Phylogeny, Taxonomy, and Biogeography of Pterocarya (Juglandaceae)
Abstract
1. Introduction
2. Results
2.1. RAD-seq and Data Matrices for Phylogenetic Inference
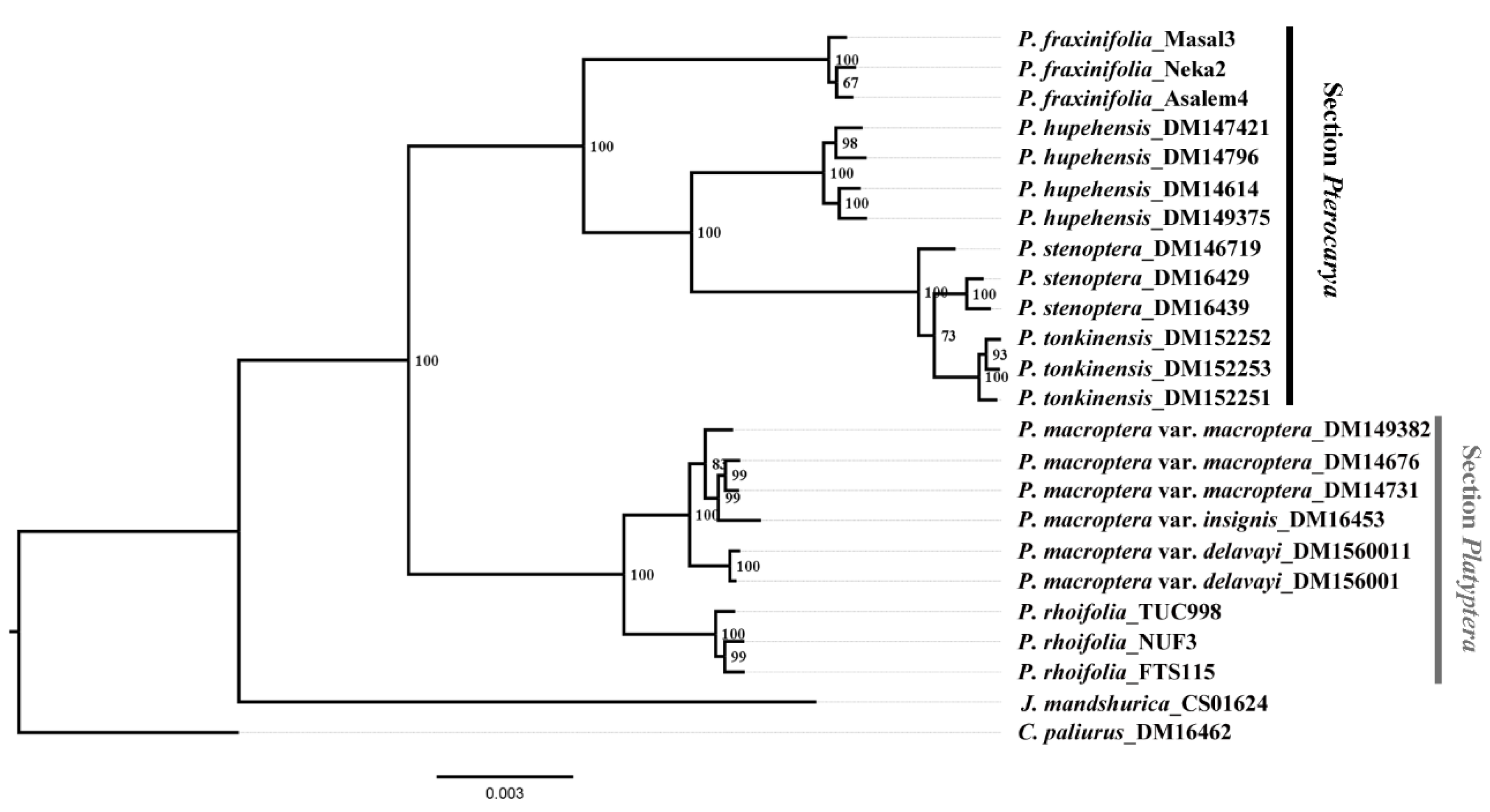
2.2. RAD-seq Phylogenetic Reconstruction
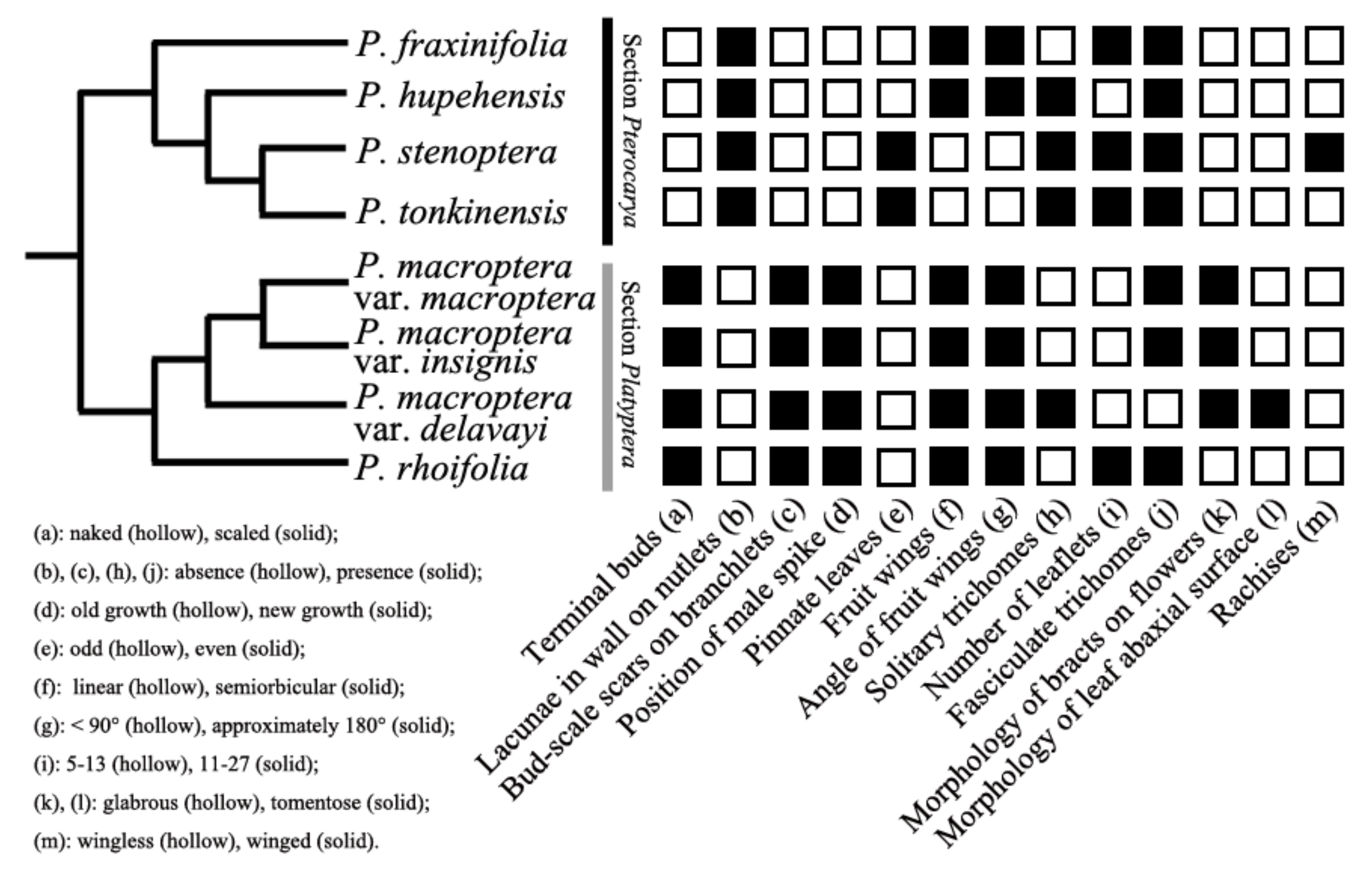
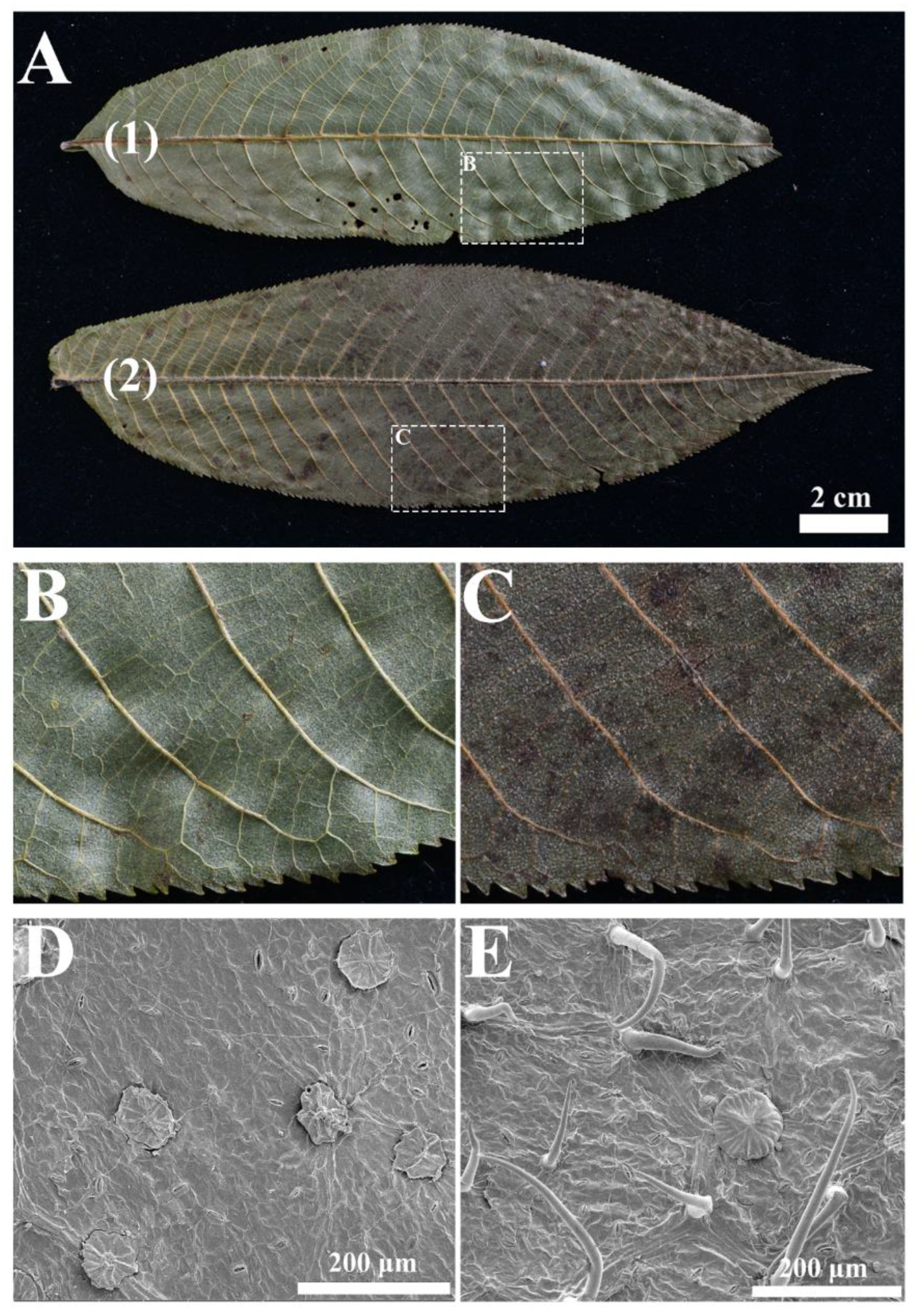
2.3. Morphological Traits and Taxonomic Conclusions
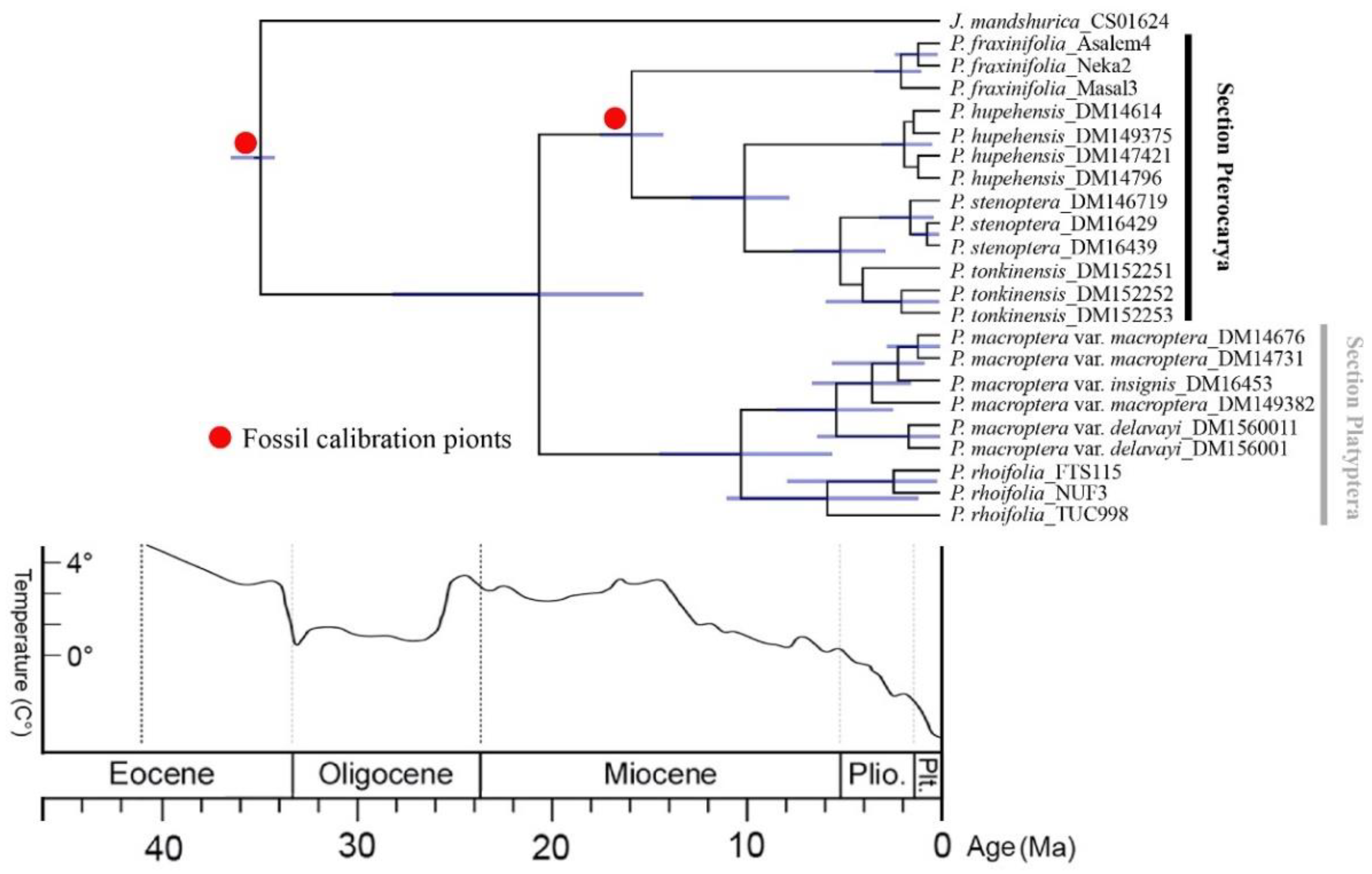
2.4. Estimation of Divergence Times
3. Discussion
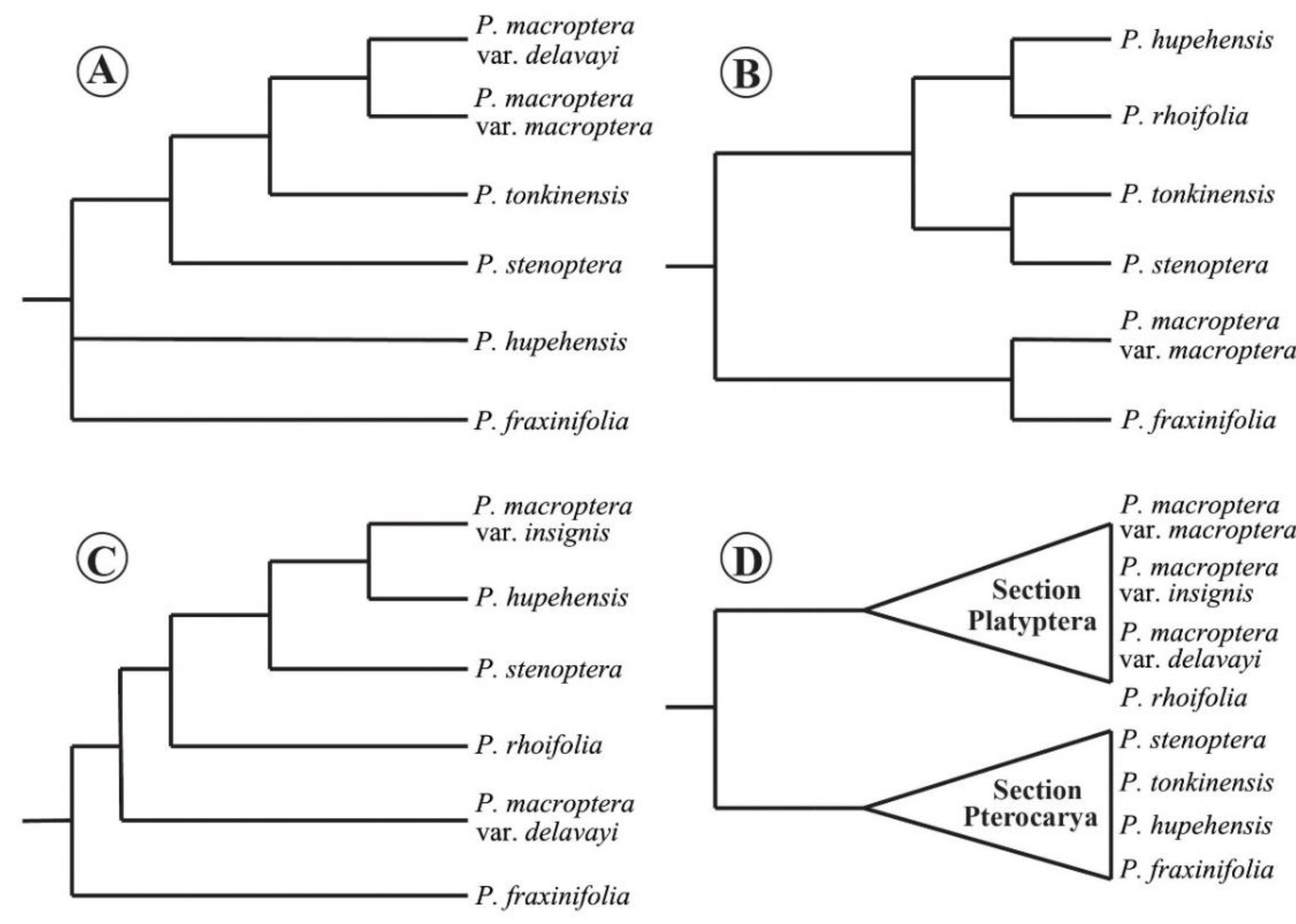
3.1. Phylogenetic Hypothesis for Pterocarya
3.2. Taxonomic Implications and Evolutionary Importance of Morphological Features
3.3. East Asian versus Southern European/West Asian Disjunctions of Relict Trees: The Importance of the Gobi Desert’s Formation and Climatic Cooling after the Middle Miocene Epoch
3.4. Late Miocene Diversification in the East Asian Refugium
4. Materials and Methods
4.1. Taxon Sampling and DNA Extraction
4.2. RAD-seq Library Preparation
4.3. Processing and Clustering RAD-seq Data
4.4. Morphological Evaluations and Data Sets
4.5. Phylogenetic Reconstruction
4.6. Fossil Constraints and Estimations of Divergence Times
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Donoghue, M.J.; Bell, C.D.; Li, J.H. Phylogenetic patterns in Northern Hemisphere plant geography. Int. J. Plant Sci. 2001, 162, S41–S52. [Google Scholar] [CrossRef]
- Milne, R.I.; Abbott, R.J. The origin and evolution of tertiary relict floras. Adv. Bot. Res. 2002, 38, 281–314. [Google Scholar]
- Mao, K.S.; Hao, G.; Liu, J.Q.; Adams, R.P.; Milne, R.I. Diversification and biogeography of Juniperus (Cupressaceae): Variable diversification rates and multiple intercontinental dispersals. New Phytol. 2010, 188, 254–272. [Google Scholar] [CrossRef]
- Raven, P.H. Plant species disjunctions: A summary. Ann. Mo. Bot. Gard. 1972, 59, 234–246. [Google Scholar] [CrossRef]
- Thorne, R.F. Major disjunctions in the geographic ranges of seed plants. Q. Rev. Biol. 1972, 47, 365–411. [Google Scholar] [CrossRef]
- Tiffney, B.H. The Eocene North Atlantic land bridge: Its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J. Arnold Arbor. 1985, 66, 243–273. [Google Scholar] [CrossRef]
- Milne, R.I. Northern hemisphere plant disjunctions: A window on tertiary land bridges and climate change? Ann. Bot. 2006, 98, 465–472. [Google Scholar] [CrossRef]
- Wen, J. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Evol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]
- Wen, J. Evolution of eastern Asian-eastern North American biogeographic disjunctions: A few additional issues. Int. J. Plant Sci. 2001, 162, S117–S122. [Google Scholar] [CrossRef]
- Xiang, Q.Y.; Soltis, D.E.; Soltis, P.S.; Manchester, S.R.; Crawford, D.J. Timing the eastern Asian-Eastern North American floristic disjunction: Molecular clock corroborates paleontological estimates. Mol. Phylogenet. Evol. 2000, 15, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.Y.; Zhang, W.H.; Ricklefs, R.E. Regional differences in rates of plant speciation and molecular evolution: A comparison between eastern Asia and eastern North America. Evolution 2004, 58, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Nie, Z.L.; Ickert-Bond, S.M. Advances in biogeography in the age of a new modern synthesis. J. Syst. Evol. 2019, 57, 543–546. [Google Scholar] [CrossRef]
- Wu, Z. On the significance of Pacific intercontinental discontinuity. Ann. Mo. Bot. Gard. 1983, 70, 577–590. [Google Scholar]
- Wen, J.; Nie, Z.L.; Ickert-Bond, S.M. Intercontinental disjunctions between eastern Asia and western North America in vascular plants highlight the biogeographic importance of the Bering land bridge from late Cretaceous to Neogene. J. Syst. Evol. 2016, 54, 469–490. [Google Scholar] [CrossRef]
- Xiang, Q.Y.; Soltis, D.E.; Soltis, P.S. The eastern Asian and eastern and western North America floristic disjunction: Congruent phylogenetic patterns in seven diverse genera. Mol. Phylogenet. Evol. 1998, 10, 178–190. [Google Scholar] [CrossRef]
- Wen, J.; Ickert-Bond, S.M. Evolution of the Madren-Tethyan disjunctions and the North and South American amphitropical disjunctions in plants. J. Syst. Evol. 2009, 47, 331–348. [Google Scholar] [CrossRef]
- Donoghue, M.J.; Smith, S.A. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1633–1644. [Google Scholar] [CrossRef]
- Manchester, S.R. Biogeographical relationships of North American tertiary floras. Ann. Mo. Bot. Gard. 1999, 82, 472–522. [Google Scholar] [CrossRef]
- Ickert-Bond, S.M.; Wen, J. Phylogeny and biogeography of Altingiaceae: Evidence from combined analysis of five non-coding chloroplast regions. Mol. Phylogenet. Evol. 2006, 39, 512–528. [Google Scholar] [CrossRef]
- Li, J.H.; Tredici, P.D. The Chinese Parrotia: A sibling species of the Persian Parrotia. Arnoldia 2008, 66, 2–9. [Google Scholar]
- Li, J.; Stukel, M.; Bussies, P.; Skinner, K.; Lemmon, A.R.; Lemmon, E.M.; Brown, K.; Bekmetjev, A.; Swenson, N.G. Maple phylogeny and biogeography inferred from phylogenomic data. J. Syst. Evol. 2019, 57, 594–606. [Google Scholar] [CrossRef]
- Naciri, Y.; Christe, C.; Betrisey, S.; Song, Y.G.; Deng, M.; Garfi, G.; Kozlowski, G. Species delimation in the East Asian species of the relict tree genus Zelkova (Ulmaceae): A complex history of diversification and admixture among species. Mol. Phylogenet. Evol. 2019, 134, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.X.; Fu, C.X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Zhou, Z.; Hong, D.Y.; Niu, Y.; Li, G.D.; Nie, Z.L.; Wen, J.; Sun, H. Phylogenetic and biogeographic analyses of the Sino-Himalayan endemic genus Cyananthus (Campanulaceae) and implications for the evolution of its sexual system. Mol. Phylogenet. Evol. 2013, 68, 482–497. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.X.; Cheng, S.M.; Tian, S.; Li, B.; Fan, D.M.; Chen, Y.J.; Soltis, D.E.; Soltis, P.S.; Zhang, Z.Y. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J. Biogeogr. 2016, 43, 351–360. [Google Scholar] [CrossRef]
- Du, F.K.; Hou, M.; Wang, W.T.; Mao, K.S.; Hampe, A. Phylogeography of Quercus aquifolioides provides novel insights into the Neogene history of a major global hotspot of plant diversity in south-west China. J. Biogeogr. 2017, 44, 294–307. [Google Scholar] [CrossRef]
- Qiu, Y.X.; Guan, B.C.; Fu, C.X.; Comes, H.P. Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol. Phylogenet. Evol. 2009, 51, 281–293. [Google Scholar] [CrossRef]
- Qiu, Y.X.; Sun, Y.; Zhang, X.P.; Lee, J.; Fu, C.X.; Comes, H.P. Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to Quaternary climate change and landbridge configurations. New Phytol. 2009, 183, 480–495. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.S.; Chen, C.; Comes, H.P.; Sakaguchi, S.; Liu, Y.H.; Tanaka, N.; Sakio, H.; Qiu, Y.X. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol. 2012, 196, 617–630. [Google Scholar] [CrossRef]
- Bai, W.N.; Wang, W.T.; Zhang, D.Y. Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytol. 2016, 209, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.Q.; Hu, H.Y.; Xie, C.; Lai, S.P.; Yang, M.; He, X.J.; Zhou, S.D. Molecular phylogeny, biogeography and ecological niche modelling of Cardiocrinum (Liliaceae): Insights into the evolutionary history of endemic genera distributed across the Sino-Japanese floristic region. Ann. Bot. 2017, 119, 59–72. [Google Scholar] [CrossRef]
- Ye, J.W.; Bai, W.N.; Bao, L.; Wang, T.M.; Wang, H.F.; Ge, J.P. Sharp genetic discontinuity in the aridity-sensitive Lindera obtusiloba (Lauraceae): Solid evidence supporting the Tertiary floral subdivision in East Asia. J. Biogeogr. 2017, 44, 2082–2095. [Google Scholar] [CrossRef]
- Kuang, K.R.; Zheng, S.X.; Li, P.Q.; Lu, A.M. Myricaceae, Juglandaceae and Betulaceae. In Flora of China (Chinese Version); Wu, Z.Y., Ed.; Science Press: Beijing, China, 1979; Volume 21, pp. 21–30. [Google Scholar]
- Lu, A.M.; Donald, E.S.; Grauke, L.J. Juglandaceae. In Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press & Missouri Botanical Garden Press: Beijing, China; St. Louis, MO, USA, 1999; Volume 4, pp. 277–285. [Google Scholar]
- Maharramova, E.; Huseynova, I.; Kolbaia, S.; Gruenstaeudl, M.; Borsch, T.; Muller, L.A. Phylogeography and population genetics of the riparian relict tree Pterocarya fraxinifolia (Juglandaceae) in the South Caucasus. Syst. Biodivers. 2018, 16, 14–27. [Google Scholar] [CrossRef]
- Manchester, S.R. The Fossil History of the Juglandaceae. Mongraphs in Systematic Botany from the Missouri Botanical Garden; Allen Press: Lawrence, KS, USA, 1987; Volume 21, pp. 1–137. [Google Scholar]
- Kozlowski, G.; Betrisey, S.; Song, Y.G. Wingnuts (Pterocarya) and Walnut Family: Relict Trees: Linking the Past, Present and Future; Natural History Museum Fribourg: Fribourg, Switzerland, 2018; pp. 1–59. [Google Scholar]
- Song, Y.G.; Fragnière, Y.; Meng, H.H.; Li, Y.; Bétrisey, S.; Corrales, A.; Manchester, S.; Deng, M.; Jasińska, A.K.; Sâm, H.V.; et al. Global biogeographic synthesis and priority conservation regions of the relict tree family Juglandaceae. J. Biogeogr. 2020, 47, 643–657. [Google Scholar] [CrossRef]
- Tkach, N.V.; Hoffmann, M.H.; Roser, M.; Korobkov, A.A.; Von Hagen, K.B. Parallel evolutionary patterns in multiple lineages of arctic Artemisia L. (Asteraceae). Evolution 2008, 62, 184–198. [Google Scholar] [CrossRef]
- Schwery, O.; Onstein, R.E.; Bouchenak-Khelladi, Y.; Xing, Y.W.; Carter, R.J.; Linder, H.P. As old as the mountains: The radiations of the Ericaceae. New Phytol. 2015, 207, 355–367. [Google Scholar] [CrossRef]
- Massatti, R.; Reznicek, A.A.; Knowles, L.L. Utilizing RADseq data for phylogenetic analysis of challenging taxonomic groups: A case study in Carex sect. Racemosae. Am. J. Bot. 2016, 103, 337–347. [Google Scholar] [CrossRef]
- Manos, P.S.; Stone, D.E. Evolution, phylogeny, and systematics of the Juglandaceae. Ann. Mo. Bot. Gard. 2001, 88, 231–269. [Google Scholar] [CrossRef]
- Xiang, X.G.; Wang, W.; Li, R.Q.; Lin, L.; Liu, Y.; Zhou, Z.K.; Li, Z.Y.; Chen, Z.D. Large-scale phylogenetic analyses reveal fagalean diversification promoted by the interplay of diaspores and environments in the Paleogene. Perspect. Plant Ecol. 2014, 16, 101–110. [Google Scholar] [CrossRef]
- Xing, Y.W.; Onstein, R.E.; Carter, R.J.; Stadler, T.; Linder, H.P. Fossils and a large molecular phylogeny show that the evolution of species richness, generic diversity, and turnover rates are disconnected. Evolution 2014, 68, 2821–2832. [Google Scholar] [CrossRef]
- Mostajeran, F.; Yousefzadeh, H.; Davitashvili, N.; Kozlowski, G.; Akbarinia, M. Phylogenetic relationships of Pterocarya (Juglandaceae) with an emphasis on the taxonomic status of Iranian populations using ITS and trnH-psbA sequence data. Plant Biosyst. 2016, 151, 1012–1021. [Google Scholar] [CrossRef]
- Miller, M.R.; Dunham, J.P.; Amores, A.; Cresko, W.A.; Johnson, E.A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007, 17, 240–248. [Google Scholar] [CrossRef]
- Rowe, H.C.; Renaut, S.; Guggisberg, A. RAD in the realm of next-generation sequencing technologies. Mol. Ecol. 2011, 20, 3499–3502. [Google Scholar] [CrossRef]
- Hipp, A.L.; Eaton, D.A.R.; Cavender-Bares, J.; Fitzek, E.; Nipper, R.; Manos, P.S. A framework phylogeny of the American oak clade based on sequenced RAD data. PLoS ONE 2014, 9, e93975. [Google Scholar] [CrossRef]
- Razkin, O.; Sonet, G.; Breugelmans, K.; Madeira, M.J.; Gomez-Moliner, B.J.; Backeljau, T. Species limits, interspecific hybridization and phylogeny in the cryptic land snail complex Pyramidula: The power of RADseq data. Mol. Phylogenet. Evol. 2016, 101, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.A.R.; Ree, R.H. Inferring Phylogeny and Introgression using RADseq Data: An Example from Flowering Plants (Pedicularis: Orobanchaceae). Syst. Biol. 2013, 62, 689–706. [Google Scholar] [CrossRef]
- Cruaud, A.; Gautier, M.; Galan, M.; Foucaud, J.; Saune, L.; Genson, G.; Dubois, E.; Nidelet, S.; Deuve, T.; Rasplus, J.Y. Empirical Assessment of RAD Sequencing for Interspecific Phylogeny. Mol. Biol. Evol. 2014, 31, 1272–1274. [Google Scholar] [CrossRef]
- Liu, L.X.; Jin, X.J.; Chen, N.; Li, X.; Li, P.; Fu, C.X. Phylogeny of Morella rubra and its relatives (Myricaceae) and genetic resources of Chinese bayberry using RAD sequencing. PLoS ONE 2015, 10, e0139840. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Mu, X.Y.; Tong, L.; Sun, M.; Zhu, Y.X.; Wen, J.; Lin, Q.W.; Liu, B. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol. Phylogenet. Evol. 2020, 147, 106802. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Low, S.L.; Song, Y.G.; Nurainas; Kozlowski, G.; Do, T.V.; Li, L.; Zhou, S.S.; Tan, Y.H.; Cao, G.L.; et al. Shining a light on species delimitation in the tree genus Engelhardia Leschenault ex Blume (Juglandaceae). Mol. Phylogenet. Evol. 2020, 152, 106918. [Google Scholar] [CrossRef]
- Wolfe, J.A. Some aspects of plant geography of Northern Hemisphere during late Cretaceous and Tertiary. Ann. Mo. Bot. Gard. 1975, 62, 264–279. [Google Scholar] [CrossRef]
- Kozlowski, G.; Gratzfeld, J. Zelkova—An Ancient Tree: Global Status and Conservation Action; Natural History Museum Fribourg: Fribourg, Switzerland, 2013; pp. 1–60. [Google Scholar]
- Ickert-Bond, S.M.; Pigg, K.B.; Wen, J. Comparative infructescence morphology in Liquidambar (Altingiaceae) and its evolutionary significance. Am. J. Bot. 2005, 92, 1234–1255. [Google Scholar] [CrossRef]
- Jia, D.R.; Bartish, I.V. Climatic changes and orogeneses in the Late Miocene of Eurasia: The main triggers of an expansion at a continental scale? Front. Plant Sci. 2018, 9, 1400. [Google Scholar] [CrossRef]
- Lu, H.Y.; Wang, X.Y.; Wang, X.Y.; Chang, X.; Zhang, H.Z.; Xu, Z.W.; Zhang, W.C.; Wei, H.Z.; Zhang, X.J.; Yi, S.W.; et al. Formation and evolution of Gobi Desert in central and eastern Asia. Earth Sci. Rev. 2019, 194, 251–263. [Google Scholar] [CrossRef]
- Bosboom, R.E.; Abels, H.A.; Hoorn, C.; Van den Berg, B.C.J.; Guo, Z.; Dupont-Nivet, G. Aridification in continental Asia after the Middle Eocene Climatic Optimum (MECO). Earth Planet. Sci. Lett. 2014, 389, 34–42. [Google Scholar] [CrossRef]
- Carrapa, B.; DeCelles, P.G.; Wang, X.; Clementz, M.T.; Mancin, N.; Stoica, M.; Kraatz, B.; Meng, J.; Abdulov, S.; Chen, F.H. Tectono-climatic implications of Eocene Paratethys regression in the Tajik basin of central Asia. Earth Planet. Sci. Lett. 2015, 424, 168–178. [Google Scholar] [CrossRef]
- Zheng, H.B.; Wei, X.C.; Tada, R.J.; Clift, P.D.; Wang, B.; Jourdan, F.; Wang, P.; He, M.Y. Late Oligocene-early Miocene birth of the Taklimakan Desert. Proc. Natl. Acad. Sci. USA 2015, 112, 7662–7667. [Google Scholar] [CrossRef]
- Li, J.X.; Yue, L.P.; Roberts, A.P.; Hirt, A.M.; Pan, F.; Guo, L.; Xu, Y.; Xi, R.G.; Guo, L.; Qiang, X.K.; et al. Global cooling and enhanced Eocene Asian mid-latitude interior aridity. Nat. Commun. 2018, 9, 3026. [Google Scholar] [CrossRef]
- Cao, Y.N.; Comes, H.P.; Sakaguchi, S.; Chen, L.Y.; Qiu, Y.X. Evolution of East Asia’s Arcto-Tertiary relict Euptelea (Eupteleaceae) shaped by Late Neogene vicariance and Quaternary climate change. BMC Evol. Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Eaton, D.A.R. PyRAD: Assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 2014, 30, 1844–1849. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genome-sequencing projects. Mol. Biol. Evol. 2008, 25, 2409–2419. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Deng, M.; Hipp, A.; Song, Y.G.; Li, Q.S.; Coombes, A.; Cotton, A. Leaf epidermal features of Quercus subgenus Cyclobalanopsis (Fagaceae) and their systematic significance. Biol. J. Linn. Soc. 2014, 176, 224–259. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Brummond, A.T. Tracer v1.5. 2007. Available online: http://tree.bio.ed.ac.uk/software/tracer/ (accessed on 11 October 2019).
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]

| Summary Statistic | Raw Reads | Clean Reads | Total Length of Clean Reads (Gbp) | Clean Data Percentage (%) | Q30 Percentage (%) | GC Percentage (%) |
|---|---|---|---|---|---|---|
| Average | 11,055,000 | 9,947,083 | 1.54 | 84.30 | 91.54 | 47.10 |
| Maximum | 18,580,000 | 17,290,000 | 2.36 | 90.02 | 92.39 | 58.57 |
| Minimum | 4,780,000 | 3,790,000 | 0.69 | 76.16 | 88.55 | 43.03 |
| SD | 3,232,889 | 3,250,324 | 0.37 | 3.64 | 0.88 | 3.91 |
| Summary Statistic | RAD Tags (R1) | Total Clusters (R1) | Mean Depth of Clusters | H | E | Consensus Loci | Loci in Final Data Set |
|---|---|---|---|---|---|---|---|
| Average | 5,502,955 | 1,728,343 | 15.67 | 0.0413 | 0.0103 | 102,981 | 9287 |
| Maximum | 8,591,043 | 3,985,579 | 17.75 | 0.0526 | 0.0136 | 204,925 | 13,650 |
| Minimum | 2,495,755 | 769,873 | 13.04 | 0.0350 | 0.0075 | 38,695 | 4222 |
| SD | 1,316,019 | 733,868 | 1.11 | 0.0043 | 0.0016 | 39,940 | 2668 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.-G.; Li, Y.; Meng, H.-H.; Fragnière, Y.; Ge, B.-J.; Sakio, H.; Yousefzadeh, H.; Bétrisey, S.; Kozlowski, G. Phylogeny, Taxonomy, and Biogeography of Pterocarya (Juglandaceae). Plants 2020, 9, 1524. https://doi.org/10.3390/plants9111524
Song Y-G, Li Y, Meng H-H, Fragnière Y, Ge B-J, Sakio H, Yousefzadeh H, Bétrisey S, Kozlowski G. Phylogeny, Taxonomy, and Biogeography of Pterocarya (Juglandaceae). Plants. 2020; 9(11):1524. https://doi.org/10.3390/plants9111524
Chicago/Turabian StyleSong, Yi-Gang, Ying Li, Hong-Hu Meng, Yann Fragnière, Bin-Jie Ge, Hitoshi Sakio, Hamed Yousefzadeh, Sébastien Bétrisey, and Gregor Kozlowski. 2020. "Phylogeny, Taxonomy, and Biogeography of Pterocarya (Juglandaceae)" Plants 9, no. 11: 1524. https://doi.org/10.3390/plants9111524
APA StyleSong, Y.-G., Li, Y., Meng, H.-H., Fragnière, Y., Ge, B.-J., Sakio, H., Yousefzadeh, H., Bétrisey, S., & Kozlowski, G. (2020). Phylogeny, Taxonomy, and Biogeography of Pterocarya (Juglandaceae). Plants, 9(11), 1524. https://doi.org/10.3390/plants9111524









