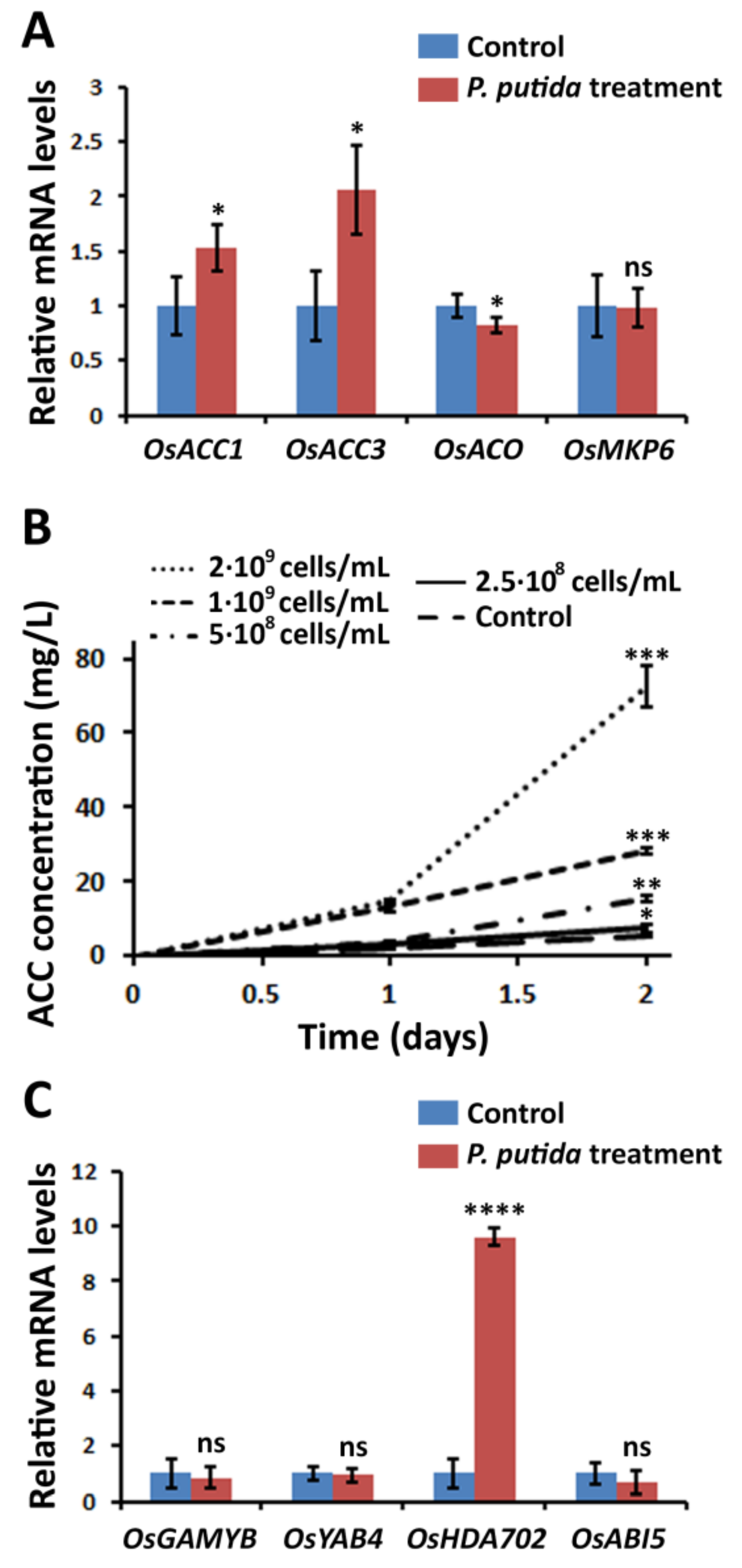
2.2. P. putida Induces ACC Release from Rice Plants into the Medium
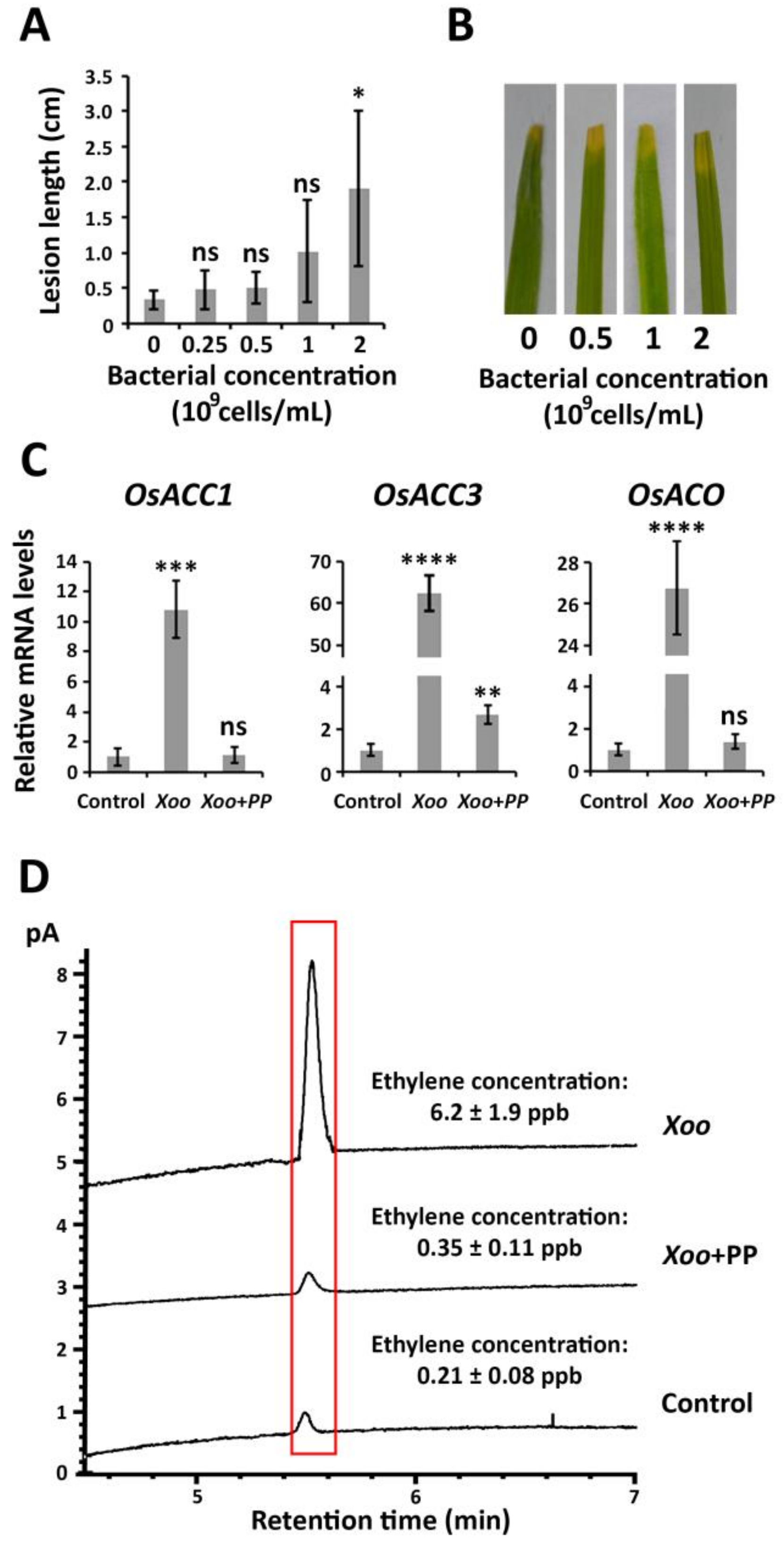
2.3. P. putida Inhibits ET Biosynthesis in Response to X. oryzae pv. oryzae Causing Bacterial Blight
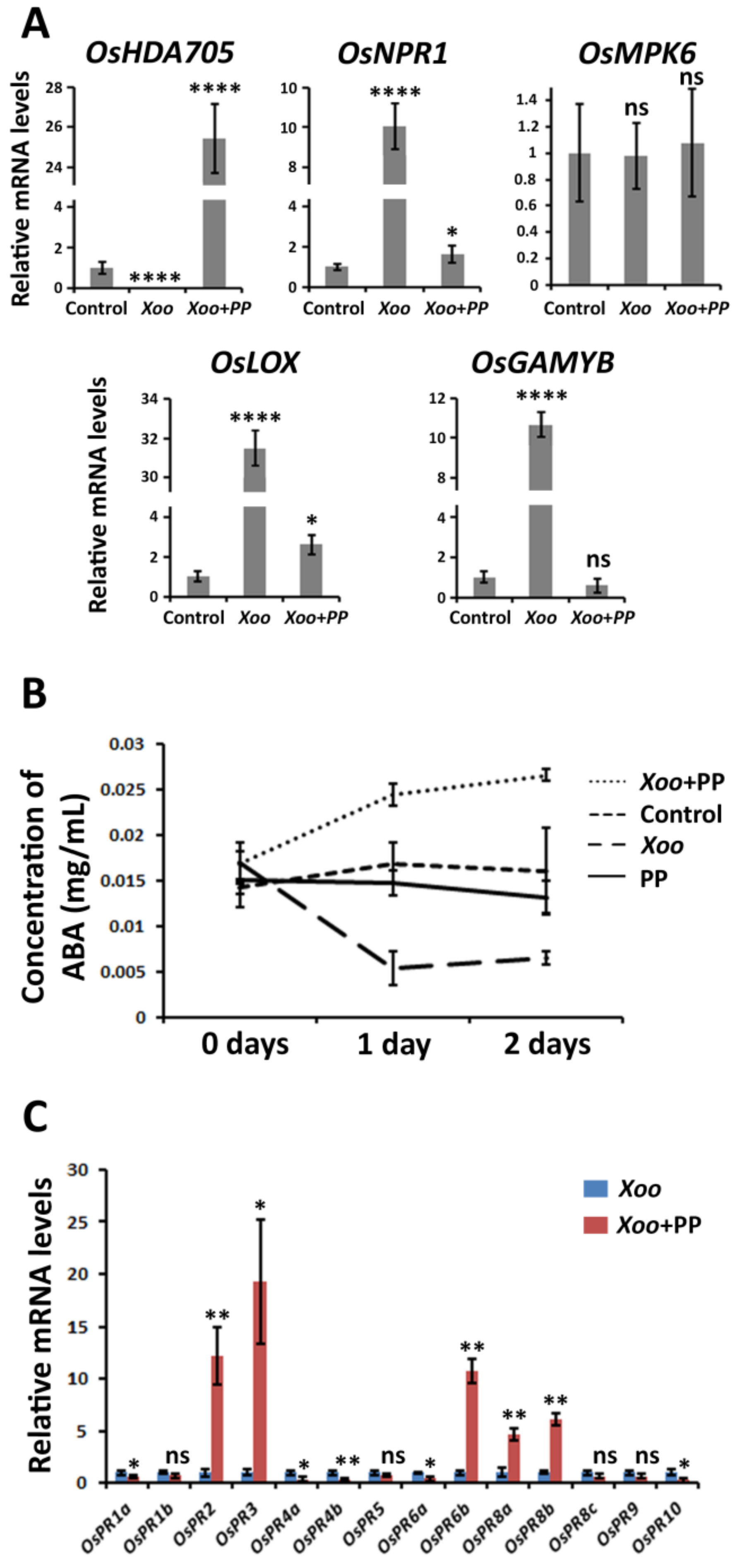
2.4. P. putida Activates an Alternative Mechanism of Defense in Rice Plants Possibly Based on ABA Accumulation
2.5. P. putida Activates the Expression of Alternative Pathogenesis-Related Genes (PRs)
3. Discussion
4. Materials and Methods
4.1. General Information and Strains
4.2. Cultivation and Treatment of Rice Plants with P. putida
4.3. Microscope Observation of P. putida Colonizing Rice Root
4.4. Measurement of Root and Stem Elongation
4.5. Detection of mRNA Levels of Key Genes in Rice Plants
4.6. Detection and Quantification of ACC in the Culture Medium of Rice Plants
4.7. Inoculation of X. oryzae pv. oryzae in Rice Plants
4.8. Measurement of ET Levels in Rice Plants
4.9. Detection and Quantification of ABA in Rice Leaves
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef]
- Denance, N.; Sanchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Barnawal, D.; Pandey, S.S.; Bharti, N.; Pandey, A.; Ray, T.; Singh, S.; Chanotiya, C.S.; Kalra, A. ACC deaminase-containing plant growth promoting rhizobacteria protect Papaver somniferum from downy mildew. J. Appl. Microbiol. 2017, 122, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant-pathogens interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef]
- Gietler, M.; Fidler, J.; Labudda, M.; Nykiel, M. Abscisic acid-enemy or savior in the response of cereals to abiotic and biotic stresses? Int. J. Mol. Sci. 2020, 21, 4607. [Google Scholar] [CrossRef]
- Wang, S.Y.; Shi, X.C.; Liu, F.Q.; Laborda, P. Chromatographic methods for detection and quantification of carbendazim in food. J. Agric. Food Chem. 2020, 68, 11880–11894. [Google Scholar] [CrossRef]
- Meftaul, I.M.; Venkateswarlu, K.; Dharmarajan, R.; Annamalai, P.; Megharaj, M. Pesticides in the urban environment: A potential threat that knocks at the door. Sci. Total Environ. 2020, 711, 134612. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N.; Barcelo, D. Persistence of pesticides-based contaminants in the environment and their effective degradation using laccase-assisted biocatalytic systmes. Sci. Total Environ. 2020, 695, 133896. [Google Scholar] [CrossRef]
- Tonelli, M.L.; Figueredo, M.S.; Rodriguez, J.; Fabra, A.; Ibañez, F. Induced systemic resistance-like responses elicited by rhizobia. Plant Soil 2020, 448, 1–14. [Google Scholar] [CrossRef]
- Wang, X.D.; Bi, W.S.; Gao, J.; Yu, X.M.; Wang, H.Y.; Liu, D.Q. Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 2018, 17, 2468–2477. [Google Scholar] [CrossRef]
- Wilkinson, S.W.; Magerøy, M.H.; Sánchez, A.L.; Smith, L.M.; Furci, L.; Cotton, T.E.A.; Krokene, P.; Ton, J. Surviving in a hostile world: Plant strategies to resist pests and diseases. Annu. Rev. Phytopathol. 2019, 57, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef]
- Vacheron, J.; Combes-Meynet, E.; Walker, V.; Gouesnard, B.; Muller, D.; Moenne-Loccoz, Y.; Prigent-Combaret, C. Expression on roots and contribution to maize phytostimulation of 1-aminocyclopropane-1-decarboxylate deaminase gene acdS in Pseudomonas fluorescens F113. Plant Soil 2016, 407, 187–202. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Du, H.; Xu, F.Y.; Ding, Y.X.; Gui, Y.; Zhang, J.H.; Xu, W.F. Root-bacteria associations boost rhizosheath formation in moderately dry soil through ethylene responses. Plant Physiol. 2020, 183, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant-bacterial interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Moffatt, B.A.; Glick, B.R. Determination of 1-aminocycopropane-1-carboxylic acid (ACC) to assess the effects of ACC deaminase-containing bacteria on roots of canola seedlings. Can. J. Microbiol. 2001, 47, 77–80. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Laborda, P.; Chen, X.; Wu, G.; Wang, S.; Lu, X.; Ling, J.; Li, K.; Liu, F. Lysobacter gummosus OH17 induces systemic resistance in Oryza sativa ‘Nipponbare’. Plant Pathol. 2020, 69, 838–848. [Google Scholar] [CrossRef]
- Beris, D.; Theologidis, I.; Skandalis, N.; Vassilakos, N. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci. Rep. 2018, 8, 10320. [Google Scholar] [CrossRef]
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef]
- Vives-Peris, V.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingovium sp. Plant Cell Rep. 2018, 37, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, C.; Laborda, P.; Zhao, Y.; Palmer, I.; Fu, Q.F.; Qiu, J.; Liu, F. Melatonin treatment inhibits the growth of Xanthomonas oryzae pv. oryzae. Front. Microbiol. 2018, 9, 2280. [Google Scholar] [CrossRef] [PubMed]
- Laborda, P.; Ling, J.; Chen, X.; Liu, F.Q. ACC deaminase from Lysobacter gummosus OH17 can promote root growth in Oryza sativa Nipponbare plants. J. Agric. Food Chem. 2018, 66, 3675–3682. [Google Scholar] [CrossRef]
- Chung, P.J.; Kim, Y.S.; Jeong, J.S.; Park, S.H.; Nahm, B.H.; Kim, J.K. The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J. 2009, 59, 764–776. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Tran, G.B.; Nguyen, N.H. Homeostasis of histone acetylation is critical for auxin signaling and root morphogenesis. Plant Mol. Biol. 2020, 103, 1–7. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, H.; Chen, F.; Liu, Y. The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 2014, 84, 125–133. [Google Scholar] [CrossRef]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Ivanchenko, M.G.; Muday, G.K. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 2008, 55, 175–187. [Google Scholar] [CrossRef]
- Lapidot, D.; Dror, R.; Vered, E.; Mishli, O.; Levy, D.; Helman, Y. Disease protection and growth promotion of potatoes (Solanum tuberosum L.) by Paenibacillus dendritiformis. Plant Pathol. 2015, 64, 545–551. [Google Scholar] [CrossRef]
- Sharma, S.; Chen, C.; Navathe, S.; Chand, R.; Pandey, S.P. A halotolerant growth promoting rhizobacteria triggers induced systemic resistance in plants and defends against fungal infection. Sci. Rep. 2019, 9, 4054. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.; Seshadri, S.; Kim, K.; Lee, G.; Sa, T. Ethylene emission and PR protein synthesis in ACC deaminase producing Methylobacterium spp. inoculated tomato plants (Lycopersicon esculentum Mill.) challenged with Ralstonia solanacearum under greenhouse conditions. Plant Physiol. Biochem. 2013, 67, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, X.; Luo, M.; Yang, S.; Wu, K. Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef]
- Fu, W.; Wu, K.; Duan, J. Sequence and expression analysis of histone deacetylases in rice. Biochem. Biophys. Res. Commun. 2007, 356, 843–850. [Google Scholar] [CrossRef]
- Jiang, D.G.; Zhou, L.Y.; Chen, W.T.; Ye, N.H.; Xia, J.X.; Zhuang, C.X. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef]
- Zhou, C.; Li, F.; Xie, Y.; Zhu, L.; Xiao, X.; Ma, Z.Y.; Wang, J.F. Involvement of abscisic acid in microbe-induced saline-alkaline resistance in plants. Plant Signal. Behav. 2017, 12, e1367465. [Google Scholar] [CrossRef]
- Victoria Salomon, M.; Bottini, R.; De Souza Filho, G.A.; Cohen, A.C.; Moreno, D.; Gil, M.; Piccoli, P. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant 2014, 151, 359–374. [Google Scholar] [CrossRef]
- Etesami, H.; Glick, B.R. Halotolerant plant growth-promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 2020, 178, 104124. [Google Scholar] [CrossRef]
- Lakkis, S.; Trotel-Aziz, P.; Rabenoelina, F.; Schwarzenberg, A.; Nguema-Ona, E.; Clement, C.; Aziz, A. Strengthening grapevine resistance by Pseudomonas fluorescens PTA-CT2 relies on distinct defense pathways in susceptible and partially resistant genotypes to downy mildew and gray mold diseases. Front. Plant Sci. 2019, 10, 1112. [Google Scholar] [CrossRef]
- Attia, M.S.; El-Sayyad, G.S.; Abd Elkodous, M.; El-Batal, A.I. The effective antagonistic potential of plant growth-promoting rhizobacteria against Alternaria solani-causing early blight disease in tomato plant. Sci Hortic. 2020, 266, 109289. [Google Scholar] [CrossRef]
- Wang, S.Y.; Shi, X.C.; Wang, R.; Wang, H.L.; Liu, F.; Laborda, P. Melatonin in fruit production and postharvest preservation: A review. Food Chem. 2020, 320, 126642. [Google Scholar] [CrossRef]
- Shinshi, H. Ethylene-regulated transcription and crosstalk wth jasmonic acid. Plant Sci. 2008, 175, 18–23. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Strader, L.C. Auxin-abscisic acid interactions in plant growth and development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Jha, P.N. The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 2017, 8, 1945. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS ONE 2017, 12, e0184523. [Google Scholar] [CrossRef]
- Wang, X.L.; Wang, Y.; Wang, L.; Liu, H.; Zhang, B.; Cao, Q.; Liu, X.; Lv, Y.; Bi, S.; Zhang, S.; et al. Arabidopsis PCaP2 functions as a linker between ABA and SA signals in plant water deficit tolerance. Front. Plant Sci. 2018, 9, 578. [Google Scholar] [CrossRef] [PubMed]
- Ponciano, G.; Yoshikawa, M.; Lee, J.L.; Ronald, P.C.; Whalen, M.C. Pathogenesis-related gene expression in rice is correlated with developmentally controlled Xa21-mediated resistance against Xanthomonas oryzae pv. oryzae. Physiol. Mol. Plant Pathol. 2006, 69, 131–139. [Google Scholar] [CrossRef]
- Chen, X.; Laborda, P.; Liu, F. Exogenous melatonin enhances rice plant resistance against Xanthomonas oryzae pv. oryzae. Plant Dis. 2020, 104, 1701–1708. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Wang, H.-L.; Tang, R.-P.; Sun, M.-Y.; Chen, T.-M.; Duan, X.-C.; Lu, X.-F.; Liu, D.; Shi, X.-C.; Laborda, P.; et al. Pseudomonas putida Represses JA- and SA-Mediated Defense Pathways in Rice and Promotes an Alternative Defense Mechanism Possibly through ABA Signaling. Plants 2020, 9, 1641. https://doi.org/10.3390/plants9121641
Wang R, Wang H-L, Tang R-P, Sun M-Y, Chen T-M, Duan X-C, Lu X-F, Liu D, Shi X-C, Laborda P, et al. Pseudomonas putida Represses JA- and SA-Mediated Defense Pathways in Rice and Promotes an Alternative Defense Mechanism Possibly through ABA Signaling. Plants. 2020; 9(12):1641. https://doi.org/10.3390/plants9121641
Chicago/Turabian StyleWang, Rui, Hai-Lin Wang, Rui-Ping Tang, Meng-Ying Sun, Tang-Min Chen, Xu-Chu Duan, Xiao-Feng Lu, Dong Liu, Xin-Chi Shi, Pedro Laborda, and et al. 2020. "Pseudomonas putida Represses JA- and SA-Mediated Defense Pathways in Rice and Promotes an Alternative Defense Mechanism Possibly through ABA Signaling" Plants 9, no. 12: 1641. https://doi.org/10.3390/plants9121641
APA StyleWang, R., Wang, H.-L., Tang, R.-P., Sun, M.-Y., Chen, T.-M., Duan, X.-C., Lu, X.-F., Liu, D., Shi, X.-C., Laborda, P., & Wang, S.-Y. (2020). Pseudomonas putida Represses JA- and SA-Mediated Defense Pathways in Rice and Promotes an Alternative Defense Mechanism Possibly through ABA Signaling. Plants, 9(12), 1641. https://doi.org/10.3390/plants9121641






