Functional Analysis of StPHT1;7, a Solanum tuberosum L. Phosphate Transporter Gene, in Growth and Drought Tolerance
Abstract
1. Introduction
2. Results
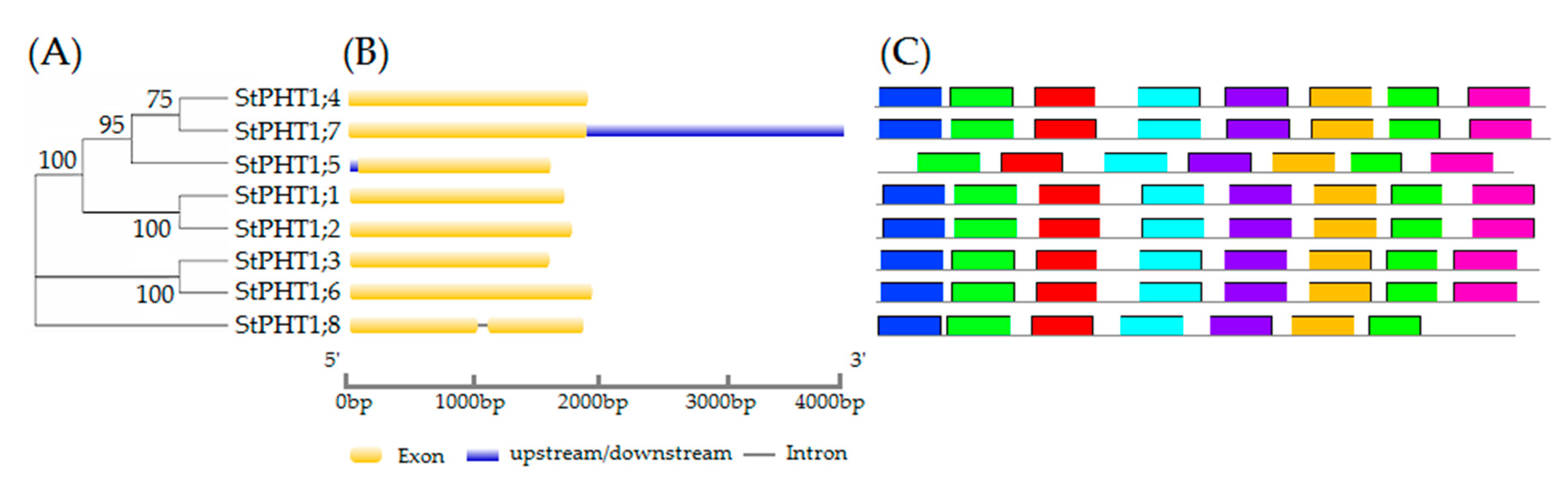
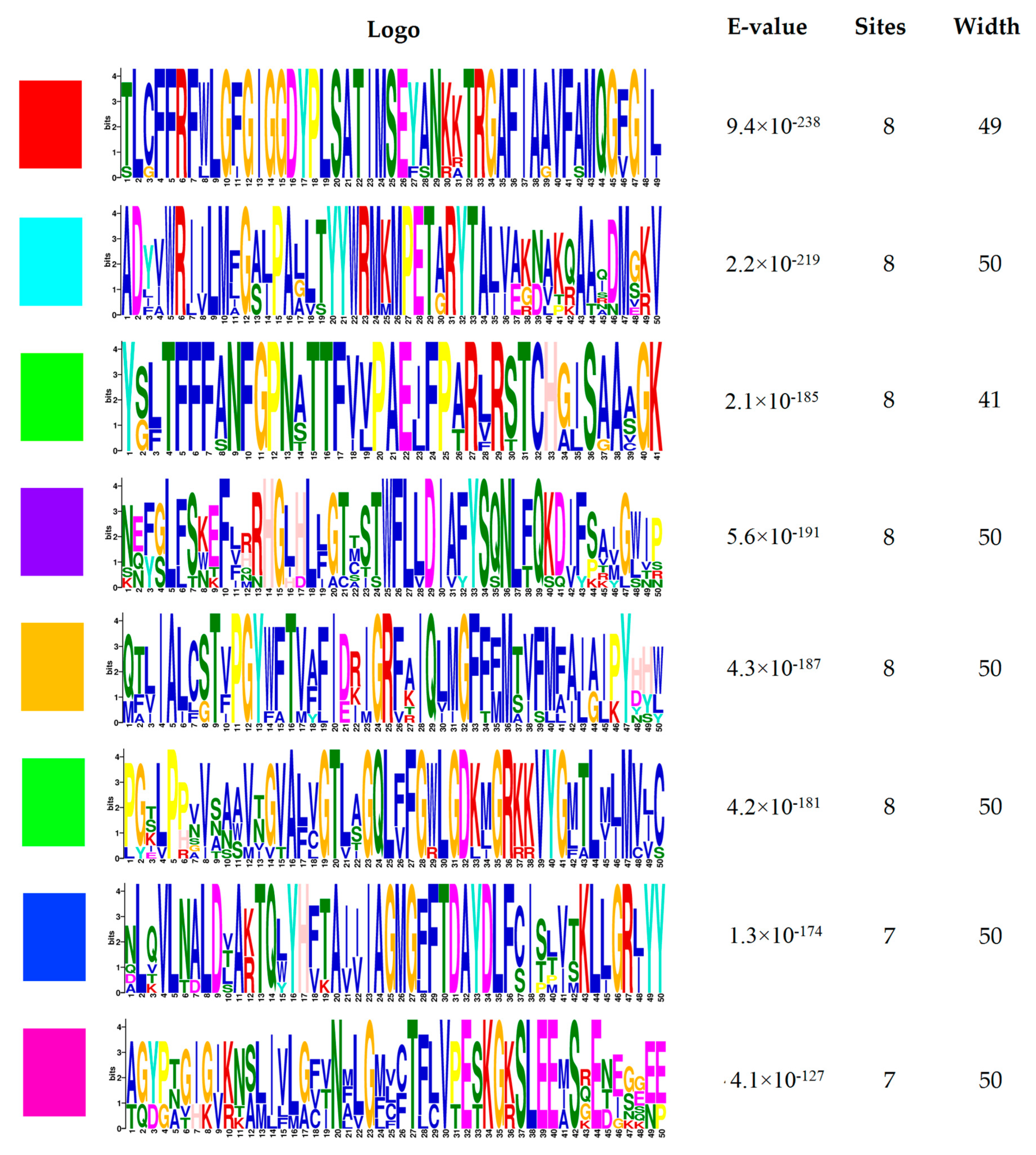
2.1. Gene Structure, Conservative Motifs, and Multiple Sequence Alignment of the Potato PHT1 Gene Family
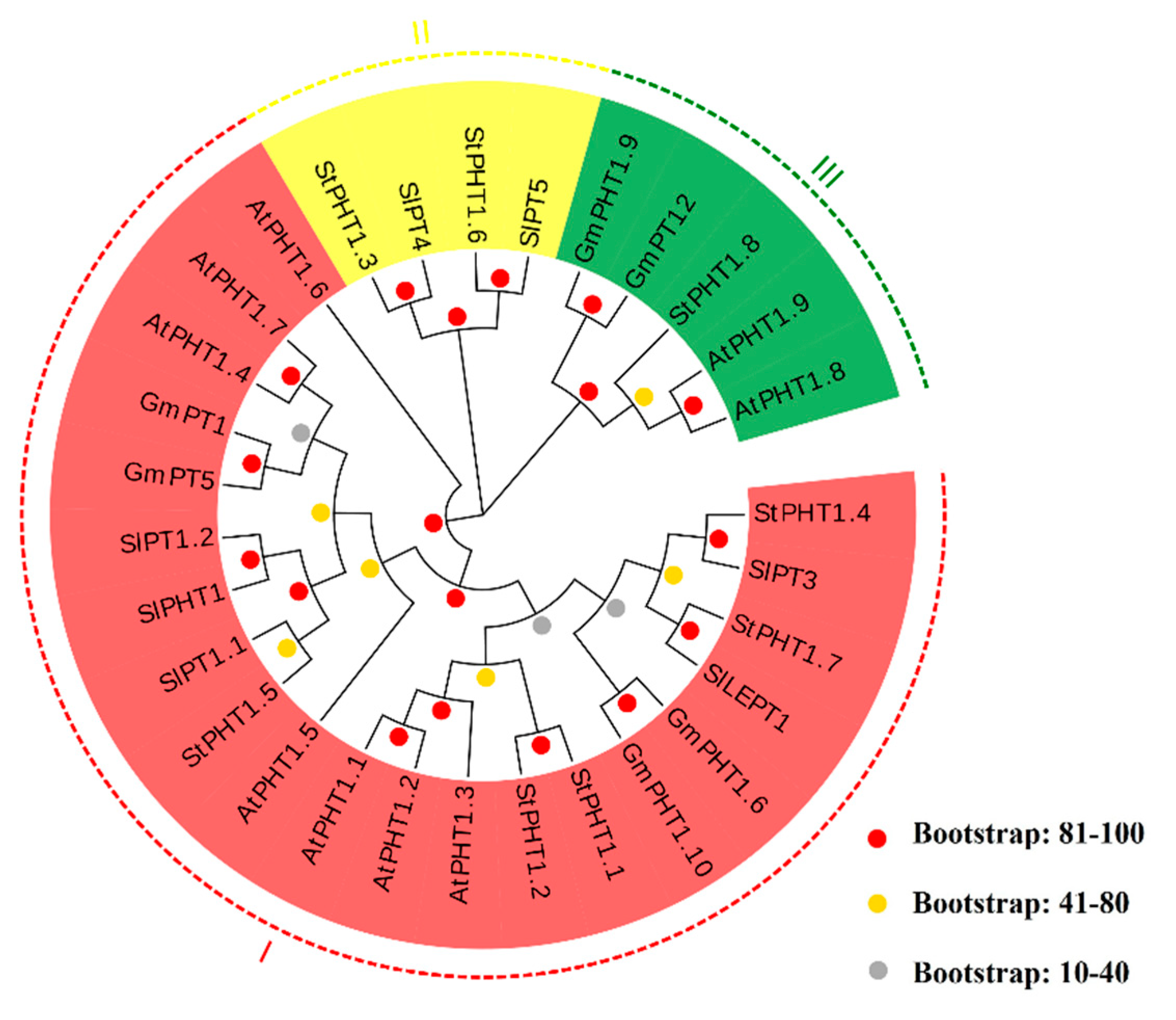
2.2. The Phylogenetic Tree of Potato PHT1 Genes
2.3. Potato PHT1 Protein Is Located on the Cell Membrane
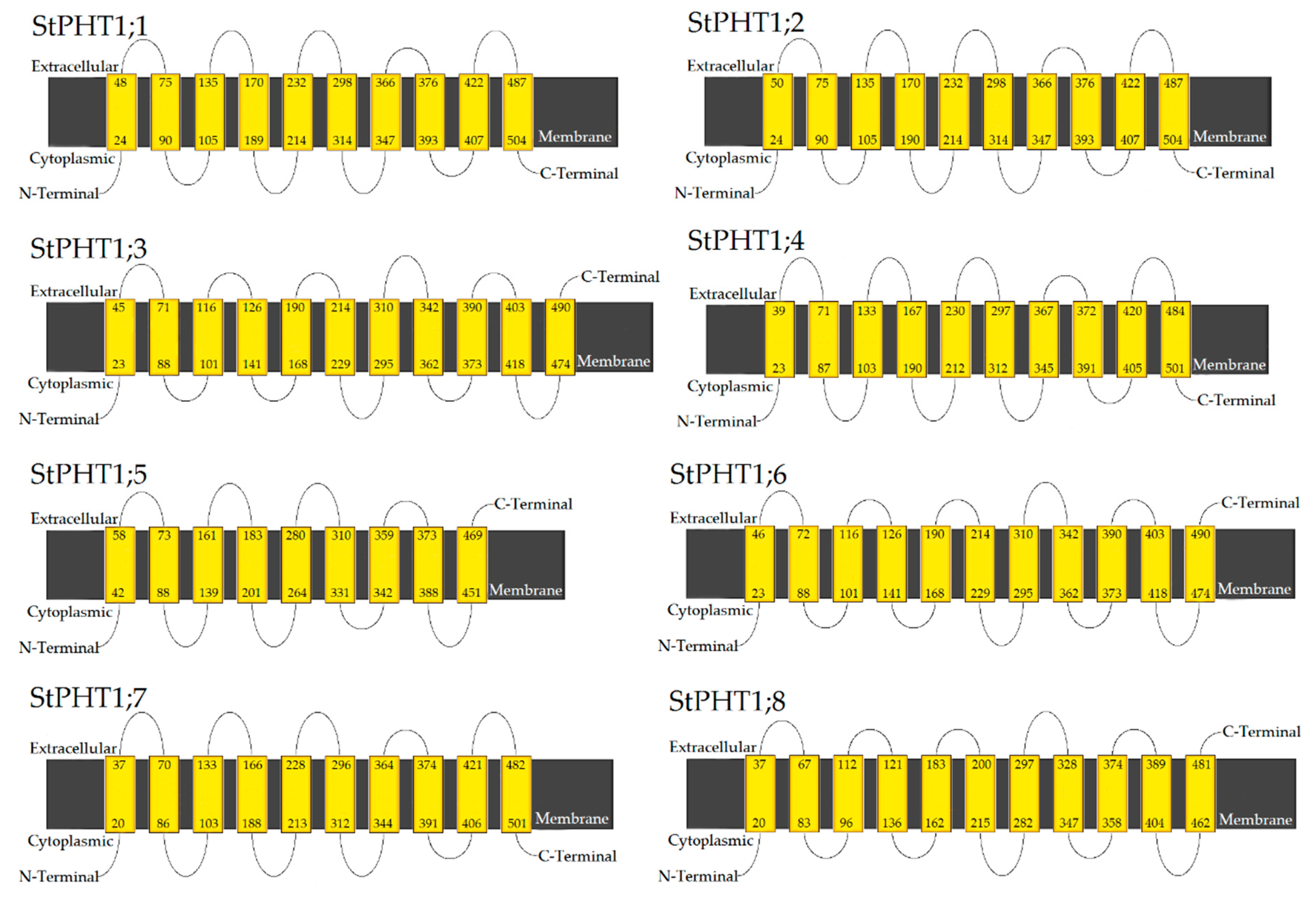
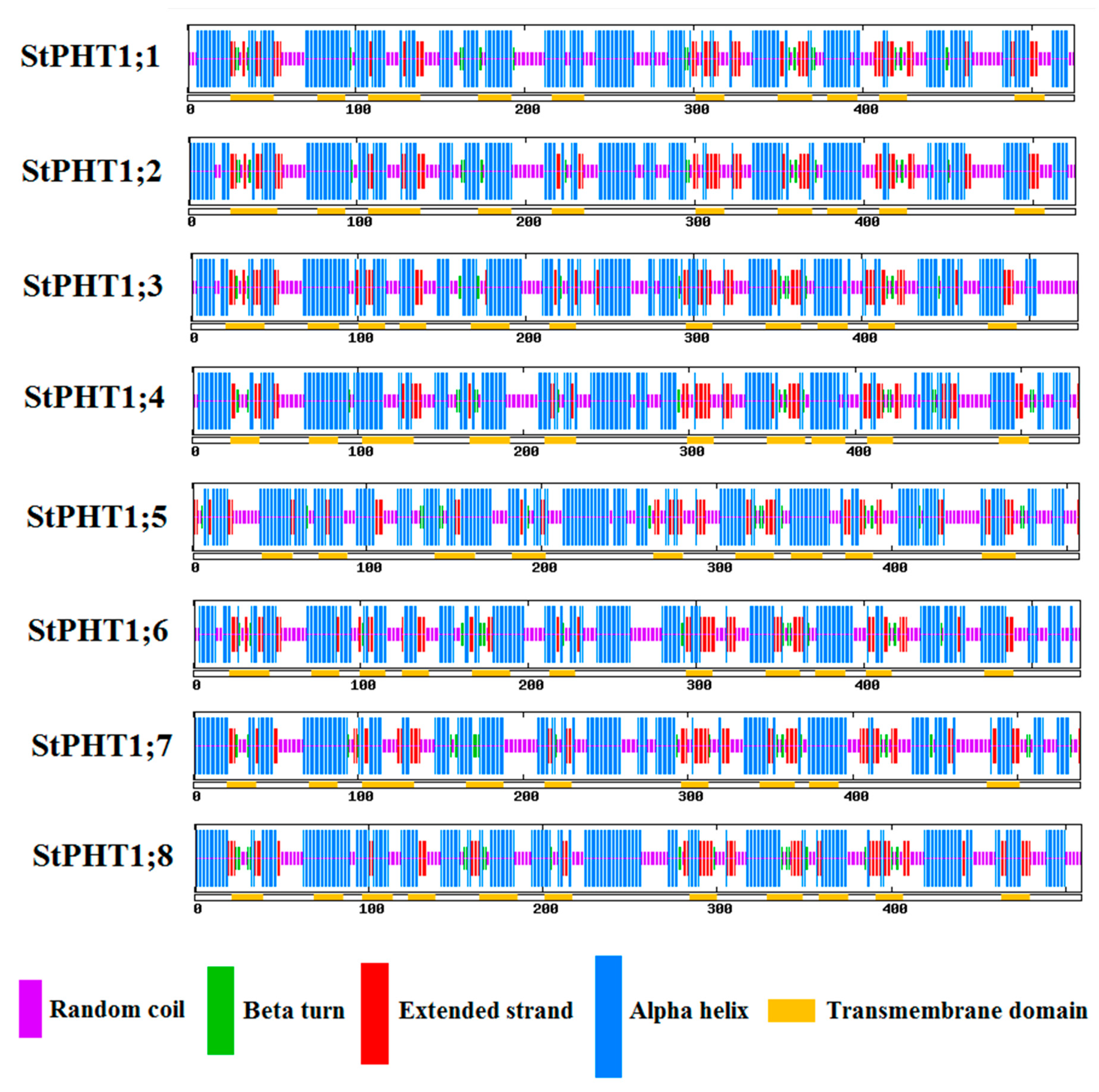
2.4. Potato PHT1 Family Members Have Similar Protein Secondary Structure
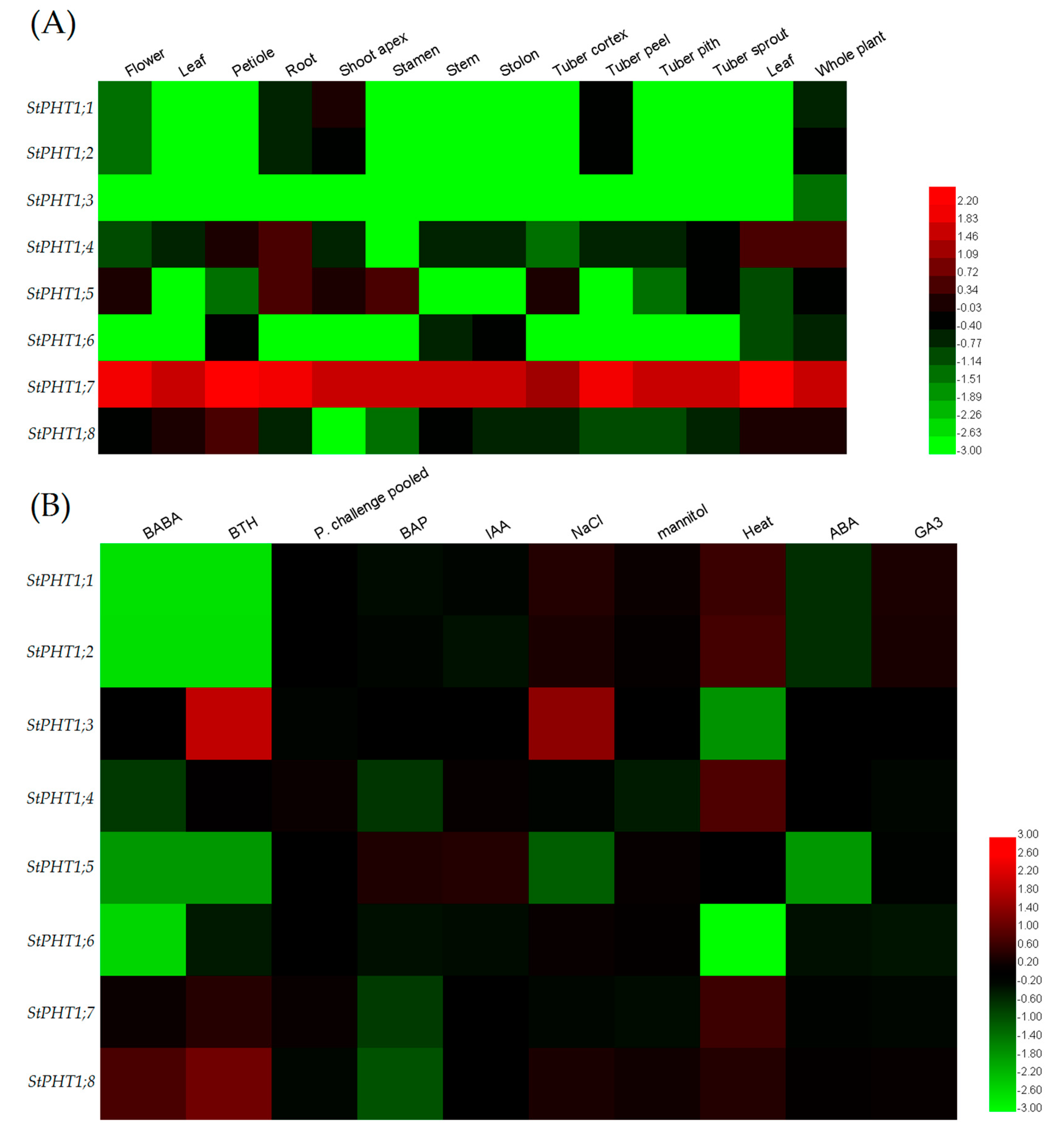
2.5. Expression Patterns of the Potato PHT1 Gene
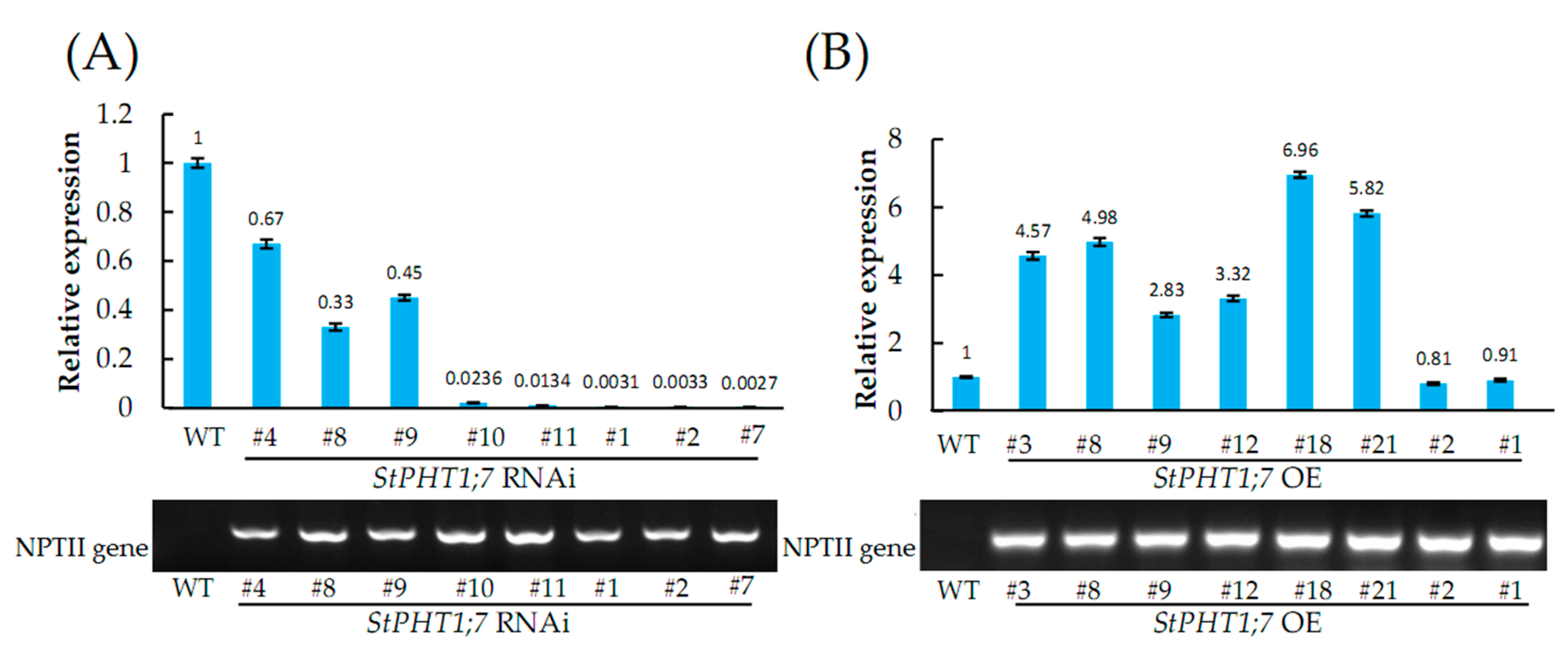
2.6. Silencing and Overexpression of StPHT1;7
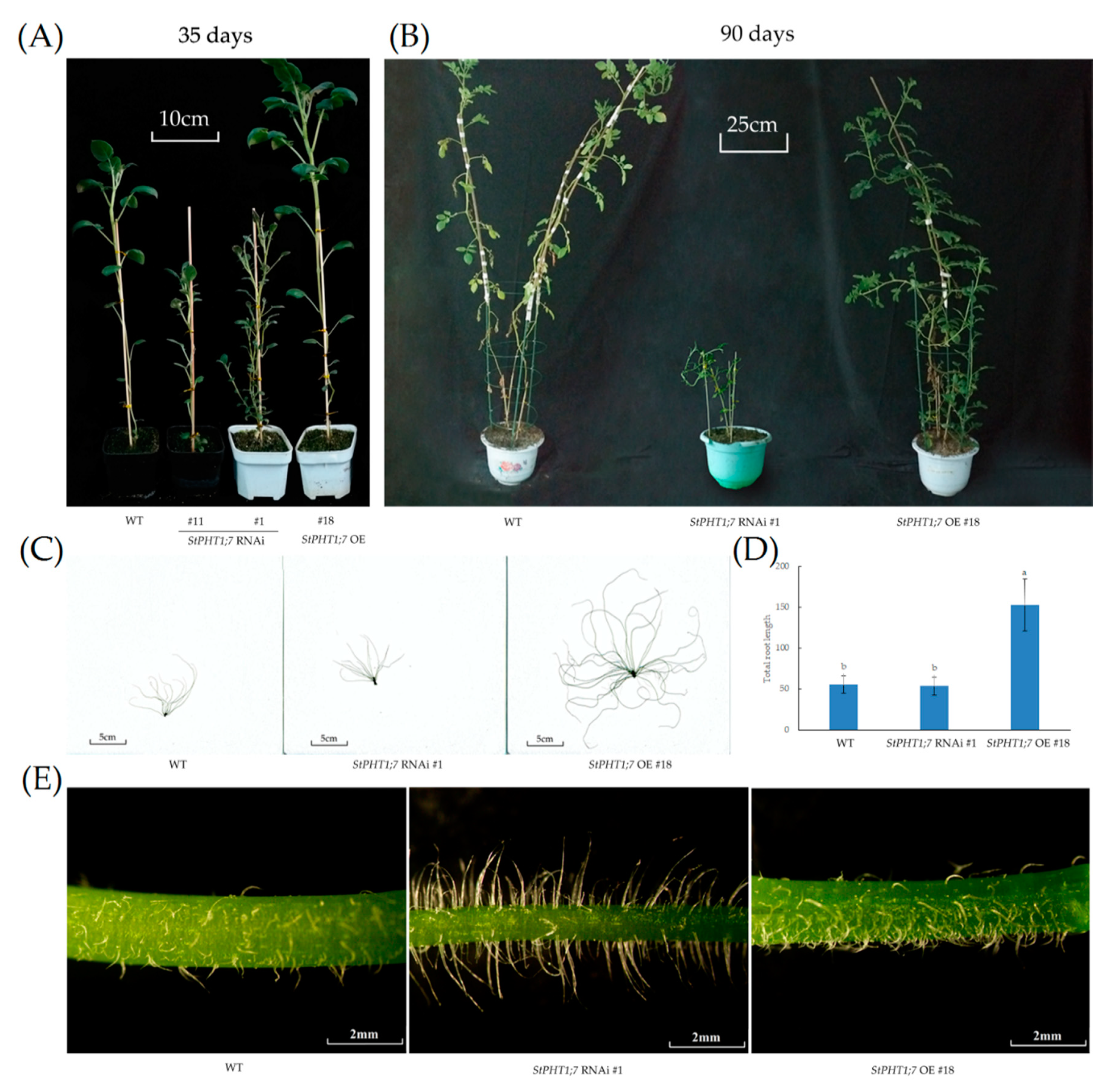
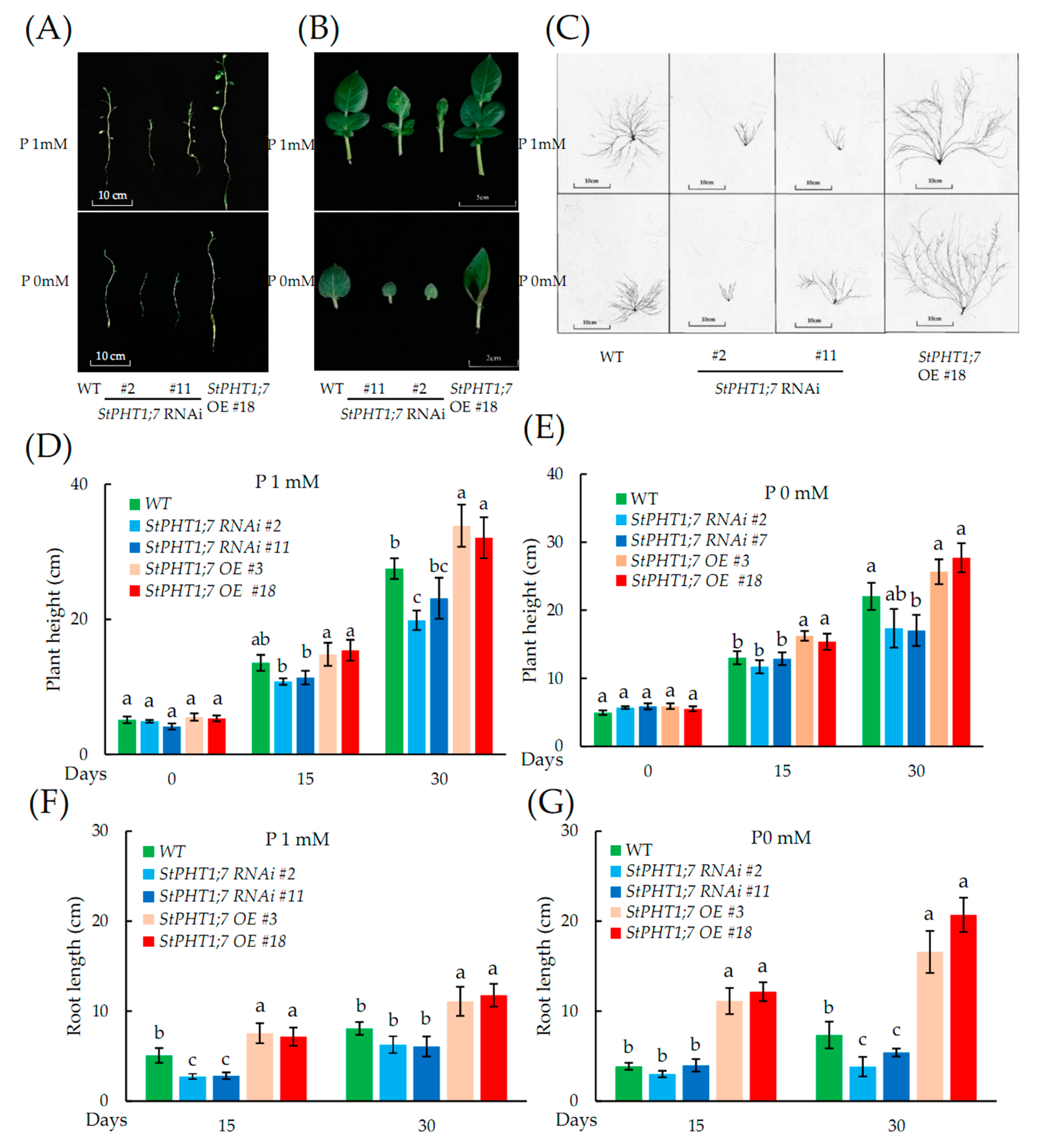
2.7. Phenotypic Characteristics of Transgenic Plants
2.8. Responses of the Transgenic Plant to Phosphorus Stress
2.9. Response of Transgenic Plants to Dehydration Treatment
2.10. Response of Transgenic Plants to Drought Stress
3. Discussion
3.1. Bioinformatics Analysis of Potato PHT1 Family
3.2. StPHT1;7 Expression Affected Potato Plants Growth
3.3. Phosphorus Stress Had Different Effects on Transgenic Plants
3.4. StPHT1;7 Affect Potato Plants Drought Tolerance
4. Materials and Methods
4.1. Identification of StPHT1s and Analysis of Their Physical and Chemical Parameters
4.2. Construction of Phylogenetic Tree of PHT1 Gene Family
4.3. Analysis of Potato PHT1 Gene Structures and Conserved Domains
4.4. Analysis of the Primary and Spatial Structure of Potato PHT1 Proteins
4.5. Tissue Expression and Stress Treatment Expression Analysis of the Potato PHT1 Genes
4.6. Growth Status of Plant Materials
4.7. Silence and Overexpression of StPHT1;7
4.8. Phenotype Identification of the Transgenic Potato
4.9. Transgenic Plant Phosphorus Stress Treatment
4.10. Transgenic Plant PEG6000 Stress Treatment
4.11. Transgenic Plant Drought Stress Treatment
4.12. Determination of Relevant Physiological Indicators
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marschner, H. Adaptation of Plants to Adverse Chemical Soil Conditions. In Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: Cambridge, MA, USA, 1995; pp. 596–680. [Google Scholar]
- Liu, C.; Rubæk, G.H.; Liu, F.; Andersen, M.N. Effect of partial root zone drying and deficit irrigation on nitrogen and phosphorus uptake in potato. Agric. Water Manag. 2015, 159, 66–76. [Google Scholar] [CrossRef]
- Tulu, S.; Zewide, I.; Mohammed, A. Effect of different rates of nitrogen and phosphorus on yield and yield components of potato (Solanum tuberosum L.) at Masha District, Southwestern Ethiopia. Int. J. Soil Sci. 2012, 7, 146–156. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. N. Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Lynch, J.P.; Brown, K.M. Root strategies for phosphorus acquisition. Plant Ecophysiol. 2008, 7, 83–116. [Google Scholar]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef]
- Bieleski, R.L. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Nielsen, K.L.; Eshel, A.; Lynch, J.P. The effect of phosphorus availability on the carbon economy of contrasting common bean (Phaseolus vulgaris L.) genotypes. J. Exp. Bot. 2001, 52, 329–339. [Google Scholar] [PubMed]
- Van Der Werf, A.; Kooijman, A.; Welschen, R.; Lambers, H. Respiratory energy costs for the maintenance of biomass, for growth and for ion uptake in roots of Carex diandra and Carex acutiformis. Physiol. Plant 1988, 72, 483–491. [Google Scholar] [CrossRef]
- Lynch, J.P. Roots of the second green revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Singh, D.K.; Sale, P.W.G.; Pallaghy, C.K.; McKenzie, B. Phosphorus concentrations in the leaves of defoliated white clover affect abscisic acid formation and transpiration in drying soil. N. Phytol. 2000, 146, 249–259. [Google Scholar] [CrossRef]
- Jin, J.; Wang, G.; Liu, X.; Pan, X.; Herbert, S.J.; Tang, C. Interaction between phosphorus nutrition and drought on grain yield, and assimilation of phosphorus and nitrogen in two soybean cultivars differing in protein concentration in grains. J. Plant Nutr. 2006, 29, 1433–1449. [Google Scholar] [CrossRef]
- Radin, J.W.; Eidenbock, M.P. Hydraulic conductance as a factor limiting leaf expansion of phosphorus-deficient cotton plants. Plant Physiol. 1984, 75, 372–377. [Google Scholar] [CrossRef]
- Jin, J.; Wang, G.; Liu, X.; Pan, X.; Herbert, S.J. Phosphorus application affects the soybean root response to water deficit at the initial flowering and full pod stages. Soil Sci. Plant Nutr. 2005, 51, 953–960. [Google Scholar] [CrossRef]
- Cramer, M.D.; Hawkins, H.-J.; Verboom, G.A. The importance of nutritional regulation of plant water flux. Oecologia 2009, 161, 15–24. [Google Scholar] [CrossRef]
- Ackerson, R.C. Osmoregulation in cotton in response to water stress. Plant Physiol. 1985, 77, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Premachandra, G.S.; Saneoka, H.; Fujita, K.; Ogata, S. Cell membrane stability and leaf water relations as affected by phosphorus nutrition under water stress in maize. Soil Sci. Plant Nutr. 1990, 36, 661–666. [Google Scholar] [CrossRef]
- Faustino, L.; Bulfe, N.M.; Pinazo, M.A.; Monteoliva, S.; Graciano, C. Dry weight partitioning and hydraulic traits in young Pinus taeda trees fertilized with nitrogen and phosphorus in a subtropical area. Tree Physiol. 2013, 33, 241–251. [Google Scholar] [CrossRef]
- Cooper, J.; Carliell-Marquet, C. A substance flow analysis of phosphorus in the UK food production and consumption system. Resour. Conserv. Recycl. 2013, 74, 82–100. [Google Scholar] [CrossRef]
- Bucher, M.; Rausch, C.; Daram, P. Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant Nutr. Soil Sci. 2015, 164, 209–217. [Google Scholar] [CrossRef]
- Smith, F.W.; Mudge, S.R.; Rae, A.L.; Glassop, D. Phosphate transport in plants. Plant Soil 2003, 248, 71–83. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, S.; Wu, X.; Wang, X.; Nan, Y.; Wang, N.; Chen, Q. Identification and characterization of phosphate transporter genes in potato. J. Biotechnol. 2017, 264, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. N. Phytol. 2008, 177, 889–898. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Lin, S.-I.; Chiou, T.-J. Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 2009, 60, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G.; Karthikeyan, A.S. Phosphate acquisition. Plant Soil 2005, 274, 37–49. [Google Scholar] [CrossRef]
- Gordon-Weeks, R.; Tong, Y.; Davies, T.G.E.; Leggewie, G. Restricted spatial expression of a high-affinity phosphate transporter in potato roots. J. Cell Sci. 2003, 116, 3135–3144. [Google Scholar] [CrossRef]
- Lapis-Gaza, H.R.; Jost, R.; Finnegan, P.M. Arabidopsis Phosphate Transporter1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 2014, 14, 334. [Google Scholar] [CrossRef]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lü, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186. [Google Scholar] [CrossRef]
- Yan, W.; Chen, G.-H.; Yang, L.-F.; Gai, J.-Y.; Zhu, Y. Overexpression of the rice phosphate transporter gene OsPT6 enhances tolerance to low phosphorus stress in vegetable soybean. Sci. Hortic. 2014, 177, 71–76. [Google Scholar] [CrossRef]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.-H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A Constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Piñeros, M.A.; Wang, Z.; Wang, W.; Li, C.; Wu, Z.; Kochian, L.V.; Wu, P. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 2013, 37, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Robredo, A.; Pérez-López, U.; De La Maza, H.S.; González-Moro, B.; Lacuesta, M.; Mena-Petite, A.; Muñoz-Rueda, A. Elevated CO2 alleviates the impact of drought on barley improving water status by lowering stomatal conductance and delaying its effects on photosynthesis. Environ. Exp. Bot. 2007, 59, 252–263. [Google Scholar] [CrossRef]
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.T.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Leggewie, G.; Willmitzer, L.; Riesmeier, J.W. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: Identification of phosphate transporters from higher plants. Plant Cell. 1997, 9, 381–392. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karandashov, V.; Bucher, M. Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 2005, 10, 22–29. [Google Scholar] [CrossRef]
- Su, S.; Wu, F.; Liu, D.; Wu, L.; Gao, S. Cloning and Characterization of a Phosphate Transporter Gene of Pht1 Family in Maize. J. Nucl. Agric. Sci. 2013, 27, 0885–0894. [Google Scholar]
- Potato Genome Sequencing Consortium; Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Parra-Almuna, L.; Pontigo, S.; Larama, G.; Cumming, J.R.; Pérez-Tienda, J.; Ferrol, N.; De la Luz Mora, M. Expression analysis and functional characterization of two PHT1 family phosphate transporters in ryegrass. Planta 2019, 251, 6. [Google Scholar] [CrossRef]
- Bai, S.; Song, Q.; Liu, G.; Jiang, Y.; Lin, S. The Analysis of Salt Tolerance of Transgenic Populus simonii × P. nigra Pollen plantlets with betA Gene. Mol. Plant Breed. 2006, 4, 41–44. [Google Scholar] [CrossRef]
- Liu, G.; Cheng, G.; Jiang, J.; Bai, S.; Yu, Y.; Cai, Z.; Dong, J. The Transformation of betA Gene into the Pollen Plantlets of Populus simonii×P. nigra. J. Plant Physiol. Mol. Biol. 2006, 32, 163–168. [Google Scholar] [CrossRef]
- Föhse, D.; Claassen, N.; Jungk, A. Phosphorus efficiency of plants: I. External and internal P requirement and P uptake efficiency of different plant-species. Plant Soil 1988, 110, 101–109. [Google Scholar] [CrossRef]
- Chen, H.; Chen, H. Effect of balanced nutrition of phosphorus and potassium on anatomical structure of cabbage (Brassica Chinensis L.). Plant Nutr. Fertil. Sci. 1996, 4, 7. [Google Scholar]
- Deng, Z.; Xu, J.; Duan, S.; Liu, J.; Bian, C.; Pang, W.; Jin, L. Effect on Growth Indicators of 11 Potato Cultivars in vitro under PEG-8000 Stress. Acta Agric. Boreali-Sin. 2014, 29, 99–106. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Boil. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Lu, W.; Li, Z.; Lei, X.; Bai, Y.; Yan, L.; Zhang, L.; Ru, J. Effect of PEG- 6000- induced water stress on in vitro plantlets of potato. J. Northeast. Agric. Univ. 2015, 46, 1–9. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhang, H.; Wang, S.; Ye, X.; Shi, L.; Xu, F.; Ding, G. Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS ONE 2019, 25, e0220374. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, H.; Zhang, X.; Chen, H.; Tang, M. Characterization of six PHT1 members in Lycium barbarum and their response to arbuscular mycorrhiza and water stress. Tree Physiol. 2017, 1, 351–366. [Google Scholar] [CrossRef]
- Molla, M.; Nasiruddin, K.; Amin, M.A.; Haque, M.S. Standardization of protocol for Agrobacterium-mediated transformation in potato (Solanum tuberosum L.). Bangladesh J. Agr. Res. 2012, 37, 185–194. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar] [PubMed]
- Tiffin, L.O. Mineral nutrition of plants: Principles and perspectives. Soil Sci. Soc. Am. J. 1972, 36, 36. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.-K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]

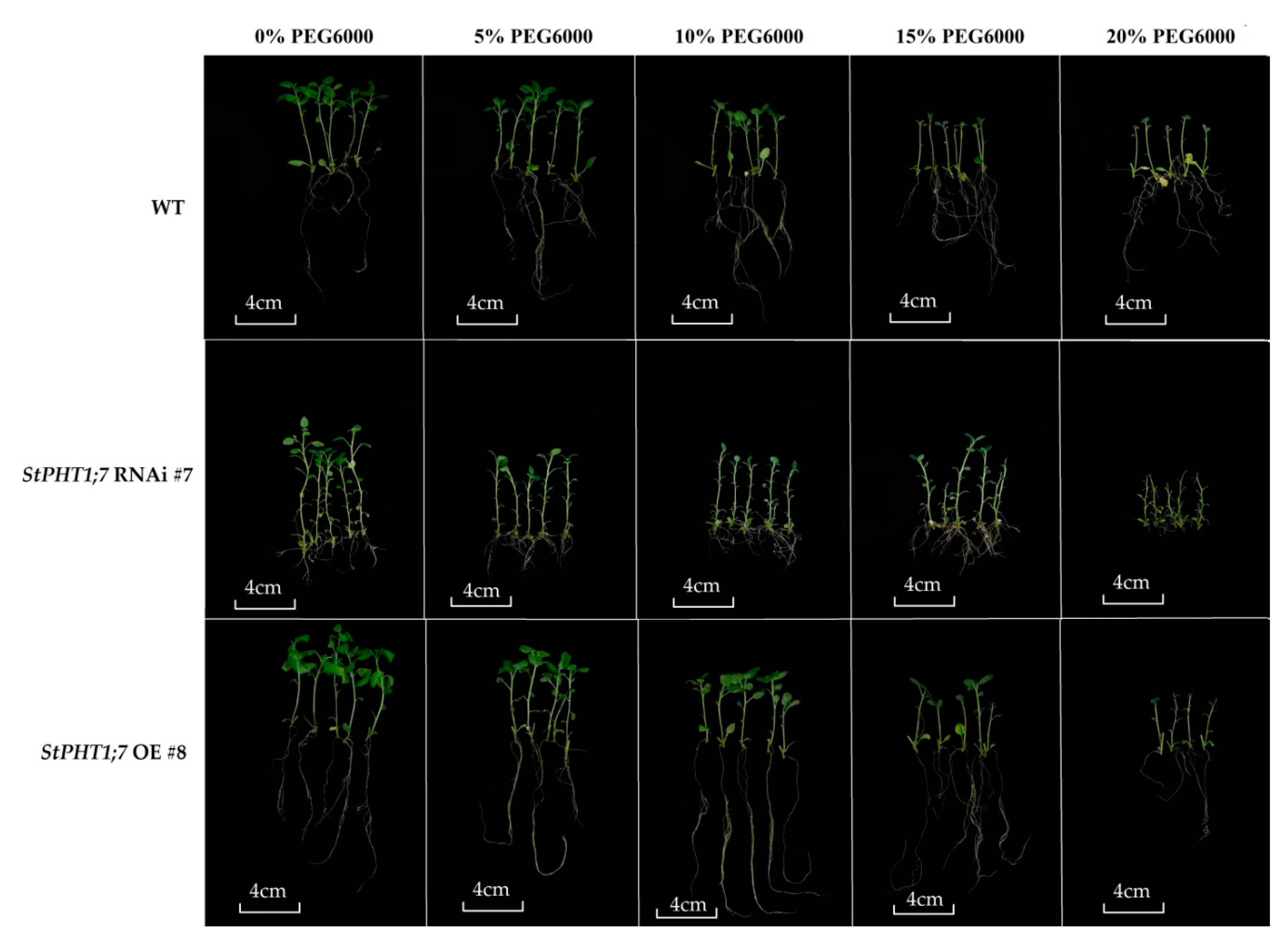

| Line | Growth Parameter (%) | ||||||
|---|---|---|---|---|---|---|---|
| Relative Plant Height | Relative Stem Fresh Weight | Relative Stem Dry Weight | Relative Root Length | Relative Root Fresh Weight | Relative Root Dry Weight | Relative Leaf Area | |
| WT | 70.21a | 44.84a | 75.57a | 58.65b | 127.58a | 193.75a | 46.14a |
| StPHT1;7 RNAi #2 | 62.90a | 38.24b | 55.15b | 94.12ab | 81.80b | 113.16b | 35.25b |
| StPHT1;7 RNAi #7 | 57.40b | 49.44a | 69.78a | 94.94ab | 105.20b | 119.66b | 49.28a |
| StPHT1;7 OE #21 | 53.54b | 31.17b | 44.10b | 53.38b | 106.97b | 129.63b | 17.56c |
| StPHT1;7 OE #8 | 62.51a | 43.81a | 73.35a | 123.09a | 84.61b | 103.77b | 51.04a |
| Mean | 61.31 | 41.50 | 63.59 | 84.83 | 101.23 | 131.99 | 39.85 |
| SD | 10.23 | 13.87 | 20.13 | 38.58 | 49.06 | 75.97 | 26.13 |
| CV | 16.68 | 33.42 | 31..66 | 45.48 | 48.46 | 57.55 | 65.57 |
| Medium | pH | Composition |
|---|---|---|
| Solid MS2 | 5.8 | Murashige and Skoog (MS) powder 4.42 g/L; sucrose: 20 g/L; agar: 8 g/L |
| Liquid MS2 | 5.8 | MS powder 4.42 g/L; sucrose: 20 g/L |
| CIM | 5.8 | MS powder 4.42 g/L; glucose: 1.6 g/L; MES: 0.5 g/L; agar: 6.5 g/L; 6-benzylaminopurine (6-BA): 0.1 mg/L; 1-naphthylacetic acid (NAA): 5 mg/L; kanamycin (Kan): 50 mg/L; timentin (TM): 200 mg/L |
| SIM | 5.8 | MS powder 4.42 g/L; glucose: 1.6 g/L; MES: 0.5 g/L; agar: 6.5 g/L; gibberellin A3 (GA3): 0.1 mg/L; NAA: 0.02 mg/L; zeatin (ZT): 2 mg/L; Kan: 50 mg/L; TM: 200 mg/L |
| SM | 5.8 | MS powder 4.42 g/L; sucrose: 20 g/L; agar: 8 g/L; Kan: 50 mg/L |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, M.; Liu, H.; Zhang, C.; Wang, D.; Liu, X.; Chen, Q. Functional Analysis of StPHT1;7, a Solanum tuberosum L. Phosphate Transporter Gene, in Growth and Drought Tolerance. Plants 2020, 9, 1384. https://doi.org/10.3390/plants9101384
Cao M, Liu H, Zhang C, Wang D, Liu X, Chen Q. Functional Analysis of StPHT1;7, a Solanum tuberosum L. Phosphate Transporter Gene, in Growth and Drought Tolerance. Plants. 2020; 9(10):1384. https://doi.org/10.3390/plants9101384
Chicago/Turabian StyleCao, Minxuan, Hengzhi Liu, Chao Zhang, Dongdong Wang, Xiaofang Liu, and Qin Chen. 2020. "Functional Analysis of StPHT1;7, a Solanum tuberosum L. Phosphate Transporter Gene, in Growth and Drought Tolerance" Plants 9, no. 10: 1384. https://doi.org/10.3390/plants9101384
APA StyleCao, M., Liu, H., Zhang, C., Wang, D., Liu, X., & Chen, Q. (2020). Functional Analysis of StPHT1;7, a Solanum tuberosum L. Phosphate Transporter Gene, in Growth and Drought Tolerance. Plants, 9(10), 1384. https://doi.org/10.3390/plants9101384





