Screening of Salt Stress Responsive Genes in Brachypodium distachyon (L.) Beauv. by Transcriptome Analysis
Abstract
1. Introduction
2. Results
2.1. Measurement of Biochemical Parameters
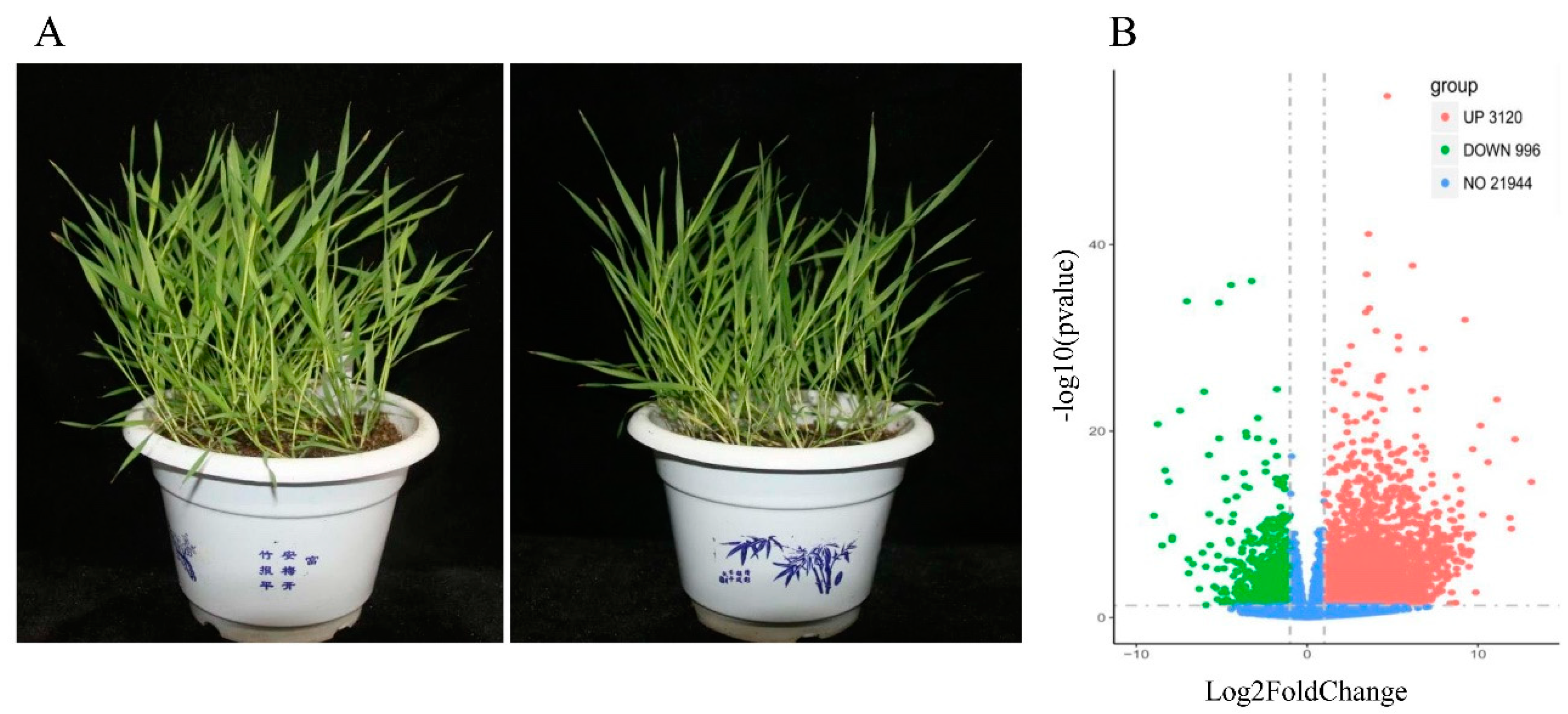
2.2. Transcriptome Profiling of B. distachyon
2.3. DEGs in B. distachyon
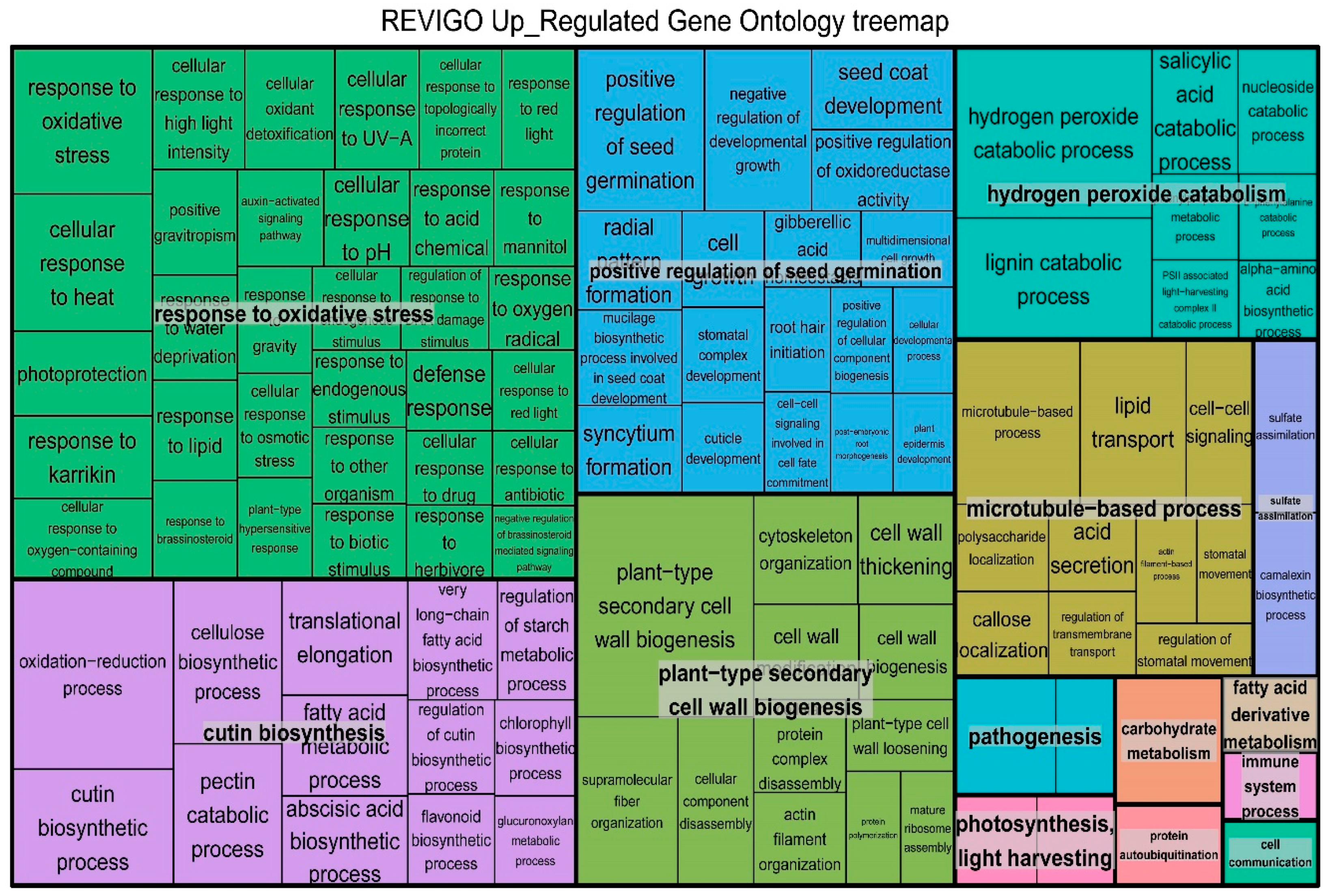
2.4. Gene Ontology (GO) Enrichment Results of DEGs in B. distachyon
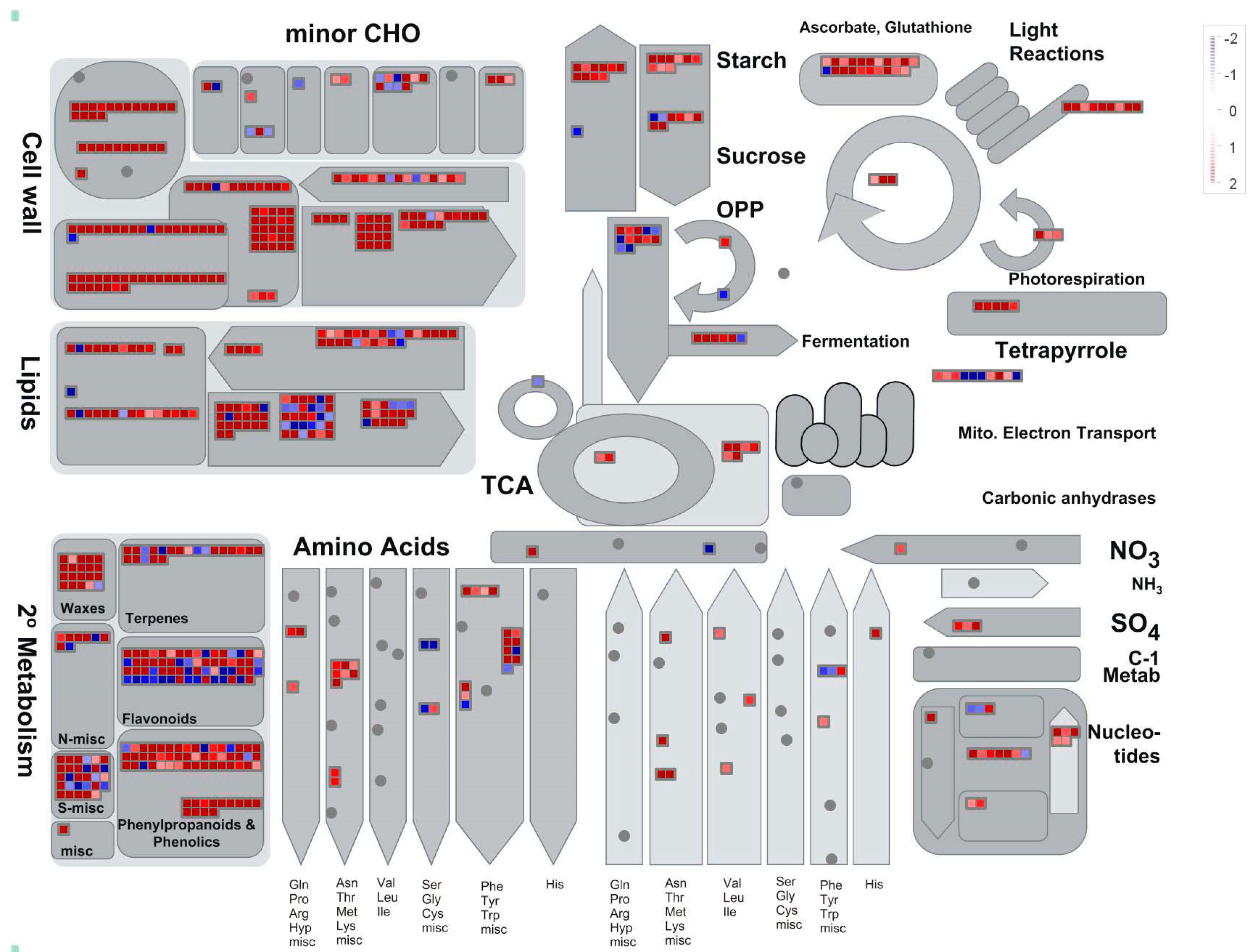
2.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) and MapMan Enrichment Results of DEGs in B. distachyon
2.6. The Differentially Expressed TFs
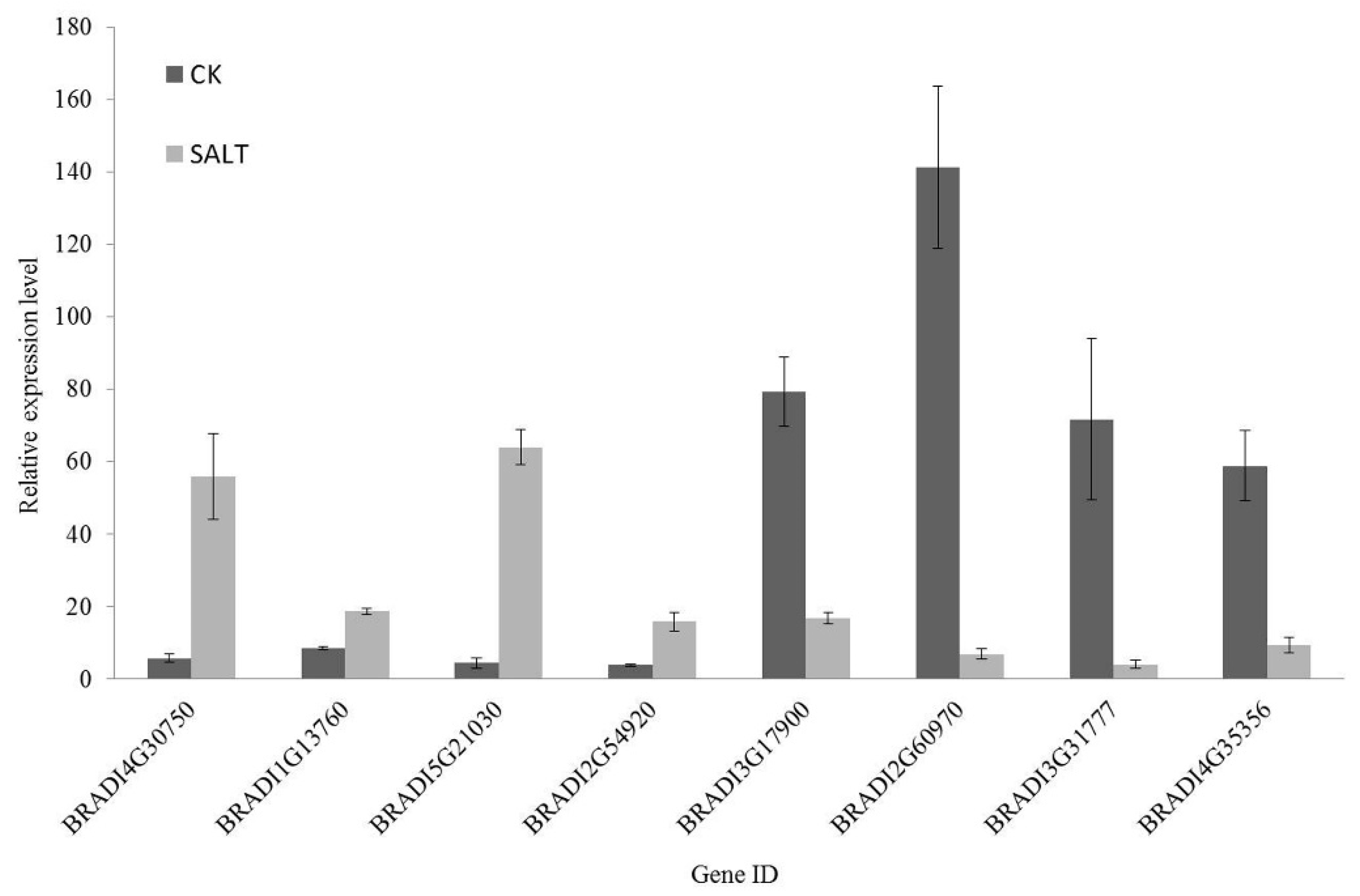
2.7. Quantitative Real-Time PCR (qRT-PCR) Validation
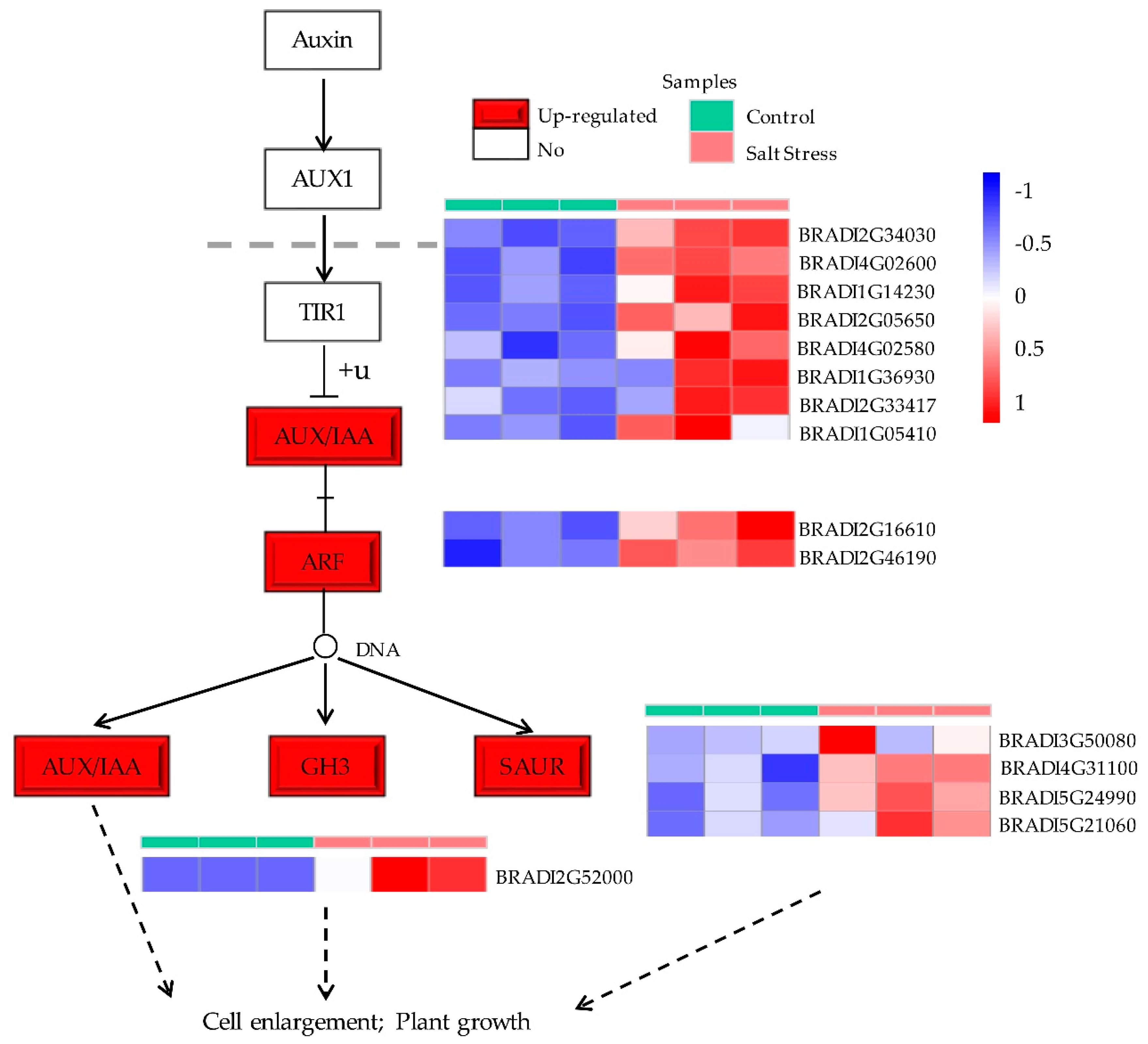
3. Discussion
4. Materials and Methods
4.1. Material Preparation and Salt Stress Treatment
4.2. Measurement of Biochemical Parameters
4.3. Transcriptional Profiling
4.4. Measurement of Gene Expression and Differential Expression Analysis in B. distachyon
4.5. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment and Differentially Expressed TFs Analysis
4.6. Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, R.; Wang, X.; Liu, K.; Zhang, X.; Zhang, L.; Fan, S. Comparative transcriptome analysis of halophyte Zoysia macrostachya in response to salinity stress. Plants 2020, 9, 458. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Qian, Z.; Luo, M.; Fan, S.; Zhang, X.; Zhang, L. Identification of salt stress responding genes using transcriptome analysis in green alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 3359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yao, Y.; Li, X.; Zhang, L.; Fan, S. Transcriptomic analysis identifes novel genes and pathways for salt stress responses in Suaeda salsa leaves. Sci. Rep. 2020, 10, 4236. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lyu, M.-J.A.; Leng, B.; Zhu, X.; Wang, B. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 2016, 91, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Shimazaki, T.; Gulzar, S.; Kikuchi, A.; Gul, B.; Khan, M.A.; Koyro, H.W.; Huchzermeyer, B.; Watanabe, K.N. The influence of genes regulating transmembrane transport of Na+ on the salt resistance of Aeluropus lagopoides. Funct. Plant Biol. 2013, 40, 860–871. [Google Scholar] [CrossRef]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 2020, 71, 24.01–24.31. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Sudhir, P.; Murthy, S.D.S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Donaldsona, L.; Ludidib, N.; Knightc, M.R.; Gehringb, C.; Denbya, K. Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 2004, 569, 317–320. [Google Scholar] [CrossRef]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific responses to drought, salt and cold in the Arabidopsis root. Plant J. 2000, 23, 267–278. [Google Scholar] [CrossRef]
- Gobert, A.; Isayenkov, S.; Voelker, C.; Czempinski, K.; Maathuis, F.J.M. The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc. Natl. Acad. Sci. USA 2007, 104, 10726–10731. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by Calcium-Dependent Protein Kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef] [PubMed]
- Hadi, M.R.; Karimi, N. The role of calcium in plant’s salt tolerance. J. Plant Nutr. 2012, 35, 2037–2054. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, S.; Li, C.; Zhang, P.; Zhang, P. Transcriptome sequencing of Antarctic moss under salt stress emphasizes the important roles of the ROS-scavenging system. Gene 2019, 969, 122–134. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Bing, J.; Zhang, G. The role analysis of APX gene family in the growth and developmental processes and in response to abiotic stresses in Arabidopsis thaliana. Hereditas (Beijing) 2019, 41, 534–547. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 81337. [Google Scholar] [CrossRef]
- Kito, K.; Yamane, K.; Yamamori, T.; Matsuhira, H.; Tanaka, Y.; Takabe, T. Isolation, functional characterization and stress responses of raffinose synthase genes in sugar beet. J. Plant Biochem. Biotechnol. 2018, 27, 36–45. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Kalra, A. Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. 2017, 161, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, M.; Shen, J.; Chen, D.; Zheng, Y.; Zhang, W. ZmOST1 mediates ABA regulation of guard cell ion channels and drought stress responses. J. Integr. Plant Biol. 2018, 61, 478–491. [Google Scholar] [CrossRef]
- Askari-Khorasgani, O.; Pessarakli, M. Chapter 19 Phytohormone homeostasis and crosstalk effects in response to osmotic stress. In Handbook of Plant and Crop Stress; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- WasKiewicz, A.; Beszterda, M.; Goliński, P. Salt Stress in Plants; Ahmad, P., Azooz, M.M., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Pandey, V.; Bhatt, I.D.; Nandi, S.K. Role and regulation of auxin signaling in abiotic stress tolerance. In Plant Signaling Molecules; Khan, M.I.R., Reddy, P.S., Ferrante, A., Khan, N.A., Eds.; Woodhead Publishing: Cambridge, UK, 2019; pp. 319–331. [Google Scholar] [CrossRef]
- Li, S.; An, Y.; Hailati, S.; Zhang, J.; Yang, P. Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from Medicago sativa enhanced salt stress tolerance of Arabidopsis. J. Plant Biol. 2019, 62, 374–386. [Google Scholar] [CrossRef]
- Krishna, P.; Prasad, B.D.; Rahman, T. Brassinosteroid action in plant abiotic stress tolerance. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1564, pp. 193–202. [Google Scholar]
- Ma, J.; Gao, X.; Liu, Q.; Yun, S.; Zhang, D.; Jiang, L.; Li, C. Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 2036. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Gai, X.; Ren, J.; Liu, X.; Cai, Y.; Wang, Q.; Ren, H. Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 2015, 56, 1339–1354. [Google Scholar] [CrossRef]
- Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; Vogel, J.P.; Garvin, D.F. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Kellogg, E.A. Flowering Plants. Monocots; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Vogel, J.P.; Garvin, D.F.; Leong, O.M.; Hayden, D.M. Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult. 2006, 84, 199–211. [Google Scholar] [CrossRef]
- Draper, J.; Mur, L.A.J.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P.M. Brachypodium distachyon. A New Model System for Functional Genomics in Grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Pan, Q.; Chen, S.; Feng, C.; Hai, J.; Li, H. Comparison of Trihelix transcription factors between wheat and Brachypodium distachyon at genome-wide. BMC Genom. 2019, 20, 142. [Google Scholar] [CrossRef]
- Kang, H.; Zhang, M.; Zhou, S.; Guo, Q.; Chen, F.; Wu, J.; Wang, W. Overexpression of wheat ubiquitin gene, Ta-Ub2, improves abiotic stress tolerance of Brachypodium distachyon. Plant Sci. 2016, 248, 102–115. [Google Scholar] [CrossRef]
- Lv, D.; Subburaj, S.; Cao, M.; Yan, X.; Li, X.; Appels, R.; Sun, D.; Ma, W.; Yan, Y. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteom. 2014, 13, 632–652. [Google Scholar] [CrossRef]
- You, J.; Zhang, L.; Song, B.; Qi, X.; Chan, Z. Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS ONE 2015, 10, 0122027. [Google Scholar] [CrossRef]
- Wang, L.; Hu, W.; Sun, J.; Liang, X.; Yang, X.; Wei, S.; Wang, X.; Zhou, Y.; Xiao, Q.; Yang, G. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 2015, 237, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e0146242. [Google Scholar] [CrossRef]
- Luo, Q.; Teng, W.; Fang, S.; Li, H.; Li, B.; Chu, J.; Li, Z.; Zheng, Q. Transcriptome analysis of salt-stress response in three seedling tissues of common wheat. Crop J. 2019, 7, 378–392. [Google Scholar] [CrossRef]
- Yang, Z.; Zheng, H.; Wei, X.; Song, J.; Wang, B.; Sui, N. Transcriptome analysis of sweet Sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 2018, 430, 423–439. [Google Scholar] [CrossRef]
- Owen, H.J. Role of abscisic acid in a Ca2+ second messenger system. Physiol. Plant. 1988, 72, 637–641. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Qi, G.; Feng, H.; Zhao, S.; Wu, W. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013, 74, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Hong, M.J.; Jang, J.H.; Seo, Y.W. cDNA-AFLP analysis reveals differential gene expression in response to salt stress in Brachypodium distachyon. Genes Genom. 2012, 34, 475–484. [Google Scholar] [CrossRef]
- Sade, N.; Maria, D.M.R.W.; Ke, X.; Brotman, Y.; Wright, M.; Khan, I.; De Souza, W.; Bassil, E.; Tobias, C.M.; Thilmony, R. Salt tolerance of two perennial grass Brachypodium sylvaticum accessions. Plant Mol. Biol. 2018, 96, 305–314. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, J.; Lu, J.; Yang, Y.; Zhang, X.; Wan, D.; Liu, J. Transcriptome differences between two sister desert poplar species under salt stress. BMC Genom. 2014, 15, 337. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, R.; Ma, Y.; Song, J. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 2018, 146, 1–7. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, X.; Wang, N.; Cui, Y.; Zhang, L.; Fan, S. Transcriptome analysis of salt stress response in halophyte Atriplex centralasiatica leaves. Acta Physiol. Plant. 2020, 42, 3. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Qi, W.; Xue, B.; Wei, Z.; Dabing, X.; Yan, W.; Jun, Y.; Liang, Z.; Gang, Z. De novo assembly and analysis of tartary buckwheat (Fagopyrum tataricum Garetn.) transcriptome discloses key regulators involved in salt-stress response. Genes 2017, 8, 255. [Google Scholar] [CrossRef]
- Chan, C.; Lam, H.-M. A putative lambda class glutathione S-transferase enhances plant survival under salinity stress. Plant Cell Physiol. 2014, 55, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Jha, B.; Sharma, A.; Mishra, A. Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 2011, 38, 4823–4832. [Google Scholar] [CrossRef]
- Long, L.; Yang, W.W.; Liao, P.; Guo, Y.; Kumar, A.; Gao, W. Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci. 2019, 281, 72–81. [Google Scholar] [CrossRef]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Zhao, K. Salt Tolerance Physiology of Plants; Science and Technology of China Press: Beijing, China, 1993. [Google Scholar]
- Storey, R.; Jones, R.W. Salt stress and comparative physiology in the Gramineae. III. Effect of salinity upon ion relations and glycinebetaine and proline levels in Spartina × townsendii. Aust. J. Plant Physiol. 1978, 5, 831–838. [Google Scholar] [CrossRef]
- Amaral, M.N.D.; Arge, L.W.P.; Benitez, L.C.; Danielowski, R.; Silveira, S.F.d.S.; Farias, D.D.R.; Oliveira, A.C.D.; Maia, L.C.D.; Braga, E.J.B. Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct. Integr. Genom. 2016, 16, 567–579. [Google Scholar] [CrossRef]
- Sun, Z.; Qi, X.; Wang, Z.; Li, P.; Zhao, Y. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [CrossRef]
- Lahuta, L.B.; Górecki, R.J. Raffinose in seedlings of winter vetch (Vicia villosa Roth.) under osmotic stress and followed by recovery. Acta Physiol. Plant. 2011, 33, 725–733. [Google Scholar] [CrossRef]
- Pluskota, W.E.; Szablińska, J.; Obendorf, R.L.; Górecki, R.J.; Lahuta, L.B. Osmotic stress induces genes, enzymes and accumulation of galactinol, raffinose and stachyose in seedlings of pea (Pisum sativum L.). Acta Physiol. Plant. 2015, 37, 156. [Google Scholar] [CrossRef]
- Vardhini, B.V.; Anjum, N.A. Brassinosteroids make plant life easier under abiotic stresses mainly by modulating major components of antioxidant defense system. Front. Environ. Sci. 2015, 2, 67. [Google Scholar] [CrossRef]
- Christian, Z.; Geilfus, C.M.; Mühling, K.H.; Jutta, L.-M. The influence of salt stress on ABA and auxin concentrations in two maize cultivars differing in salt resistance. J. Plant Physiol. 2013, 170, 220–224. [Google Scholar] [CrossRef]
- Liu, W.; Li, R.; Han, T.; Cai, W.; Fu, Z.; Lu, Y. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Zhou, T.; Chen, Y.; Han, N.; Zhu, M.; Bian, H.; Wang, J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2019, 125, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Sarmad, J.; Shariati, M.; Haghjou, M.M. Relationship between endogenous abscisic acid and B-carotene synthesis in unicellular green Alga Dunaliella. Am. Eurasian J. Agric. Environ. Sci. 2007, 2, 559–564. [Google Scholar]
- Zhang, M.; Zhao, J. The Arabidopsis U-box E3 ubiquitin ligase PUB30 negatively regulates salt tolerance by facilitating BRI1 kinase inhibitor 1 (BKI1) degradation. Plant Cell Environ. 2017, 40, 2831–2843. [Google Scholar] [CrossRef]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C.; Ma, Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 2012, 426, 522–527. [Google Scholar] [CrossRef]
- Kerstiens, G.; Tych, W.; Robinson, M.F.; Mansfield, T.A. Special Issue: Stomata. Sodium-Related Partial Stomatal Closure and Salt Tolerance of Aster tripolium. New Phytol. 2002, 153, 509–515. [Google Scholar] [CrossRef]
- Kerstiens, G. Cuticular water permeability and its physiological significance. J. Exp. Bot. 1996, 47, 1813–1832. [Google Scholar] [CrossRef]
- Yan, H.; Shah, S.S.; Zhao, W.; Liu, F. Variations in water relations, stomatal characteristics, and plant growth between quinoa and pea under salt-stress conditions. Pak. J. Bot. 2020, 52, 1–7. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, S.; Mao, T.; Qu, X.; Cao, W.; Zhang, L.; Zhang, W.; He, L.; Li, S.; Ren, S.; et al. The Plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 2011, 23, 2314–2330. [Google Scholar] [CrossRef]
- Sierla, M.; Hõrak, H.; Overmyer, K.; Waszczak, C.; Yarmolinsky, D.; Maierhofer, T.; Vainonen, J.P.; Salojärvi, J.; Denessiouk, K.; Laanemets, K.; et al. The receptor-like pseudokinase GHR1 is required for stomatal closure. Plant Cell 2018, 30, 2813–2837. [Google Scholar] [CrossRef]
- Cunxu, W.; Jianbo, W.; Yifang, C.; Weidong, Z.; Guorong, S. Epicuticular wax of leaf epidermis: A functional structure for salt excretion in a halophyte Puccinellia tenuiflora. Acta Ecol. Sin. 2004, 24, 2451–2456. [Google Scholar]
- Fahn, A. Secretory tissues in vascular plants. New Phytol. 1988, 108, 229–257. [Google Scholar] [CrossRef]
- Wang, J.; Meng, Y.; Li, B.; Ma, X.; Lai, Y.; Si, E.; Yang, K.; Xu, X.; Shang, X.; Wang, H. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ. 2015, 38, 655–669. [Google Scholar] [CrossRef]
- Morel, F.; Lucarain, C. A flame spectrophotometer to determine sodium and potassium in biological samples of the nanoliter order. J. Physiol. 1967, 59, 460–461. [Google Scholar]
- Gama, P.B.S.; Tanaka, K.; Eneji, A.E.; Eltayeb, A.E.; Siddig, K.E. Salt-induced stress effects on biomass, photosynthetic rate, and reactive oxygen species-scavenging enzyme accumulation in common bean. J. Plant Nutr. 2009, 32, 837–854. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. The assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 764–775. [Google Scholar] [CrossRef]
- Tomita, K. Assay of Catalase Activity Based on the Absorption of Ultraviolet Light by H2O2; Kasanka Shishitsu Jikkenho: Tokyo, Japan, 1983; pp. 149–150. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Xie, B.; Rao, Z.; Xiang, J.; Yang, Y.; Yuan, J.; Yin, J.; Yang, Y.; Yi, M.; Cao, Y. Evaluation of uncertainty of the determination of soluble sugar in tobacco by anthrone colorimetry. China Meas. Test 2015, S1, 10–13. [Google Scholar]
- Ábrahám, E.; Hourton-Cabassa, C.; Erdei, L.; Szabados, L. Methods for determination of proline in plants. Plant Stress Toler. 2010, 639, 317–333. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Bel, M.V.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Peer, Y.V.D.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment analysis for Gene Ontology; R Package Version. 2006. Available online: https://rdrr.io/bioc/topGO/ (accessed on 27 March 2020).
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Wu, J.; Mao, X.; Tao, C.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Hong, S.Y.; Seo, P.J.; Yang, M.-S.; Xiang, F.; Park, C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]

| Sample Name | Sample Description | Total Reads | Total Mapped | Mapped Ratio of Mapped Reads | Q30 |
|---|---|---|---|---|---|
| CK_1_S | Control replication 1 | 59,206,904 | 56,875,374 | 96.06% | 95.18% |
| CK_2_S | Control replication 2 | 63,617,088 | 60,841,214 | 95.64% | 95.2% |
| CK_3_S | Control replication 3 | 57,536,132 | 55,180,960 | 95.91% | 95.26% |
| SALT_1_S | Salt stress replication 1 | 55,294,072 | 53,503,005 | 96.76% | 94.68% |
| SALT_2_S | Salt stress replication 2 | 65,989,512 | 63,143,273 | 95.69% | 95% |
| SALT_3_S | Salt stress replication 3 | 47,315,154 | 45,175,264 | 95.48% | 94.49% |
| GENE_ID | L2fc | Padj | BP Description |
|---|---|---|---|
| Up_regulated | |||
| BRADI2G30490 | 4.697967282 | 2.88 × 10−52 | |
| BRADI3G35310 | 3.591061835 | 8.98 × 10−38 | oxidation–reduction process |
| BRADI2G52155 | 6.171188364 | 1.49 × 10−34 | |
| BRADI1G34670 | 3.484127137 | 9.73 × 10−34 | |
| BRADI4G18910 | 3.643708291 | 1.86 × 10−30 | |
| BRADI1G04690 | 3.443854612 | 4.47 × 10−30 | |
| BRADI3G20677 | 9.241758235 | 2.61 × 10−29 | oxidation–reduction process |
| BRADI1G59120 | 4.055819532 | 3.72 × 10−28 | |
| BRADI2G21340 | 5.343143653 | 1.36 × 10−27 | |
| BRADI2G37720 | 2.567633743 | 1.28 × 10−26 | |
| BRADI1G44920 | 6.810598815 | 2.53 × 10−26 | |
| BRADI3G49310 | 5.355939545 | 2.80 × 10−26 | |
| BRADI3G19620 | 2.376259459 | 1.10 × 10−24 | |
| BRADI1G10150 | 1.888142734 | 5.54 × 10−24 | |
| BRADI4G01200 | 1.607591434 | 5.69 × 10−24 | metabolic process |
| Down_regulated | |||
| BRADI3G53660 | −3.245924936 | 4.07 × 10−33 | protein phosphorylation, metabolic process |
| BRADI2G44820 | −4.450443575 | 8.69 × 10−33 | metabolic process |
| BRADI3G31720 | −7.041083206 | 4.16 × 10−31 | |
| BRADI3G31727 | −5.153494722 | 5.28 × 10−31 | |
| BRADI2G19540 | −1.771707314 | 3.15 × 10−22 | protein metabolic process |
| BRADI1G74980 | −6.034665237 | 5.24 × 10−22 | |
| BRADI1G53680 | −7.436218542 | 4.34 × 10−20 | ion transmembrane transport |
| BRADI5G02750 | −2.879558911 | 2.44 × 10−19 | transmembrane transport |
| BRADI2G22520 | −8.740289045 | 9.92 × 10−19 | ion transmembrane transport |
| BRADI2G33110 | −3.576800337 | 7.04 × 10−18 | ion transmembrane transport |
| BRADI1G60250 | −3.534466753 | 1.86 × 10−17 | |
| BRADI2G44830 | −2.884382952 | 2.90 × 10−17 | protein phosphorylation, metabolic process |
| BRADI2G33950 | −5.142993438 | 2.91 × 10−17 | |
| BRADI2G19360 | −1.97415848 | 5.62 × 10−17 | |
| BRADI3G06320 | −5.743267815 | 1.17 × 10−15 | oxidation–reduction process |
| GO ID | Term | p Value |
|---|---|---|
| Up_regulated | ||
| GO:0009834 | plant-type secondary cell wall biogenesis | 4.20 × 10−9 |
| GO:0042744 | hydrogen peroxide catabolic process | 7.50 × 10−8 |
| GO:0055114 | oxidation-reduction process | 3.20 × 10−7 |
| GO:0046274 | lignin catabolic process | 8.60 × 10−6 |
| GO:0010030 | positive regulation of seed germination | 3.60 × 10−5 |
| GO:0006979 | response to oxidative stress | 4.30 × 10−5 |
| GO:0007017 | microtubule-based process | 4.40 × 10−5 |
| GO:0034605 | cellular response to heat | 8.20 × 10−5 |
| GO:0010143 | cutin biosynthetic process | 1.1 × 10−4 |
| GO:0030244 | cellulose biosynthetic process | 1.70 × 10−4 |
| GO:0048640 | negative regulation of developmental growth | 1.90 × 10−4 |
| GO:0006869 | lipid transport | 2.00 × 10−4 |
| GO:0045492 | xylan biosynthetic process | 2.20 × 10−4 |
| GO:0070592 | cell wall polysaccharide biosynthetic process | 2.30 × 10−4 |
| GO:0097435 | supramolecular fiber organization | 2.40 × 10−4 |
| Down_regulated | ||
| GO:0034605 | cellular response to heat | 2.10 × 10−7 |
| GO:0010030 | positive regulation of seed germination | 1.40 × 10−6 |
| GO:0071577 | zinc ion transmembrane transport | 7.80 × 10−5 |
| GO:0042744 | hydrogen peroxide catabolic process | 8.70 × 10−5 |
| GO:0006414 | translational elongation | 2.10 × 10−4 |
| GO:0009405 | pathogenesis | 5.60 × 10−4 |
| GO:0010106 | cellular response to iron ion starvation | 5.60 × 10−4 |
| GO:0034620 | cellular response to unfolded protein | 1.10 × 10−3 |
| GO:0071486 | cellular response to high light intensity | 1.42 × 10−3 |
| GO:0006826 | iron ion transport | 1.66 × 10−3 |
| GO:0071668 | plant-type cell wall assembly | 1.83 × 10−3 |
| GO:0000302 | response to reactive oxygen species | 2.44 × 10−3 |
| GO:0071492 | cellular response to UV-A | 2.46 × 10−3 |
| GO:0044264 | cellular polysaccharide metabolic process | 2.83 × 10−3 |
| GO:0010045 | response to nickel cation | 4.98 × 10−3 |
| TF Family | Up Gene Num. | Down Gene Num. |
|---|---|---|
| bHLH | 34 | 14 |
| MYB | 36 | 4 |
| AP2/ERF | 19 | 6 |
| HB | 15 | 3 |
| bZIP | 12 | 3 |
| NAC | 9 | 6 |
| C2C2 | 9 | 4 |
| C2H2 | 9 | 4 |
| WRKY | 7 | 4 |
| OFP | 10 | 0 |
| Tify | 8 | 0 |
| GARP | 2 | 5 |
| HSF | 1 | 6 |
| B3 | 6 | 0 |
| GRAS | 2 | 4 |
| C3H | 3 | 2 |
| MADS | 3 | 2 |
| TCP | 5 | 0 |
| Trihelix | 5 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Wang, Q.; Liu, Y.; Zhang, X.; Zhang, L.; Fan, S. Screening of Salt Stress Responsive Genes in Brachypodium distachyon (L.) Beauv. by Transcriptome Analysis. Plants 2020, 9, 1522. https://doi.org/10.3390/plants9111522
Guo X, Wang Q, Liu Y, Zhang X, Zhang L, Fan S. Screening of Salt Stress Responsive Genes in Brachypodium distachyon (L.) Beauv. by Transcriptome Analysis. Plants. 2020; 9(11):1522. https://doi.org/10.3390/plants9111522
Chicago/Turabian StyleGuo, Xiuxiu, Qingjun Wang, Yuan Liu, Xuejie Zhang, Luoyan Zhang, and Shoujin Fan. 2020. "Screening of Salt Stress Responsive Genes in Brachypodium distachyon (L.) Beauv. by Transcriptome Analysis" Plants 9, no. 11: 1522. https://doi.org/10.3390/plants9111522
APA StyleGuo, X., Wang, Q., Liu, Y., Zhang, X., Zhang, L., & Fan, S. (2020). Screening of Salt Stress Responsive Genes in Brachypodium distachyon (L.) Beauv. by Transcriptome Analysis. Plants, 9(11), 1522. https://doi.org/10.3390/plants9111522





