Molecular Detection of Potato Viruses in Bangladesh and Their Phylogenetic Analysis
Abstract
1. Introduction
2. Results
2.1. Detection of Potato Viruses
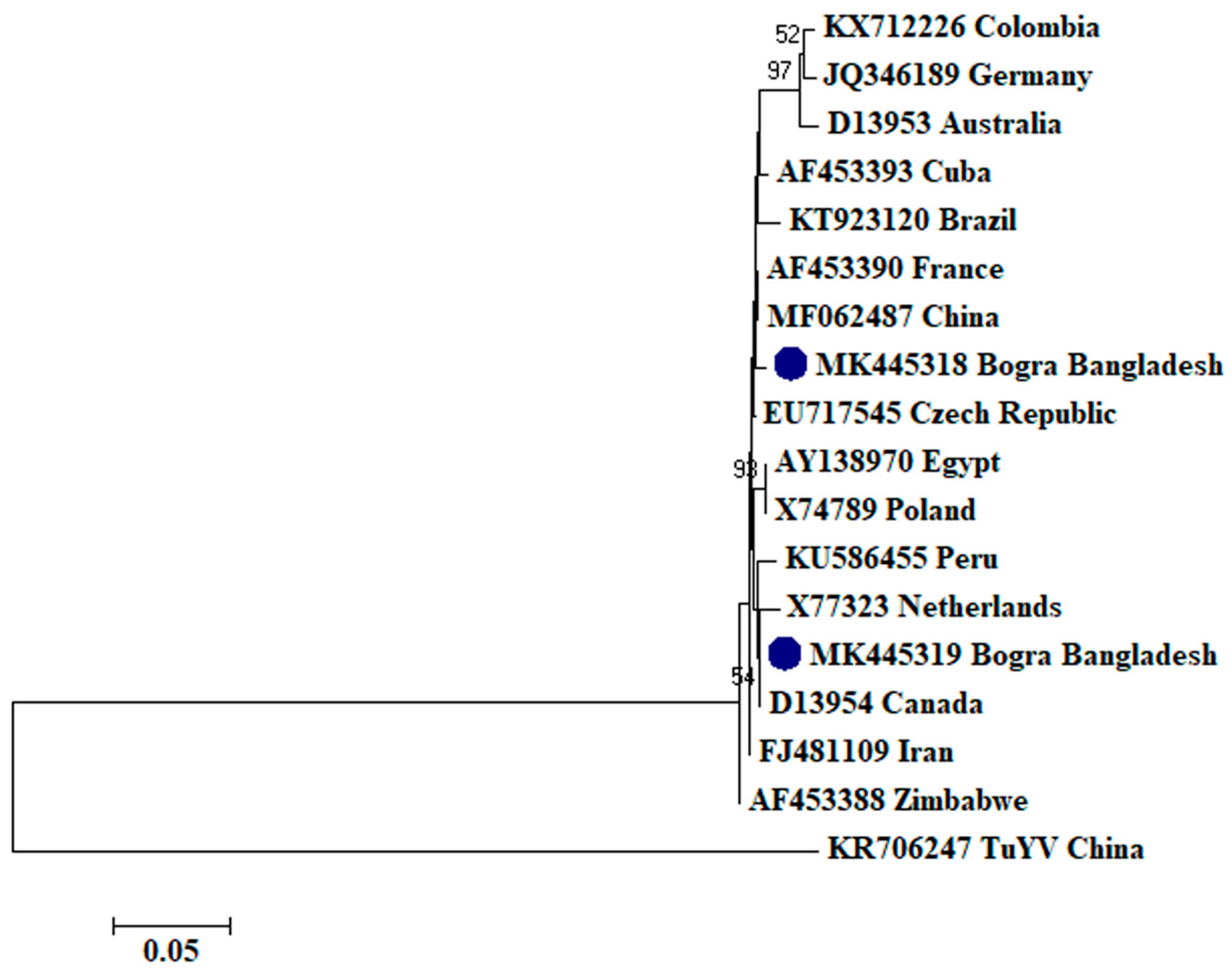
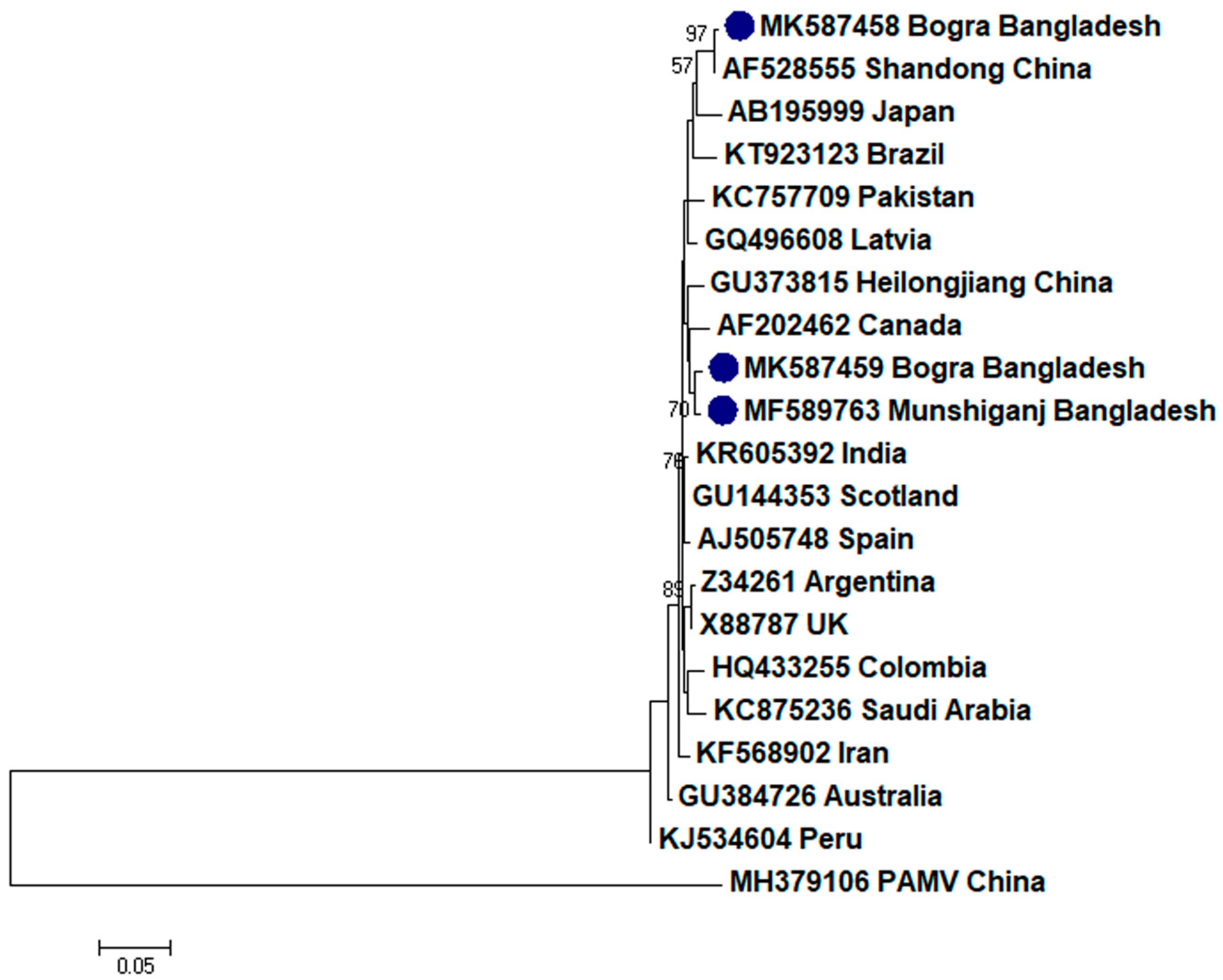
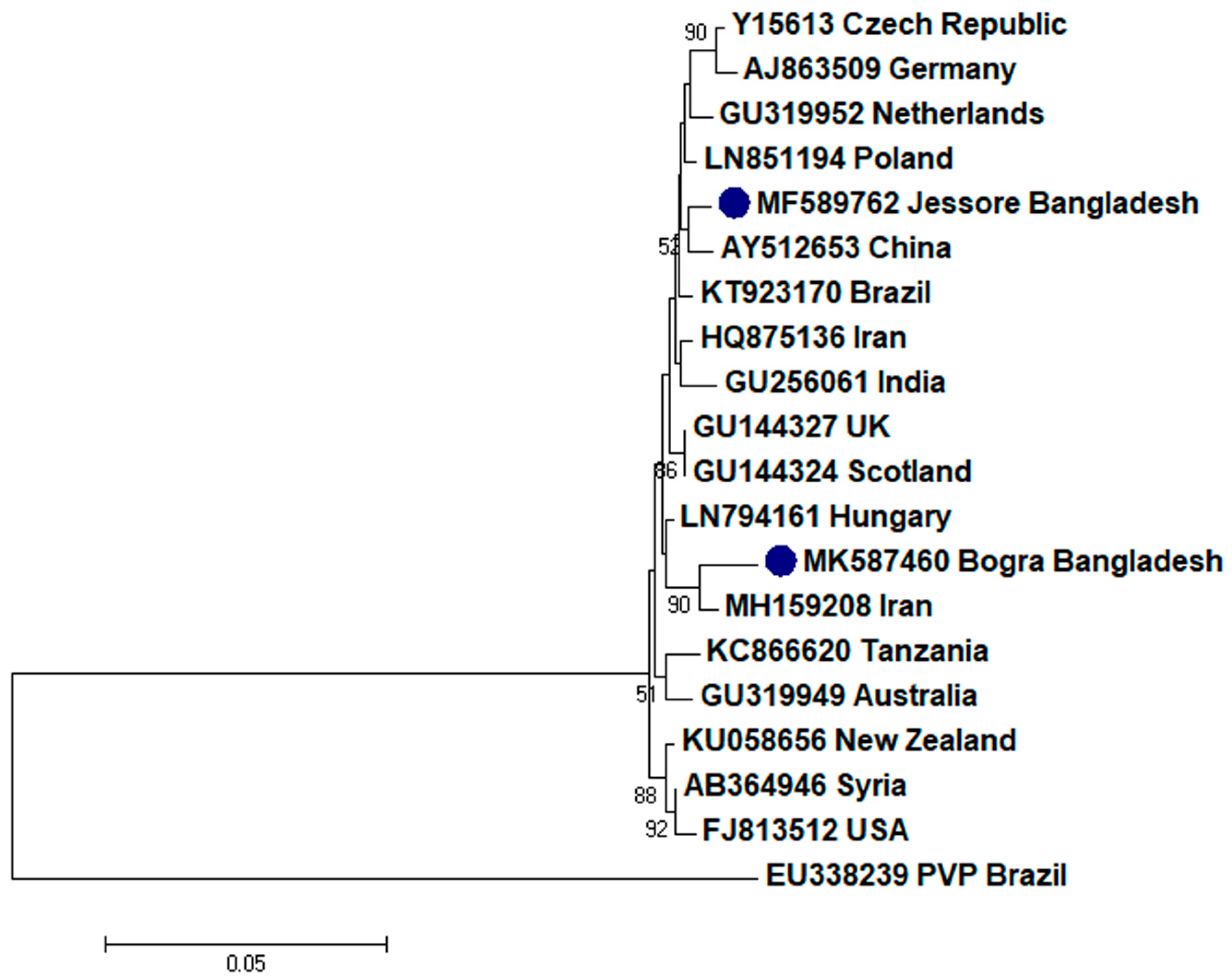
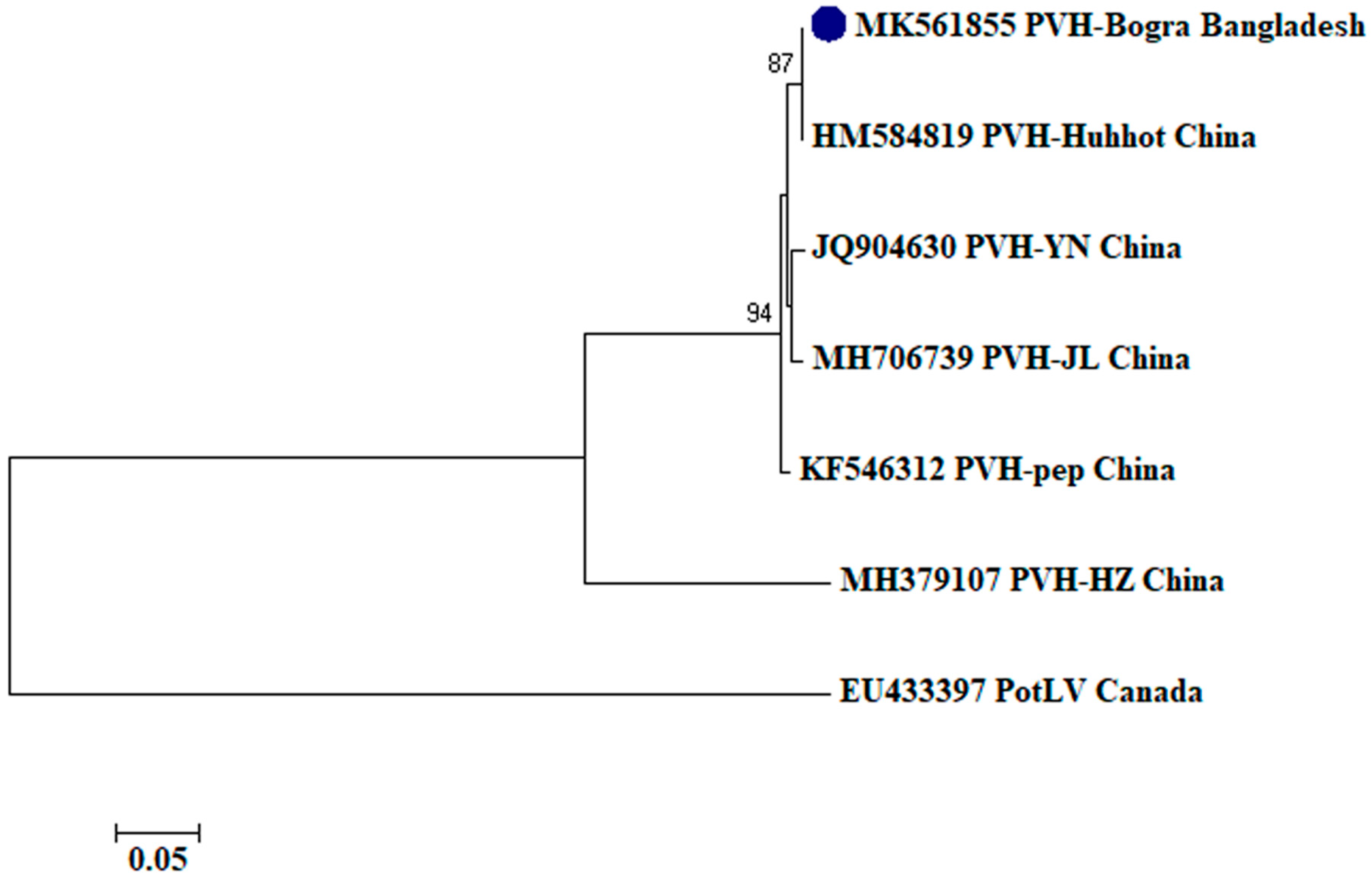
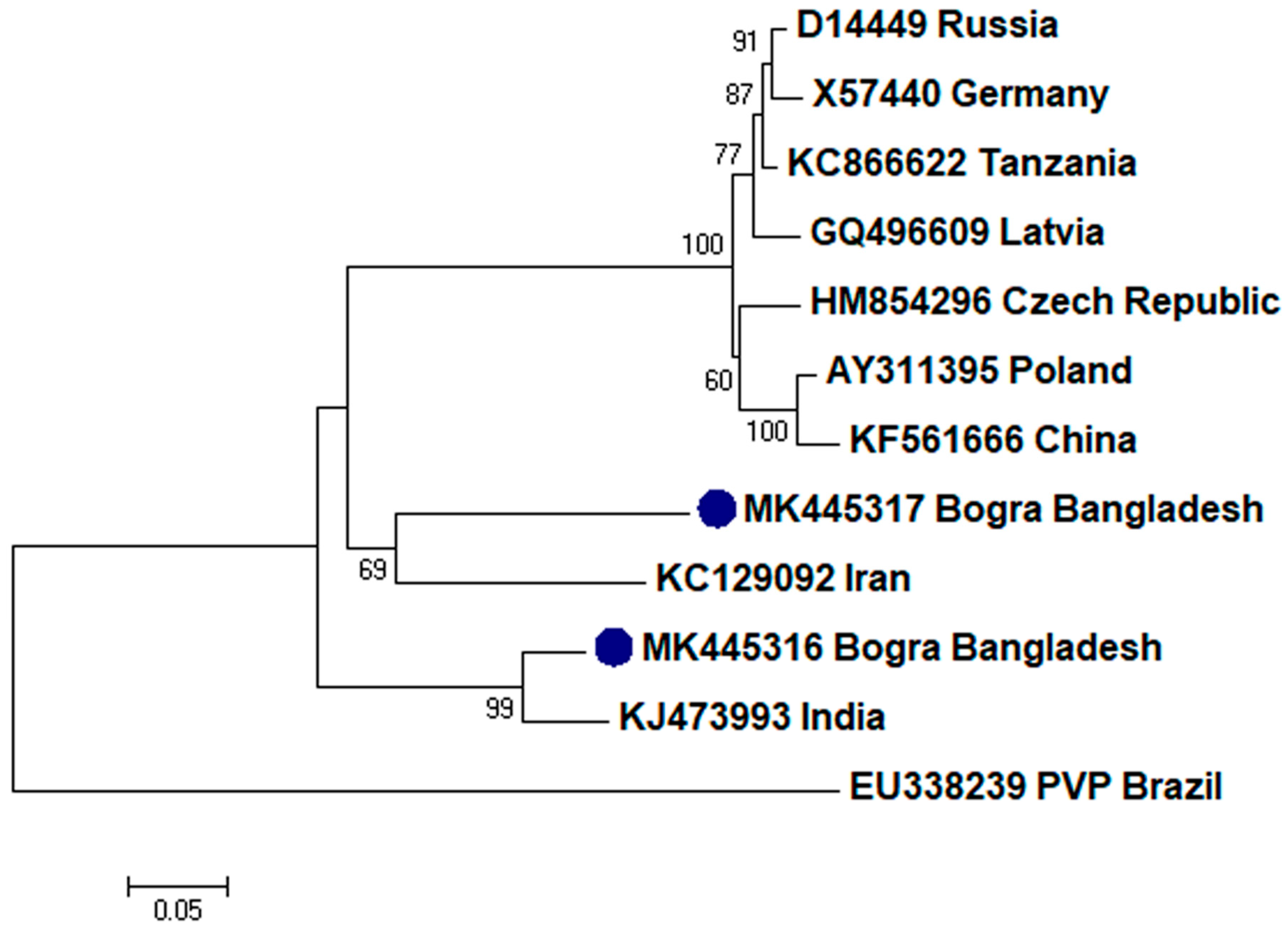
2.2. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Collection of Potato Tuber Samples
4.2. RT-PCR Detection
4.3. Cloning and Sequence Analysis
4.4. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Uddin, M.A.; Yasmin, S.; Rahman, M.L.; Hossain, S.M.B.; Choudhury, R.U. Challenges of potato cultivation in Bangladesh and developing digital databases of potato. Bangladesh J. Agril. Res. 2010, 35, 453–463. [Google Scholar] [CrossRef][Green Version]
- [FAOSTAT] Food and Agricultural Organization of the United Nations. Potato Production in 2018; Region/World/Production Quantity/Crops from Pick Lists; Statistical Division, Economic and Social Department: Rome, Italy, 2020; Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 14 August 2020).
- Awasthi, L.P.; Verma, H.N. Current status of viral diseases of potato and their ecofriendly management—A critical review. Virol. Virol. Res. Rev. 2017, 1, 1–16. [Google Scholar]
- Kerlan, C.; Moury, B. Potato Virus Y. In Encyclopaedia of Virology; Mahy, B.W.J., Regenmortel, V., Eds.; Academic Press: London, UK, 2008; pp. 287–296. [Google Scholar]
- Halterman, D.; Charkowski, A.; Verchot, J. Potato viruses and seed certification in the USA to provide healthy propagated tubers. Pest Technol. 2012, 6, 1–14. [Google Scholar]
- Kreuze, J.F.; Souza-Dias, J.A.C.; Jeevalatha, A.; Figueira, A.R.; Valkonen, J.P.T.; Jones, R.A.C. Viral diseases in potato. In The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Campos, H., Ortiz, O., Eds.; Springer: Cham, Switzerland, 2020; pp. 389–430. [Google Scholar]
- Wales, S.; Platt, H.W.; Cattlin, N. Diseases, Pests and Disorders of Potatoes: A Colour Handbook, 1st ed.; Manson Publishing Ltd.: London, UK, 2008. [Google Scholar]
- Pallás, V.; Sánchez-Navarro, J.A.; James, D. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 2018, 9, 2087. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Khan, A.L. Pathological Constraints of Seed Potato Production in Bangladesh. In Proceedings of the International Seminar on Seed Potato, Dhaka, Bangladesh, 8–10 January 1990; p. 240. [Google Scholar]
- Khan, A.L.; Ali, M.S.; Bari, M.A. A Review of the Viral Diseases of Potato in Bangladesh. In Proceedings of the First National Workshop on Tuber Crop, Gazpur, Bangladesh, 28–30 May 1991; p. 286. [Google Scholar]
- Rashid, M.; Li, Y.Y.; Wang, Y.; Han, C.G. First report of Potato virus H infecting potatoes in Bangladesh. Plant Dis. 2019, 103, 1051. [Google Scholar] [CrossRef]
- Rashid, M.; Zhang, X.Y.; Wang, Y.; Han, C.G. First report of Potato virus S infecting potatoes in Bangladesh. Plant Dis. 2019, 103, 781. [Google Scholar] [CrossRef]
- Hossain, M.D.; Mia, S.A.; Ali, M.S.; Khan, A. Effect of degrees of severity of leaf roll virus on the growth and yield of potato. Bangladesh J. Bot. 1989, 18, 15–21. [Google Scholar]
- Hossain, M.; Ali, M. Effect of potato virus Y severities on virus concentration, dilution endpoint and potato yield. Bangladesh J. Plant Pathol. 1992, 8, 27–30. [Google Scholar]
- Muqit, A.; Akanda, A.M. Research on Virus and Mycoplasma Disease Management of Bangladesh Agricultural Research Institute mandated crops. In Strategic Intervention of Plant Pathology Research in Bangladesh; BARI: Gazpur, Bangladesh, 2007; p. 344. [Google Scholar]
- Ghorai, A.K.; Kang, S.S.; Sharma, A.; Sharma, S. Occurrence of Cucumber mosaic virus on potato and its transmission to muskmelon under potato-cucurbit cropping pattern followed in Punjab, India. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 2947–2965. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Q.; Chai, M.; Liu, J.; Zhang, L.; Cheng, X. First report of Potato aucuba mosaic virus on potato in China. Plant Dis. 2018, 102, 2671. [Google Scholar] [CrossRef]
- Salazar, L.F. Virus detection and management in developing countries. In Advances in Potato Pest Biology and Management; Zehnder, G.W., Powelson, M.L., Jansson, R.K., Raman, K.B., Eds.; APS Press: St. Paul, MN, USA, 1994; pp. 643–652. [Google Scholar]
- Hull, R. Nucleic acid hybridization procedures. In Diagnosis of Plant Virus Diseases; Matthews, R.E.F., Ed.; CRC Press: Boca Raton, FL, USA, 1993; pp. 253–271. [Google Scholar]
- Mumford, R.A.; Waish, K.; Barker, L.; Boonham, N. Detection of Potato mop top virus using a multiplex-real time fluorescent reverse transcription polymerase chain reaction assay. Phytopathology 2000, 90, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Singh, R.P. A new approach for the simultaneous differentiation of biological and geographical strains of Potato virus Y by multiplex RT-PCR. J. Virol. Methods 2002, 104, 41–54. [Google Scholar] [CrossRef]
- Singh, R.P.; Valkonen, J.P.; Gray, S.M.; Boonham, N.; Jones, R.A.; Kerlan, C.; Scubert, J. Discussion paper: The naming of Potato virus Y strains infecting potato. Arch. Virol. 2008, 153, 1–13. [Google Scholar] [CrossRef]
- Dietzgen, R.G. Application of PCR in plant virology. In Plant Viruses as Molecular Pathogens; Khan, J.A., Dijkstra, J., Eds.; The Haworth Press: New York, NY, USA, 2002; pp. 471–500. [Google Scholar]
- Ju, H.J. Simple and rapid detection of Potato leafroll virus (PLRV) by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Plant Pathol. J. 2011, 27, 1–4. [Google Scholar] [CrossRef]
- Jeong, J.; Cho, S.Y.; Lee, W.H.; Lee, K.J.; Ju, H.J. Development of a rapid detection method for Potato virus X by reverse transcription loop-mediated isothermal amplification. Plant Pathol. J. 2015, 31, 219–225. [Google Scholar] [CrossRef]
- Treder, K.; Chołuj, J.; Zacharzewska, B.; Babujee, L.; Mielczarek, M.; Burzyński, A.; Rakotondrafara, A.M. Optimization of a magnetic capture RT-LAMP assay for fast and real-time detection of Potato virus Y and differentiation of N and O serotypes. Arch. Virol. 2018, 163, 447–458. [Google Scholar] [CrossRef]
- Adams, I.; Fox, A. Diagnosis of plant viruses using next-generation sequencing and metagenomic analysis. In Current Research Topics in Plant Virology; Wang, A., Zhou, X., Eds.; Springer: Cham, Switzerland, 2016; pp. 323–335. [Google Scholar]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral diagnostics in plants using next generation sequencing: Computational analysis in practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef]
- Bhat, A.I.; Rao, G.P. Next-generation sequencing for diagnosis of virus. In Characterization of Plant Viruses; Bhat, A.I., Rao, G.P., Eds.; Humana Press: New York, NY, USA, 2020; pp. 389–395. [Google Scholar]
- Koenig, R.; Lesemann, D.E.; Adam, G.; Winter, S. Diagnostic techniques: Plant viruses. In Desk Encyclopedia of Plant and Fungal Virology; Mahy, B.W., van Regenmortel, M.H., Eds.; Academic Press: London, UK, 2009; pp. 18–30. [Google Scholar]
- Faccioli, G. Control of potato viruses using meristem culture and stem-cutting cultures, thermotherapy and chemotherapy. In Virus and Virus-Like Diseases of Potatoes and Production of Seed-Potatoes; Loebenstein, G., Berger, P.H., Brunt, A., Lawson, R.H., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 365–390. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Peiman, M.; Xie, C. Sensitive detection of potato viruses, PVX, PLRV and PVS, by RT PCR in potato leaf and tuber. Australas. Plant Dis. Notes 2006, 1, 41–46. [Google Scholar] [CrossRef]
- Stevenson, W.R. Compendium of Potato Diseases, 2nd ed.; APS Press: St. Paul, MN, USA, 2001. [Google Scholar]
- Zhang, W.; Zhang, Z.; Fan, G.; Gao, Y.; Wen, J.; Bai, Y.; Qiu, C.; Zhang, S.; Shen, Y.; Men, X. Development and application of a universal and simplified multiplex RT-PCR assay to detect five potato viruses. J. Gen. Plant Pathol. 2017, 83, 33–45. [Google Scholar] [CrossRef]
- Wang, B.; Ma, Y.L.; Zhang, Z.B.; Wu, Z.M.; Wu, Y.F.; Wang, Q.C.; Li, M.F. Potato viruses in China. Crop Prot. 2011, 30, 1117–1123. [Google Scholar] [CrossRef]
- Barker, H.; Dale, M.F.B. Resistance to viruses in potato. In Natural Resistance Mechanisms of Plants to Viruses; Loebenstein, G., Carr, J.P., Eds.; Springer: Cham, Switzerland, 2006; pp. 341–366. [Google Scholar]
- Dolby, C.A.; Jones, R.A.C. Occurrence of the Andean strain of Potato virus S in imported potato material and its effects on potato cultivars. Plant Pathol. 1987, 36, 381–388. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhang, R.N.; Xiang, H.Y.; Abouelnasr, H.; Li, D.W.; Yu, J.L.; McBeath, J.H.; Han, C.G. Discovery and characterization of a novel Carlavirus infecting potatoes in China. PLoS ONE 2013, 8, e69255. [Google Scholar] [CrossRef] [PubMed]
- Yardimci, N.; Culal, K.H.; Demir, Y. Detection of PVY, PVX, PVS, PVA, and PLRV on different potato varieties in Turkey using DAS-ELISA. J. Agric. Sci. Tech. 2015, 17, 757–764. [Google Scholar]
- Hull, R. Matthews’ Plant Virology, 4th ed.; Academic Press: London, UK, 2002. [Google Scholar]
- Gibbs, A.J.; Ohshima, K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010, 48, 205–223. [Google Scholar] [CrossRef]
- Rota-Stabelli, O.; Telford, M.J. A multi criterion approach for the selection of optimal outgrous in phylogeny: Recovering some support for Mandibulata over Myriochelata using mitogenomics. Mol. Phylogenet. Evol. 2008, 48, 103–111. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Payne, A.; DeSalle, R. Random roots and lineage sorting. Mol. Phylogenet. Evol. 2012, 64, 12–20. [Google Scholar] [CrossRef]
- Graham, S.W.; Olmstead, R.G.; Barrett, S.C.H. Rooting phylogenetic trees with distant outgroups: A case study from the Commelinoid monocots. Mol. Biol. Evol. 2002, 19, 1769–1781. [Google Scholar] [CrossRef]
- Abbas, M.F.; Hameed, S.; Rauf, A.; Nosheen, Q.; Ghani, A.; Qadir, A.; Zakia, S. Incidence of six viruses in potato growing areas of Pakistan. Pak. J. Phytopathol. 2012, 24, 44–47. [Google Scholar]
- Nosheen, Q.; Hameed, S.; Mughal, S.M.; Abbas, M.F. Serological identity of Potato virus X (PVX) and PCR characterization of its coat protein (CP) gene. Int. J. Phytopathol. 2013, 2, 92–96. [Google Scholar] [CrossRef]
- Al-Saikhan, M.S.; Alhudaib, K.A.; Soliman, A.M. Detection of three potato viruses isolated from Saudi Arabia. Int. J. Virol. 2014, 10, 224–234. [Google Scholar] [CrossRef][Green Version]
- Han, C.G.; Li, D.W.; Xing, Y.M.; Zhu, K.; Tian, Z.F.; Cai, Z.N.; Yu, J.L.; Liu, Y. Wheat yellow mosaic virus widely occurring in wheat (Triticum aestivum) in China. Plant Dis. 2000, 84, 627–630. [Google Scholar] [CrossRef]
- Khine, M.O.; Michaela, B.; Liu, Y.; Kundu, J.K.; Wang, X. Molecular diversity of Barley yellow dwarf virus-PAV from China and the Czech Republic. J. Integr. Agric. 2020, 19, 2–11. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Zhao, T.Y.; Li, Y.Y.; Xiang, H.Y.; Dong, S.W.; Zhang, Z.Y.; Wang, Y.; Li, D.W.; Yu, J.L.; Han, C.G. The conserved proline18 in the polerovirus P3a is important for Brassica yellows virus systemic infection. Front. Microbiol. 2018, 9, 613. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T.A. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]


| Sample Collected Regions | No. of Collected Tuber Samples | Associated Virus Name | RT-PCR Detection | |
|---|---|---|---|---|
| No. of Infected Tubers | Infection Ratio (%) | |||
| Munshiganj | 50 | PLRV/PVX | 7 | 14 |
| PVX/PVS | 5 | 10 | ||
| PLRV/PVX/PVS | 7 | 14 | ||
| PLRV/PVX/PVY | 6 | 12 | ||
| PLRV/PVY/PVS | 6 | 12 | ||
| Jessore | 50 | PLRV/PVX | 3 | 6 |
| PVX/PVS | 5 | 10 | ||
| PVX/PAMV | 6 | 12 | ||
| PLRV/PVX/PVS | 3 | 6 | ||
| PLRV/PVY/PVS | 5 | 10 | ||
| PVX/PVH/PAMV | 4 | 8 | ||
| PLRV/PVX/PVH/PAMV | 5 | 10 | ||
| PVX/PVS/PVH/PAMV | 3 | 6 | ||
| PLRV/PVX/PVS/PVH/PAMV | 1 | 2 | ||
| Bogra | 120 | PVX/PAMV | 4 | 3.33 |
| PLRV/PVX/PVS | 5 | 4.17 | ||
| PLRV/PVX/PVY | 4 | 3.33 | ||
| PLRV/PVY/PVS | 4 | 3.33 | ||
| PVX/PVH/PAMV | 11 | 9.17 | ||
| PLRV/PVX/PVH/PAMV | 15 | 12.50 | ||
| PVX/PVS/PVH/PAMV | 7 | 5.83 | ||
| PLRV/PVX/PVS/PVH/PAMV | 4 | 3.33 | ||
| PLRV/PVX/PVS/PVH/PAMV/PVM | 10 | 8.33 | ||
| Virus Name | GenBank Accession No. | Sample Collected Regions | ||
|---|---|---|---|---|
| Bogra | Munshiganj | Jessore | ||
| PLRV | MK445318 | + | - | - |
| PLRV | MK445319 | + | + | + |
| PVX | MK587458 | + | - | - |
| PVX | MK587459 | + | - | - |
| PVX | MF589763 | - | + | + |
| PVY | MF589764 | + | + | + |
| PVS | MF589762 | - | + | + |
| PVS | MK587460 | + | - | - |
| PVH | MK561855 | + | - | + |
| PAMV | MK445320 | + | - | + |
| PAMV | MK445321 | + | - | - |
| PAMV | MK445322 | + | - | - |
| PVM | MK445316 | + | - | - |
| PVM | MK445317 | + | - | - |
| Primer | Sequence (5′ to 3′) |
|---|---|
| PLRV-CPF | ATGAGTACGGTCGTGGTTAA |
| PLRV-CPR | CTATTTGGGGTTTTGCAAAG |
| PVX-CPF | TAGCCACAACACAGGCCACAG |
| PVX-CPR | TTATGGTGGTGGGAGAGTGACAA |
| PVY-CPF | ACGTCCAAAATGAGAATGCC |
| PVY-CPR | TGGTGTTCGTGATGTGACCT |
| PVS-CPF | ATGCCGCCCAAACCGGAT |
| PVS-CPR | TCATTGGTTGATCGCATTAC |
| PVH-CPF | ATGGATGTTGCTACTTC |
| PVH-CPR | ATTGCCGCTTGTTATTC |
| PAMV-CPF | ATGGTTGATTCTAAGAAAAC |
| PAMV-CPR | TTAGGATGGTGGTGCCATGA |
| PVM-CPF | ATGGGAGATTCAACTAAGAA |
| PVM-CPR | CTTCATTTGTTATTCGACTT |
| PVA-CPF | AGCCGAAACTCTTGATGCAA |
| PVA-CPR | GCACTTCGGTTACACCCCCT |
| PVH-TGB1F | AACTGCAGATGGATGTGCTGGTAG |
| PVH-TGB1R | CGGGATCAGGCGGCGGTGAAAGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, M.-O.; Wang, Y.; Han, C.-G. Molecular Detection of Potato Viruses in Bangladesh and Their Phylogenetic Analysis. Plants 2020, 9, 1413. https://doi.org/10.3390/plants9111413
Rashid M-O, Wang Y, Han C-G. Molecular Detection of Potato Viruses in Bangladesh and Their Phylogenetic Analysis. Plants. 2020; 9(11):1413. https://doi.org/10.3390/plants9111413
Chicago/Turabian StyleRashid, Mamun-Or, Ying Wang, and Cheng-Gui Han. 2020. "Molecular Detection of Potato Viruses in Bangladesh and Their Phylogenetic Analysis" Plants 9, no. 11: 1413. https://doi.org/10.3390/plants9111413
APA StyleRashid, M.-O., Wang, Y., & Han, C.-G. (2020). Molecular Detection of Potato Viruses in Bangladesh and Their Phylogenetic Analysis. Plants, 9(11), 1413. https://doi.org/10.3390/plants9111413







