Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum
Abstract
1. Introduction
2. Results
2.1. Identification of G. hirsutum JmjC Genes
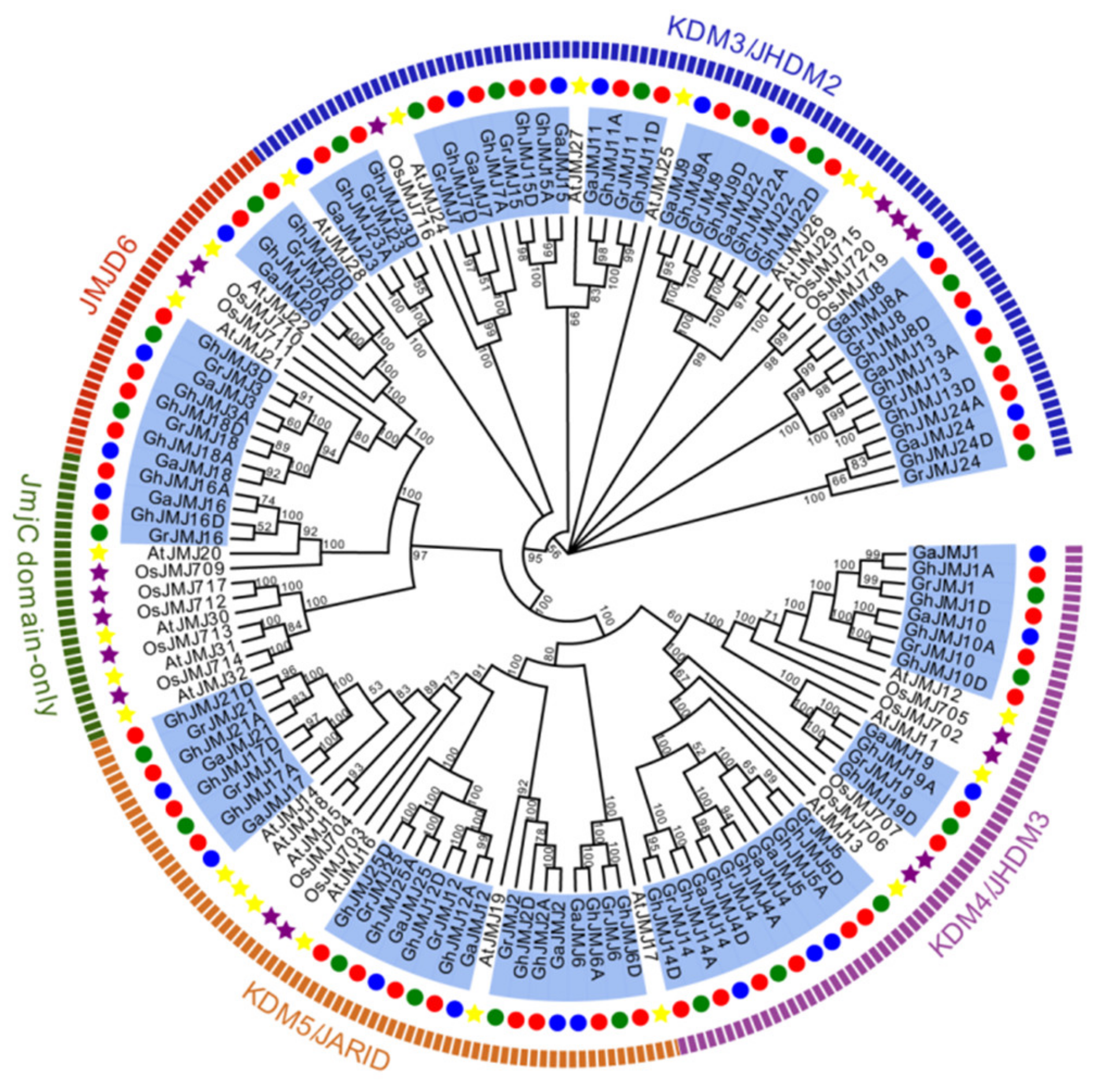
2.2. Phylogenetic Analysis of G. hirsutum JmjC Genes
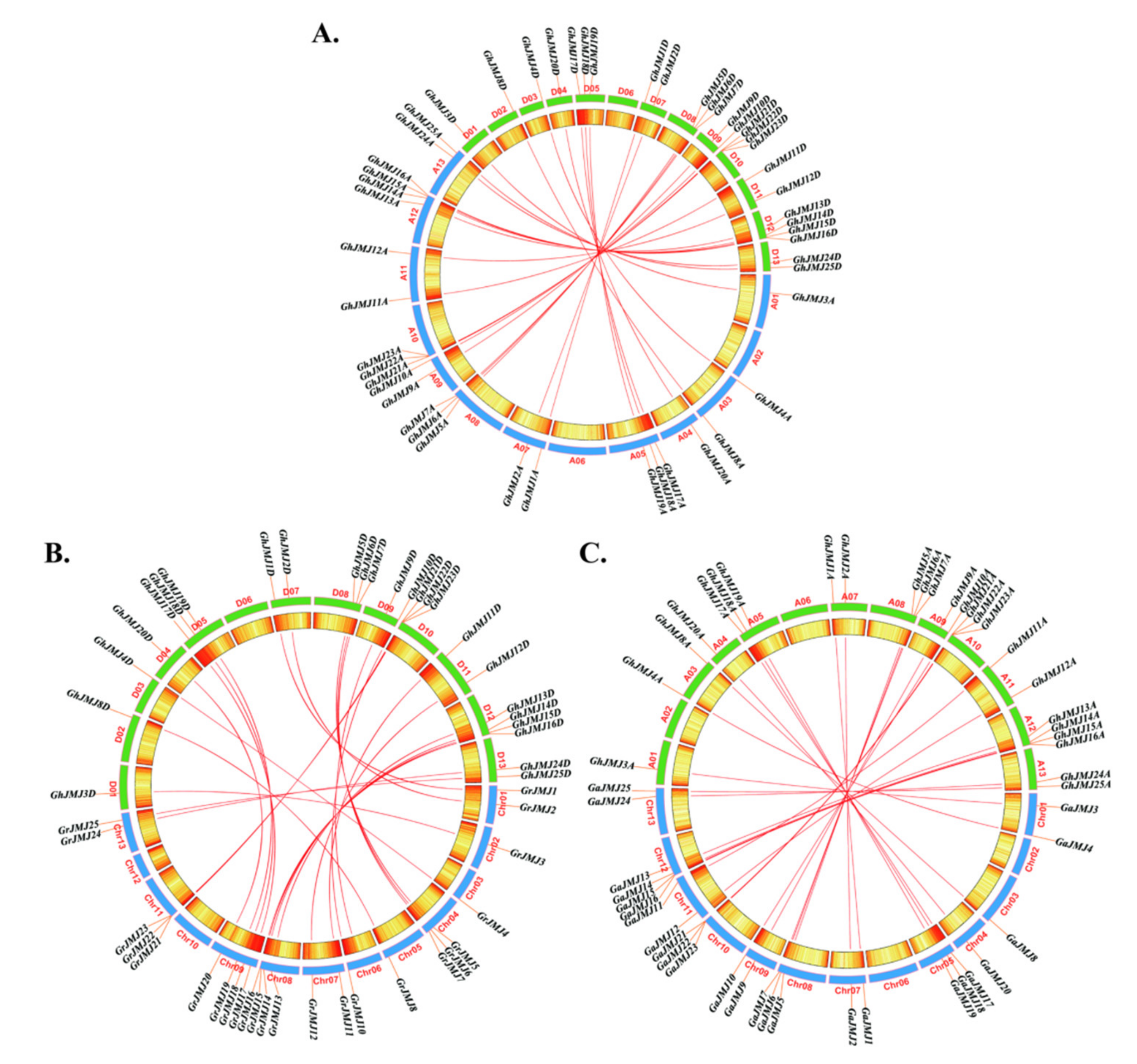
2.3. Chromosomal Localization and Collinearity of G. hirsutum JmjC Genes
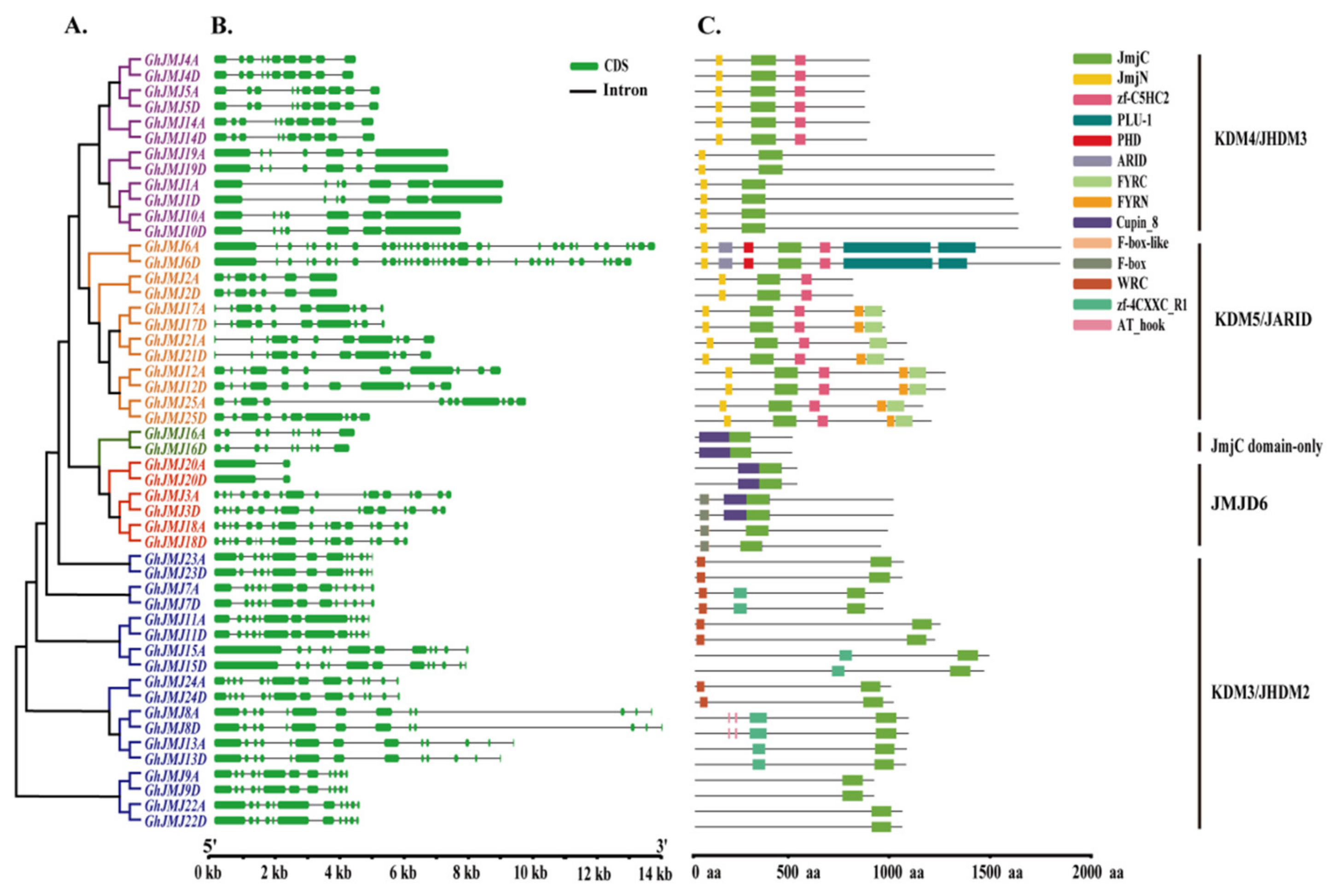
2.4. Structural Features and Conserved Domains of G. hirsutum JmjC Genes
2.5. Cis-Elements in the Promoter Regions of G. hirsutum JmjC Genes
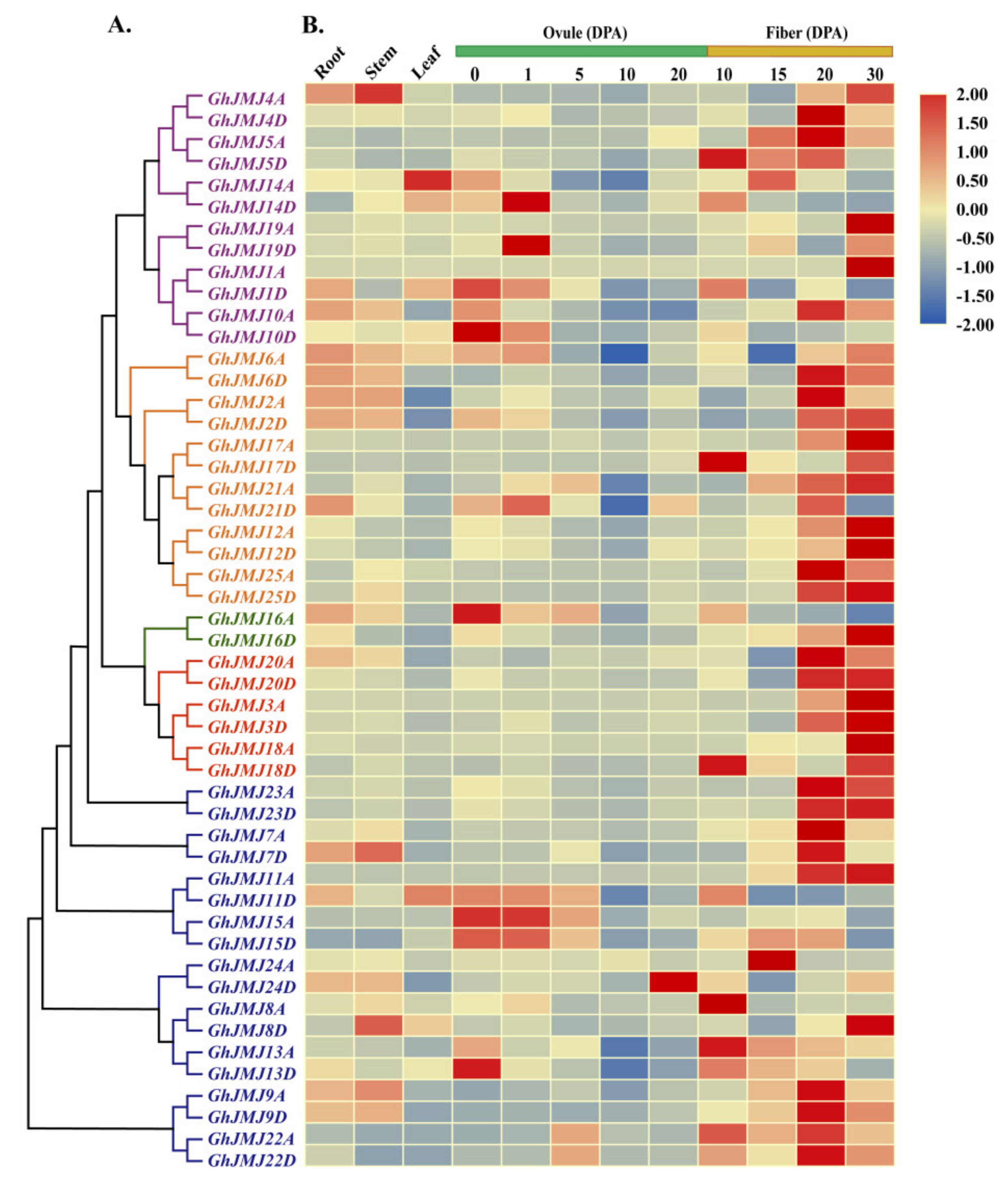
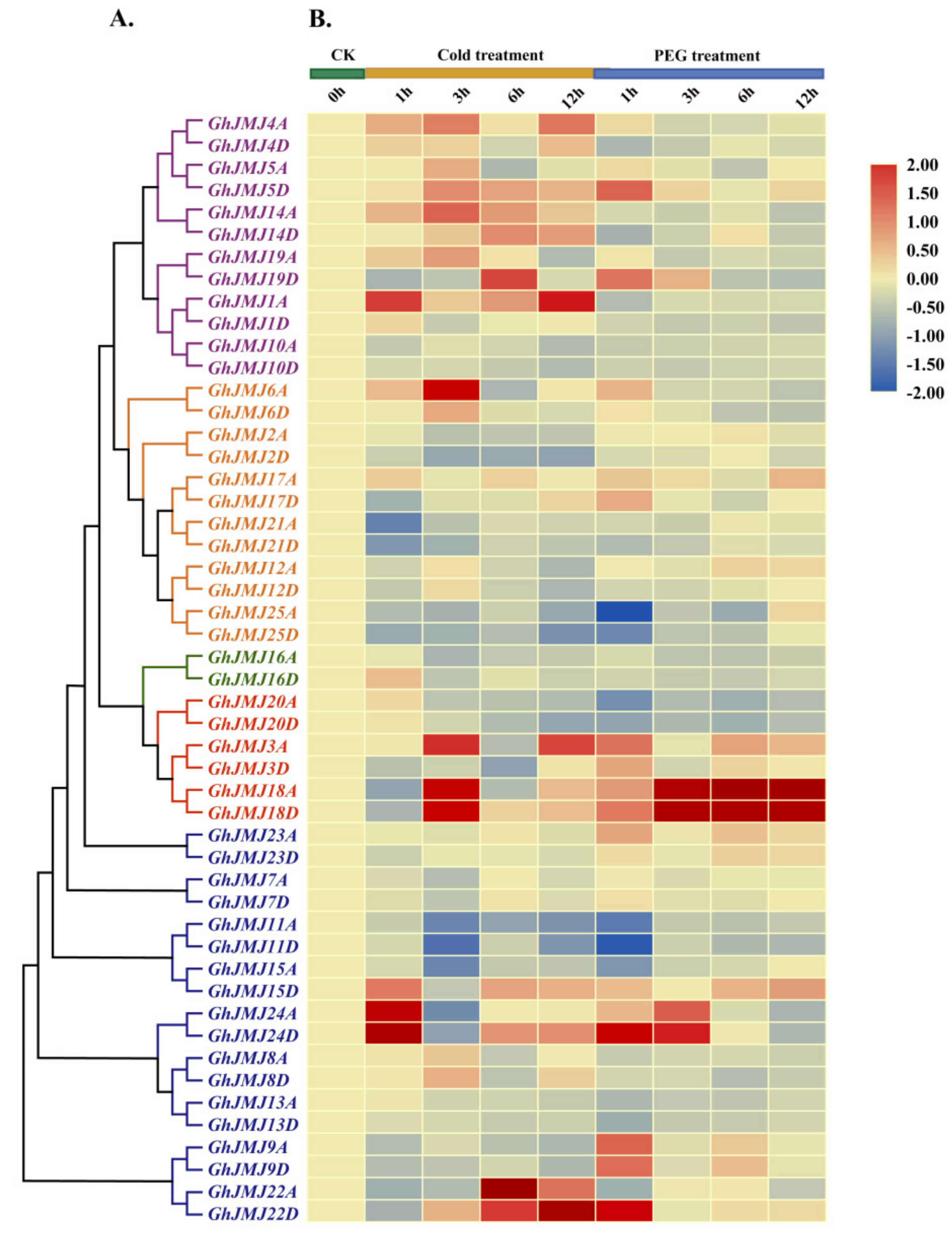
2.6. Expression Profiling of G. hirsutum JmjC Genes in Different Tissues and under Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of JmjC Genes in G. hirsutum and other Cotton Species
4.2. Phylogenetic Analysis of G. hirsutum JmjC Genes
4.3. Chromosomal Mapping and Synteny Analysis of G. hirsutum JmjC Genes
4.4. Structural Analysis of G. hirsutum JmjC Genes
4.5. Prediction of Cis-Acting Elements in Promoter Regions of G. hirsutum JmjC Genes
4.6. Plant Materials and Stress Treatments
4.7. Expression Analysis of G. hirsutum JmjC Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef]
- Cairns, B.R. The logic of chromatin architecture and remodelling at promoters. Nature 2009, 461, 193–198. [Google Scholar] [CrossRef]
- Holliday, R. DNA methylation and epigenetic defects in carcinogenesis. Mutat. Res. 1987, 181, 215–217. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Bhaumik, S.R.; Smith, E.; Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct Mol. Biol. 2007, 14, 1008–1016. [Google Scholar] [CrossRef]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef]
- Mosammaparast, N.; Shi, Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010, 79, 155–179. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef]
- Zhou, D.-X.; Hu, Y. Regulatory function of histone modifications in controlling rice gene expression and plant growth. Rice 2010, 3, 103–111. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar] [CrossRef]
- Tahiliani, M.; Mei, P.; Fang, R.; Leonor, T.; Rutenberg, M.; Shimizu, F.; Li, J.; Rao, A.; Shi, Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007, 447, 601–605. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Lu, F.; Li, G.; Cui, X.; Liu, C.; Wang, X.J.; Cao, X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008, 50, 886–896. [Google Scholar] [CrossRef]
- Luo, M.; Hung, F.-Y.; Yang, S.; Liu, X.; Wu, K. Histone lysine demethylases and their functions in plants. Plant Mol. Biol. 2014, 32, 558–565. [Google Scholar] [CrossRef]
- Yamane, K.; Toumazou, C.; Tsukada, Y.; Erdjument-Bromage, H.; Tempst, P.; Wong, J.; Zhang, Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006, 125, 483–495. [Google Scholar] [CrossRef]
- Allis, C.D.; Berger, S.L.; Cote, J.; Dent, S.; Jenuwien, T.; Kouzarides, T.; Pillus, L.; Reinberg, D.; Shi, Y.; Shiekhattar, R.; et al. New nomenclature for chromatin-modifying enzymes. Cell 2007, 131, 633–636. [Google Scholar] [CrossRef]
- Jones, M.A.; Morohashi, K.; Grotewold, E.; Harmer, S.L. Arabidopsis JMJD5/JMJ30 acts Independently of LUX ARRHYTHMO within the plant circadian clock to enable temperature compensation. Front. Plant Sci. 2019, 10, 57. [Google Scholar] [CrossRef]
- Miura, A.; Nakamura, M.; Inagaki, S.; Kobayashi, A.; Saze, H.; Kakutani, T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. Embo. J. 2009, 28, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Shiraishi, A.; Miura, A.; Kakutani, T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 2008, 319, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Noh, B.; Lee, S.H.; Kim, H.J.; Yi, G.; Shin, E.A.; Lee, M.; Jung, K.J.; Doyle, M.R.; Amasino, R.M.; Noh, Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004, 16, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Li, L.; Guo, M.; Chory, J.; Yin, Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 7618–7623. [Google Scholar] [CrossRef]
- Lu, F.; Cui, X.; Zhang, S.; Jenuwein, T.; Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011, 43, 715–719. [Google Scholar] [CrossRef]
- Lu, F.; Cui, X.; Zhang, S.; Liu, C.; Cao, X. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 2010, 20, 387–390. [Google Scholar] [CrossRef]
- Ning, Y.Q.; Ma, Z.Y.; Huang, H.W.; Mo, H.; Zhao, T.T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef]
- Yang, H.; Han, Z.; Cao, Y.; Fan, D.; Li, H.; Mo, H.; Feng, Y.; Liu, L.; Wang, Z.; Yue, Y.; et al. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 2012, 8, e1002664. [Google Scholar] [CrossRef]
- Yang, H.; Mo, H.; Fan, D.; Cao, Y.; Cui, S.; Ma, L. Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 2012, 31, 1297–1308. [Google Scholar] [CrossRef]
- Lu, S.X.; Knowles, S.M.; Webb, C.J.; Celaya, R.B.; Cha, C.; Siu, J.P.; Tobin, E.M. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011, 155, 906–915. [Google Scholar] [CrossRef]
- Shen, Y.; Silva, N.C.e.; Audonnet, L.; Servet, C.; Wei, W.; Zhou, D.-X. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 2014, 5, 290. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, A.; Jin, J.B.; Zhao, B.; Wang, T.J.; Wu, Y.; Wang, S.; Liu, Y.; Wang, J.; Guo, P.; et al. Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 2019, 223, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wu, X.; Liu, D.; Song, S.; Liu, D.; Wang, H. Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 2016, 474, 271–276. [Google Scholar] [CrossRef]
- Liu, G.; Khan, N.; Ma, X.; Hou, X. Identification, Evolution, and Expression Profiling of Histone Lysine Methylation Moderators in Brassica rapa. Plants 2019, 8, 526. [Google Scholar] [CrossRef]
- Qian, Y.; Chen, C.; Jiang, L.; Zhang, J.; Ren, Q. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genom. 2019, 20, 256. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, L.; Wang, L.; Liu, L.; Li, L.; Sun, L.; Rao, Q.; Zhang, J.; Huang, S. JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol. 2015, 15, 286. [Google Scholar] [CrossRef]
- Li, T.; Chen, X.; Zhong, X.; Zhao, Y.; Liu, X.; Zhou, S.; Cheng, S.; Zhou, D.-X. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 2013, 25, 4725–4736. [Google Scholar] [CrossRef]
- Nardeli, S.M.; Artico, S.; Aoyagi, G.M.; de Moura, S.M.; da Franca Silva, T.; Grossi-de-Sa, M.F.; Romanel, E.; Alves-Ferreira, M. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 2018, 127, 169–184. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, J.; Zuo, D.; Wang, Q.; Malik, W.; Zhang, Y.; Abid, M.A.; Cheng, H.; Yang, Q.; Song, G. Recent insights into cotton functional genomics: Progress and future perspectives. Plant Biotechnol. J. 2018, 16, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Scheffler, B.E.; Dennis, E.; Triplett, B.A.; Zhang, T.; Guo, W.; Chen, X.; Stelly, D.M.; Rabinowicz, P.D.; Town, C.D. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Li, F.G. The Gossypium raimondii genome, a huge leap forward in cotton genomics. J. Integr. Plant Biol. 2013, 55, 570–571. [Google Scholar] [CrossRef] [PubMed]
- Sunilkumar, G.; Campbell, L.M.; Puckhaber, L.; Stipanovic, R.D.; Rathore, K.S. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. USA. 2006, 103, 18054–18059. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gou, J.Y.; Li, F.G.; Shangguan, X.X.; Zhao, B.; Yang, C.Q.; Wang, L.J.; Yuan, S.; Liu, C.J.; Chen, X.Y. A cotton BURP domain protein interacts with α-expansin and their co-expression promotes plant growth and fruit production. Mol. Plant 2013, 6, 945–958. [Google Scholar] [CrossRef]
- Gelfman, S.; Cohen, N.; Yearim, A.; Ast, G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013, 23, 789–799. [Google Scholar] [CrossRef]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef]
- Cui, X.; Lu, F.; Qiu, Q.; Zhou, B.; Gu, L.; Zhang, S.; Kang, Y.; Cui, X.; Ma, X.; Yao, Q.; et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016, 48, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.L.; Kortschak, R.D.; Kalionis, B.; Saint, R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 1996, 16, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Kutateladze, T.G. PHD fingers epigenetic effectors and potential drug targets. Mol. Interv. 2009, 9, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Madsen, B.; Tarsounas, M.; Burchell, J.M.; Hall, D.; Poulsom, R.; Taylor-Papadimitriou, J. PLU-1, a transcriptional repressor and putative testis-cancer antigen, has a specific expression and localisation pattern during meiosis. Chromosoma 2003, 112, 124–132. [Google Scholar] [CrossRef]
- Zhang, S.; Tian, Z.; Li, H.; Guo, Y.; Miao, Y. Genome-wide analysis and characterization of F-box gene family in Gossypium hirsutum L. BMC Genom. 2019, 20, 993. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef]
- Cao, X.; Costa, L.M.; Biderre-Petit, C.; Kbhaya, B.; Dey, N.; Perez, P.; McCarty, D.R.; Gutierrez-Marcos, J.F.; Becraft, P.W. Abscisic acid and stress signals induce viviparous1 expression in seed and vegetative tissues of maize. Plant Physiol. 2007, 143, 720–731. [Google Scholar] [CrossRef]
- Ross, J.J.; Reid, J.B.; Swain, S.M.; Hasan, O.; Poole, A.T.; Hedden, P.; Willis, C.L. Genetic-regulation of gibberellin deactivation in pisum. Plant J. 1995, 7, 513–523. [Google Scholar] [CrossRef]
- Xu, Z.; Sun, M.; Jiang, X.; Sun, H.; Dang, X.; Cong, H.; Qiao, F. Glycinebetaine biosynthesis in response to osmotic stress depends on jasmonate signaling in watermelon suspension cells. Front. Plant Sci. 2018, 9, 1469. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; YamaguchiShinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [PubMed]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, Y.; Zhou, D.-X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta. 2011, 1809, 421–426. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, D.; Liu, C.; Shen, W.; Ruan, Y. Evolution and conservation of JmjC domain proteins in the green lineage. Mol. Genet. Genomics 2016, 291, 33–49. [Google Scholar] [CrossRef]
- Zhao, W.; Shafiq, S.; Berr, A.; Shen, W.H. Genome-wide gene expression profiling to investigate molecular phenotypes of Arabidopsis mutants deprived in distinct histone methyltransferases and demethylases. Genom Data. 2015, 4, 143–145. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Cheng, L.; Liu, Y.; Wang, H.; Ke, D.; Yuan, H.; Zhang, L.; Wang, L. Genome-wide analysis of soybean JmjC domain-containing proteins suggests evolutionary conservation following whole-genome duplication. Front. Plant Sci. 2016, 7, 1800. [Google Scholar] [CrossRef]
- Gu, T.; Han, Y.; Huang, R.; McAvoy, R.J.; Li, Y. Identification and characterization of histone lysine methylation modifiers in Fragaria vesca. Sci. Rep. 2016, 6, 23581. [Google Scholar] [CrossRef]
- Dong, Y.; Lu, J.; Liu, J.; Jalal, A.; Wang, C. Genome-wide identification and functional analysis of JmjC domain-containing genes in flower development of Rosa chinensis. Plant Mol. Biol. 2020, 102, 417–430. [Google Scholar] [CrossRef]
- Zong, W.; Zhong, X.; You, J.; Xiong, L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 2013, 81, 175–188. [Google Scholar] [CrossRef]
- Wilsker, D.; Patsialou, A.; Dallas, P.B.; Moran, E. ARID proteins: A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002, 13, 95–106. [Google Scholar] [PubMed]
- Kim, H.J.; Triplett, B.A. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001, 127, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, J.; Gao, S.; Li, Z.; Kuai, B.; Ren, G. REF6 promotes lateral root formation through de-repression of PIN1/3/7 genes. J. Integr. Plant Biol. 2019, 61, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Li, H.; Guan, H.; Zhuo, Q.; Wang, Z.; Li, S.; Si, J.; Zhang, B.; Feng, B.; Kong, L.A.; Wang, F.; et al. Genome-wide characterization of the abscisic acid-, stress- and ripening-induced (ASR) gene family in wheat (Triticum aestivum L.). Biol. Res. 2020, 53, 23. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PLoS ONE 2015, 10, e0123281. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, K.; Ren, Z.; Song, C.; Pei, X.; Liu, Y.; Wang, Z.; He, K.; Zhang, F.; Zhou, X.; et al. Molecular evolution and stress and phytohormone responsiveness of SUT genes in Gossypium hirsutum. Front. Genet. 2018, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Feng, J.; Liu, W.; Ren, Z.; Zhao, J.; Pei, X.; Liu, Y.; Yang, D.; Ma, X. Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum. Plants 2020, 9, 1617. https://doi.org/10.3390/plants9111617
Zhang J, Feng J, Liu W, Ren Z, Zhao J, Pei X, Liu Y, Yang D, Ma X. Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum. Plants. 2020; 9(11):1617. https://doi.org/10.3390/plants9111617
Chicago/Turabian StyleZhang, Jie, Junping Feng, Wei Liu, Zhongying Ren, Junjie Zhao, Xiaoyu Pei, Yangai Liu, Daigang Yang, and Xiongfeng Ma. 2020. "Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum" Plants 9, no. 11: 1617. https://doi.org/10.3390/plants9111617
APA StyleZhang, J., Feng, J., Liu, W., Ren, Z., Zhao, J., Pei, X., Liu, Y., Yang, D., & Ma, X. (2020). Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum. Plants, 9(11), 1617. https://doi.org/10.3390/plants9111617



