Development of EST-SSR Markers Linked to Flowering Candidate Genes in Elymus sibiricus L. Based on RNA Sequencing
Abstract
1. Introduction
2. Results
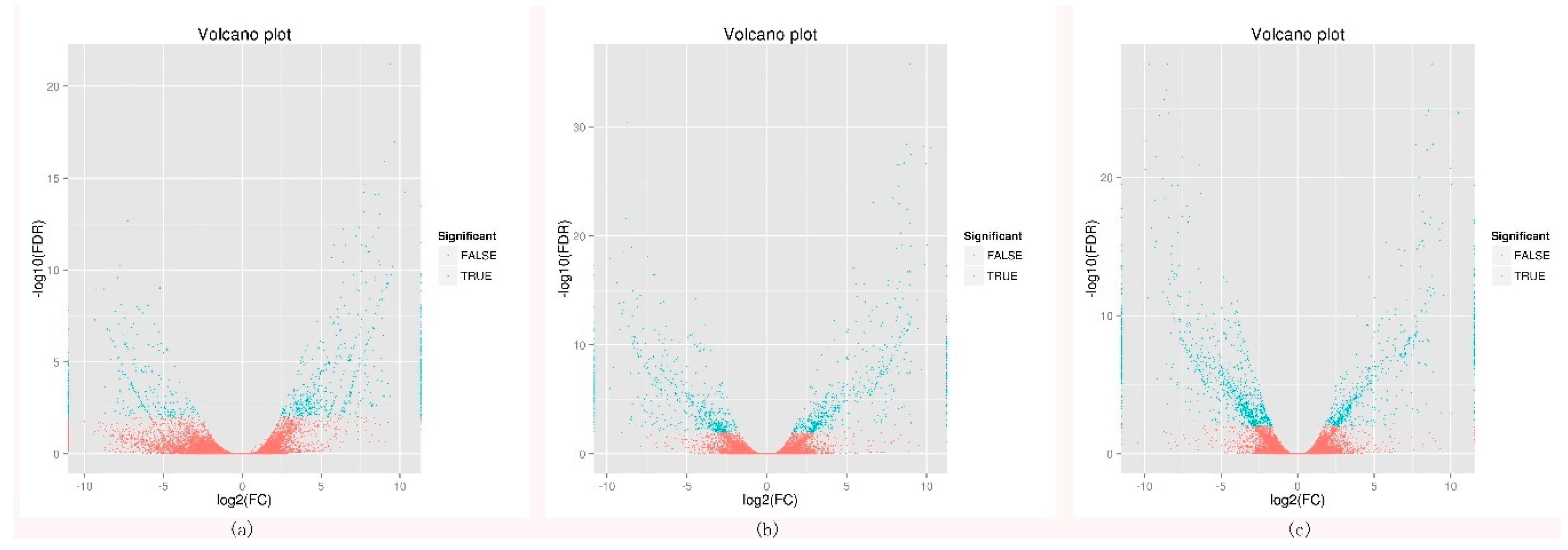
2.1. Transcriptome Sequencing, Unigenes Annotation, and Differentially Expressed Genes Analysis
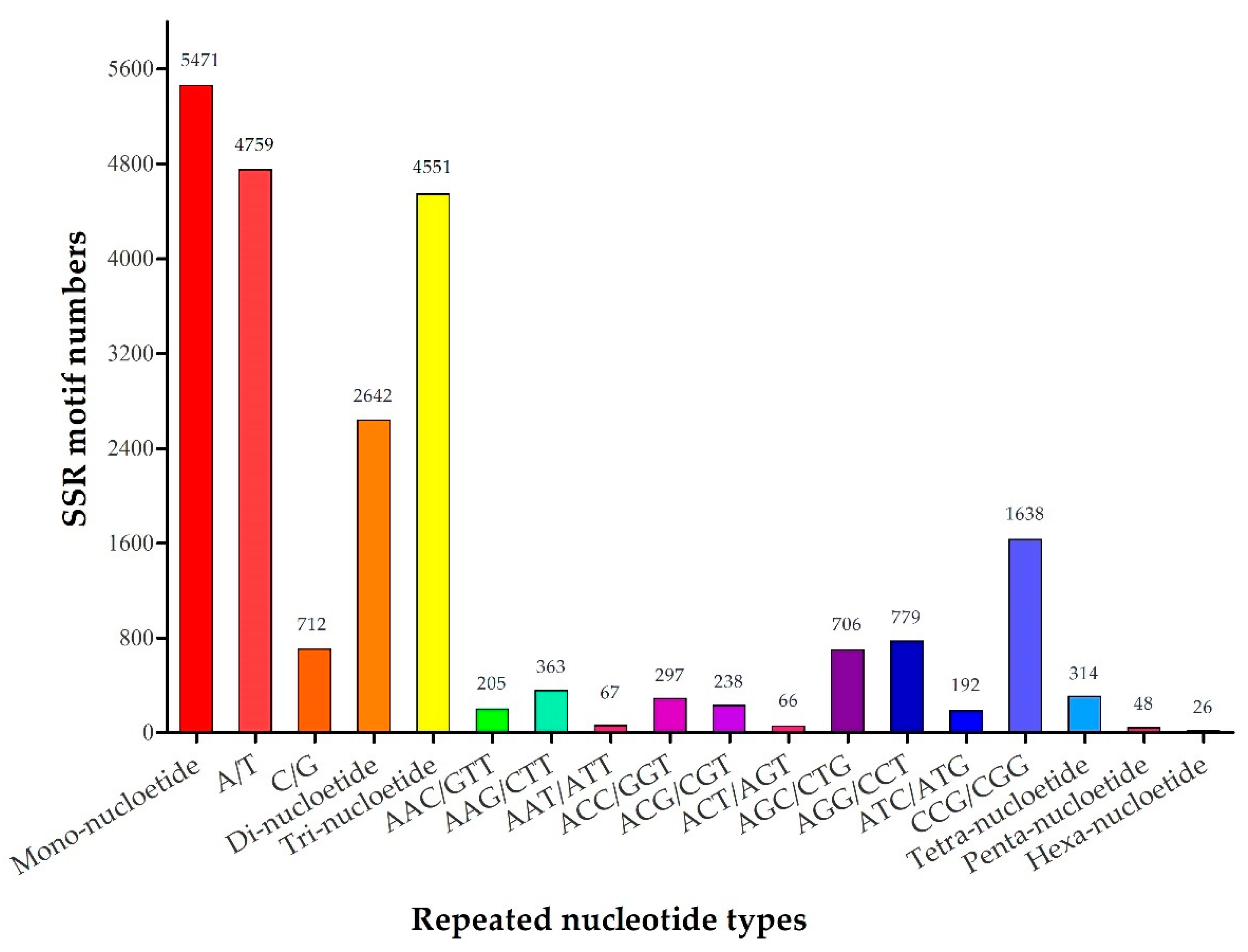
2.2. Frequency and Distribution of EST-SSR Markers
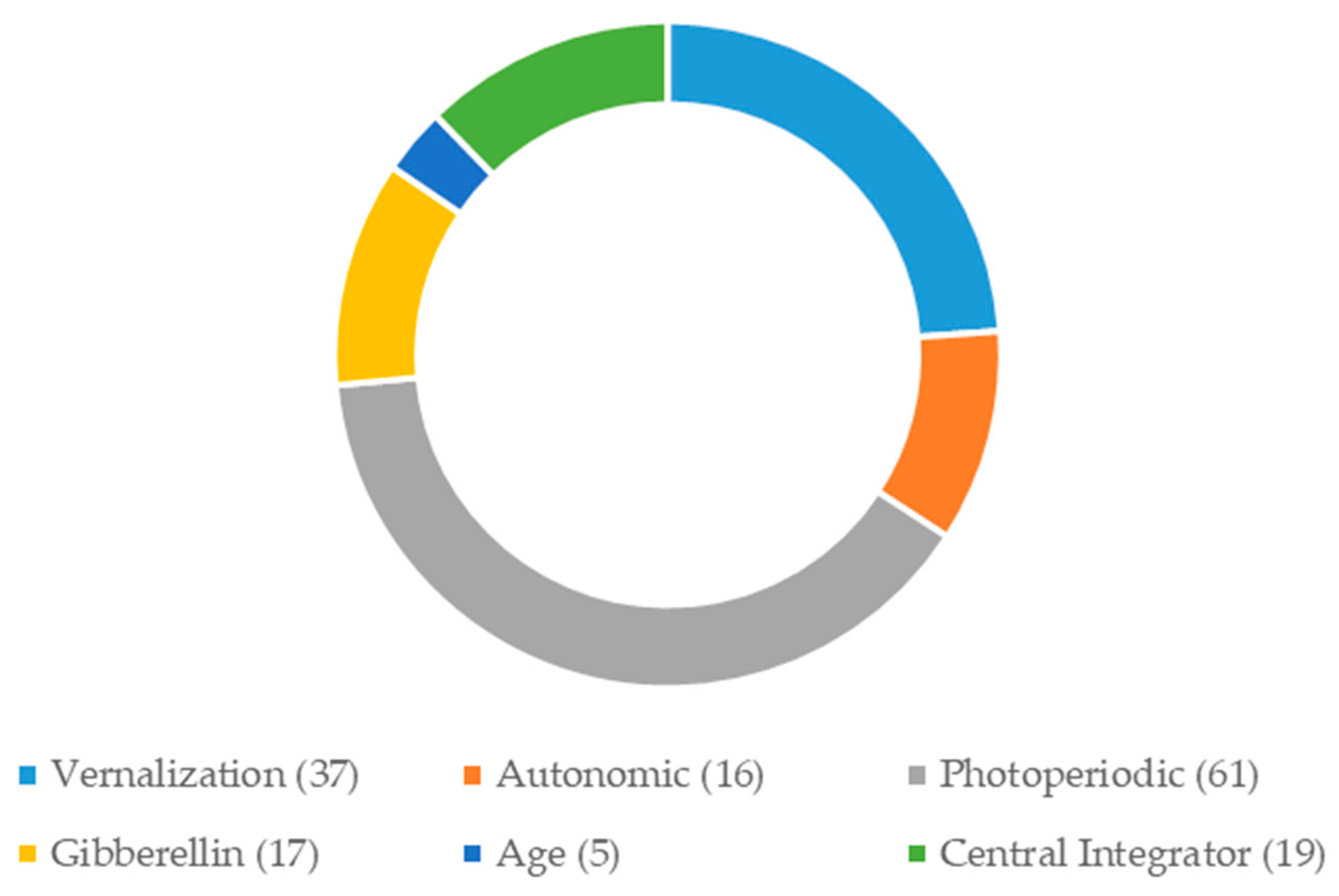
2.3. Development of Candidate Gene-Based EST-SSR Markers
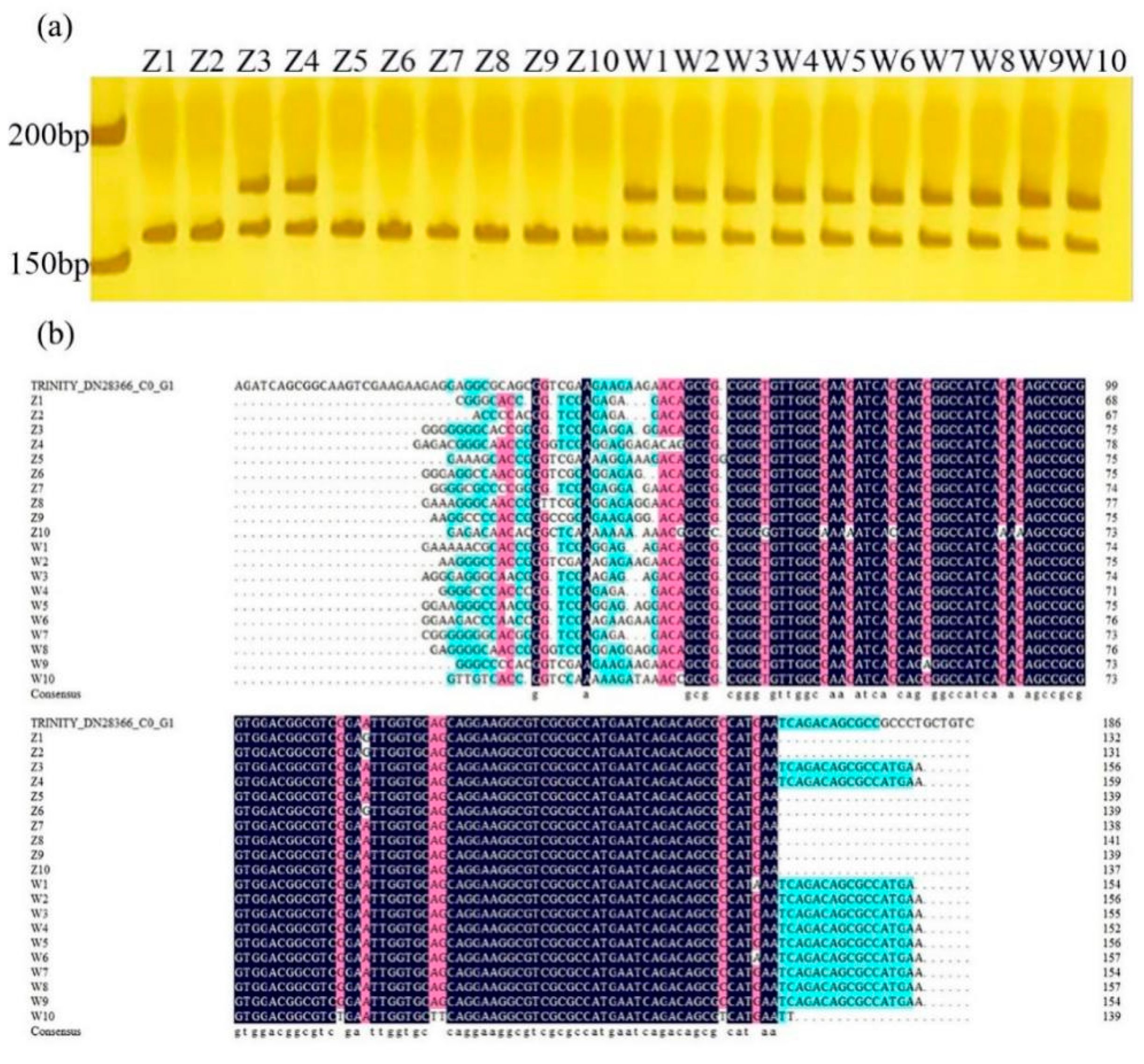
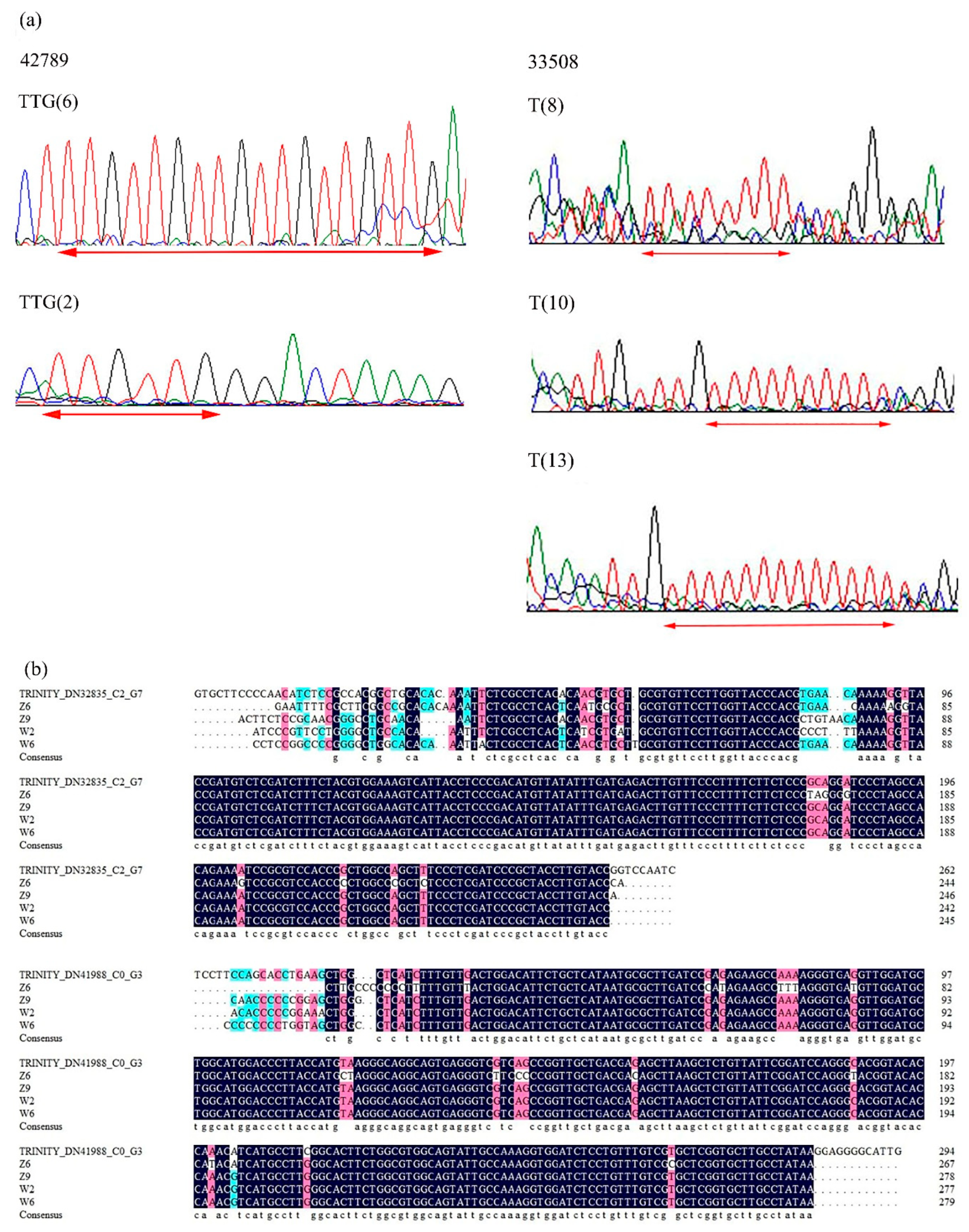
2.4. Validation of Candidate Gene-Based Specific Primers Authenticity
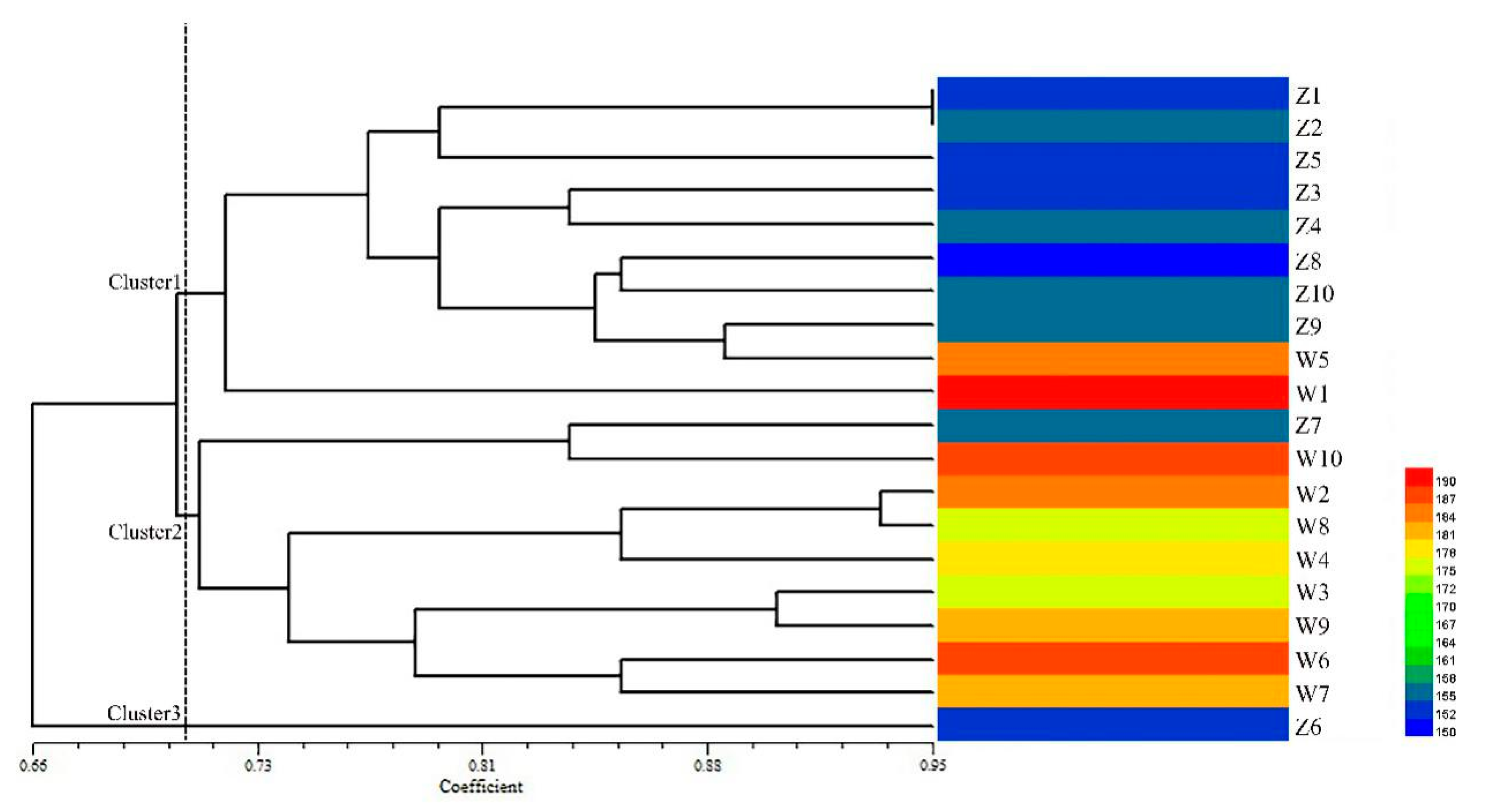
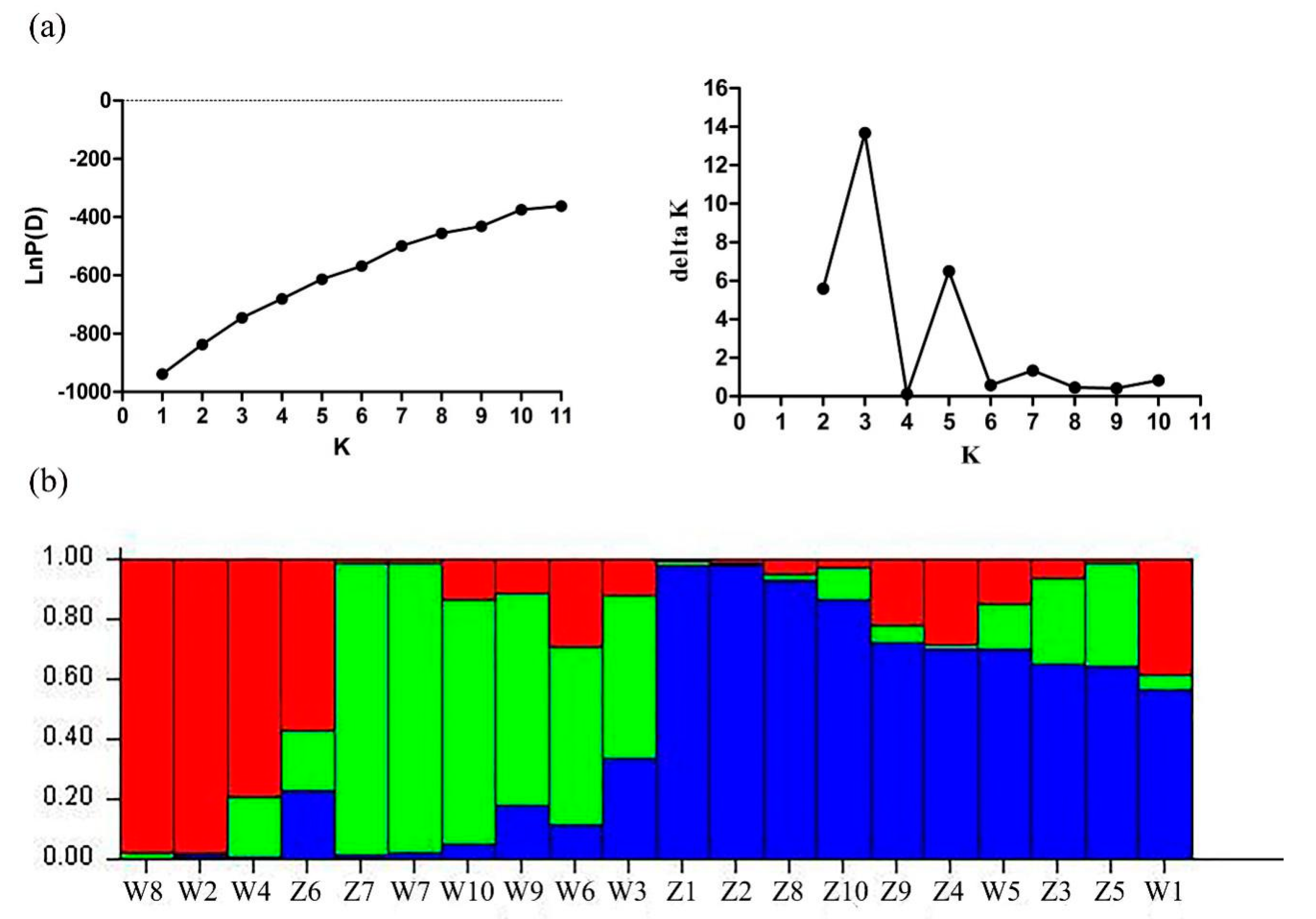
2.5. Genetic Diversity Analysis Using Candidate Gene-Based EST-SSR Markers
3. Discussion
3.1. Candidate Gene-Based EST-SSR Markers with Flowering Time for Marker-Assisted Selection
3.2. Genetic Diversity of Candidate Gene-Based EST-SSR Markers
4. Materials and Methods
4.1. Plant Materials
4.2. Development of Candidate Gene-Based EST-SSR Markers Based on RNA-Seq in E. sibiricus
4.3. DNA Extraction, Genotyping and Primer Validation
4.4. Allele Scoring and Polymorphism Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xie, W.; Zhao, X.; Zhang, J.; Wang, Y.; Liu, W. Assessment of genetic diversity of Siberian wild rye (Elymus sibiricus L.) germplasms with variation of seed shattering and implication for future genetic improvement. Biochem. Syst. Ecol. 2015, 58, 211–218. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.Q.; Zhou, Y.H.; Bai, S.Q.; Liu, W. Assessing genetic diversity of Elymus sibiricus (Poaceae: Triticeae) populations from Qinghai-Tibet Plateau by ISSR markers. Biochem. Syst. Ecol. 2008, 36, 514–522. [Google Scholar] [CrossRef]
- Xie, W.; Zhang, J.; Zhao, X.; Zhang, J.; Wang, Y. Siberian wild rye (Elymus sibiricus L.): Genetic diversity of germplasm determined using DNA fingerprinting and SCoT markers. Biochem. Syst. Ecol. 2015, 60, 186–192. [Google Scholar] [CrossRef]
- Craufurd, P.Q.; Wheeler, T.R. Climate change and the flowering time of annual crops. J. Exp. Bot. 2009, 60, 2529–2539. [Google Scholar] [CrossRef] [PubMed]
- Torabi, B.; Adibnya, M.; Rahimi, A.; Azari, A. Modeling flowering response to temperature and photoperiod in safflower. Ind. Crop. Prod. 2020, 151, 10. [Google Scholar] [CrossRef]
- Simpson, G.G.; Dean, C. Flowering—Arabidopsis, the rosetta stone of flowering time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Osnato, M.; Castillejo, C.; Matias-Hernandez, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 2001, 13, 935–941. [Google Scholar] [CrossRef]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.H.; Fornara, F.; Fan, Q.Z.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.C.; Qiao, L.F.; Chen, J.C.; Rong, Y.H.; Zhao, Y.H.; Cui, X.K.; Xu, J.P.; Hou, X.M.; Dong, C.H. Arabidopsis REM16 acts as a B3 domain transcription factor to promote flowering time via directly binding to the promoters of SOC1 and FT. Plant J. 2020. [Google Scholar] [CrossRef]
- Li, C.X.; Lin, H.Q.; Chen, A.; Lau, M.; Jernstedt, J.; Dubcovsky, J. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development 2019, 146, 11. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.C.; Zhou, Z.J.; Zho, X.S.; Yang, H.Y.; Li, M.R.; Li, H.Q. Overexpression of OsFTL10 induces early flowering and improves drought tolerance in Oryza sativa L. PeerJ 2019, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, T.; Wang, Y.M.; Zheng, J.; Li, H.F.; Hao, C.Y.; Zhang, X.Y. TaZIM-A1 negatively regulates flowering time in common wheat (Triticum aestivum L.). J. Integr. Plant Biol. 2019, 61, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.M.; Zhang, T.J.; Guo, T.; Ding, W.; Long, R.C.; Yang, Q.C.; Wang, Z. Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa). Int. J. Mol. Sci. 2019, 20, 1968. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Feng, G.Y.; Yan, H.D.; Zhang, Z.R.; Bushman, B.S.; Wang, J.P.; Bombarely, A.; Li, M.Z.; Yang, Z.F.; Nie, G.; et al. Genome assembly provides insights into the genome evolution and flowering regulation of orchardgrass. Plant Biotechnol. J. 2020, 18, 373–388. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.J.; Zhang, Q.; Qi, Y.; Guo, D.J. Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genom. 2009, 10, 10. [Google Scholar] [CrossRef]
- Der, J.P.; Barker, M.S.; Wickett, N.J.; de Pamphilis, C.W.; Wolf, P.G. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genom. 2011, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Arghavan, A.; Shekoufeh, E.; Sahar, A.; Mahsa, H.; Behrouz, S.; Hassan, J.; Hossein, F.; Sadegh, M.-F.; Fariba, R. Parallel consideration of SSRs and differentially expressed genes under abiotic stress for targeted development of functional markers in almond and related Prunus species. Sci. Hortic. 2015, 198, 462–472. [Google Scholar] [CrossRef]
- Singh, A.K.; Chaurasia, S.; Kumar, S.; Singh, R.; Kumari, J.; Yadav, M.C.; Singh, N.; Gaba, S.; Jacob, S.R. Identification, analysis and development of salt responsive candidate gene based SSR markers in wheat. BMC Plant Biol. 2018, 18, 15. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.K.; Agrawal, P.K.; Gupta, H.S.; Kumar, A.; Bhatt, J.C. Identification of candidate gene-based SSR markers for lysine and tryptophan metabolic pathways in maize (Zea mays). Plant Breed. 2012, 131, 20–27. [Google Scholar] [CrossRef]
- Jespersen, D.; Ma, X.Q.; Bonos, S.A.; Belanger, F.C.; Raymer, P.; Huang, B.R. Association of SSR and candidate gene markers with genetic variations in summer heat and drought performance for creeping bentgrass. Crop. Sci. 2018, 58, 2644–2656. [Google Scholar] [CrossRef]
- Lai, D.Y.; Li, H.Z.; Fan, S.L.; Song, M.Z.; Pang, C.Y.; Wei, H.L.; Liu, J.J.; Wu, D.; Gong, W.F.; Yu, S.X. Generation of ESTs for flowering gene discovery and SSR marker development in upland cotton. PLoS ONE 2011, 6, e28676. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.Q.; Zhang, J.C.; Zhang, Z.Y.; Xie, W.G. Elymus nutans genes for seed shattering and candidate gene-derived EST-SSR markers for germplasm evaluation. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef]
- Danilevskaya, O.N.; Meng, X.; McGonigle, B.; Muszynski, M.G. Beyond flowering time Pleiotropic function of the maize flowering hormone florigen. Plant Signal. Behav. 2011, 6, 1267–1270. [Google Scholar] [CrossRef]
- Bushman, B.S.; Robins, J.C.; Jensen, K.B. Dry matter yield, heading date, and plant mortality of Orchardgrass subspecies in a semiarid environment. Crop. Sci. 2012, 52, 745–751. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B-Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Unamba, C.I.; Nag, A.; Sharma, R.K. Next Generation Sequencing Technologies: The doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 2015, 6, 1074. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Sharma, S.; Singh, R.; Kumar, N.; Balyan, H.S. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol. Genet. Genom. 2003, 270, 315–323. [Google Scholar] [CrossRef]
- Saha, M.C.; Mian, M.A.R.; Eujayl, I.; Zwonitzer, J.C.; Wang, L.J.; May, G.D. Tall fescue EST-SSR markers with transferability across several grass species. Theor. Appl. Genet. 2004, 109, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gupta, S.; Biradar, R.S.; Gupta, P.; Dubey, S.; Singh, N.P. Association of functional markers with flowering time in lentil. J. Appl. Genet. 2018, 59, 9–21. [Google Scholar] [CrossRef]
- Li, D.M.; Wu, W.; Zhang, D.; Liu, X.R.; Liu, X.F.; Lin, Y.J. Floral transcriptome analyses of four Paphiopedilum Orchids with distinct flowering behaviors and development of simple sequence repeat markers. Plant Mol. Biol. Rep. 2015, 33, 1928–1952. [Google Scholar] [CrossRef]
- Monna, L.; Lin, H.X.; Kojima, S.; Sasaki, T.; Yano, M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor. Appl. Genet. 2002, 104, 772–778. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Armstead, I.P.; Turner, L.B.; Farrell, M.; Skot, L.; Gomez, P.; Montoya, T.; Donnison, I.S.; King, I.P.; Humphreys, M.O. Synteny between a major heading-date QTL in perennial ryegrass (Lolium perenne L.) and the Hd3 heading-date locus in rice. Theor. Appl. Genet. 2004, 108, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.U.; Davis, A.; Davis, S.J.; Quint, M. Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. 2020, 101, 1397–1410. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hao, X.Y.; Lu, Q.H.; Zhang, W.F.; Zhang, H.J.; Wang, L.; Yang, Y.J.; Xiao, B.; Wang, X.C. Genome-wide identification and expression analysis of flowering-related genes reveal putative floral induction and differentiation mechanisms in tea plant (Camellia sinensis). Genomics 2020, 112, 2318–2326. [Google Scholar] [CrossRef]
- Li, C.; Zheng, L.L.; Wang, X.N.; Hu, Z.B.; Zheng, Y.; Chen, Q.H.; Hao, X.C.; Xiao, X.; Wang, X.B.; Wang, G.D.; et al. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 2019, 285, 1–13. [Google Scholar] [CrossRef]
- Martin-Tryon, E.L.; Harmer, S.L. XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell 2008, 20, 1244–1259. [Google Scholar] [CrossRef]
- Zeng, J.; Mo, Y.L.; Chen, J.J.; Li, C.M.; Zhao, L.; Liu, Y.H. Expression and interaction proteins analysis of BjuFKF1 in stem mustard. Sci. Hortic. 2020, 269, 6. [Google Scholar] [CrossRef]
- Asp, T.; Frei, U.K.; Didion, T.; Nielsen, K.K.; Lubberstedt, T. Frequency, type, and distribution of EST-SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon, and Oryza sativa. BMC Plant Biol. 2007, 7. [Google Scholar] [CrossRef]
- Yan, Z.; Wu, F.; Luo, K.; Zhao, Y.; Yan, Q.; Zhang, Y.; Wang, Y.; Zhang, J. Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetics research. Sci. Rep. 2017, 7, 17959. [Google Scholar] [CrossRef]
- Pan, L.; Huang, T.; Yang, Z.; Tang, L.; Cheng, Y.; Wang, J.; Ma, X.; Zhang, X. EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species. Mol. Breed. 2018, 38. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, W.; Zhao, Y.; Zhang, J.; Wang, N.; Ntakirutimana, F.; Yan, J.; Wang, Y. EST-SSR marker development based on RNA-sequencing of E. sibiricus and its application for phylogenetic relationships analysis of seventeen Elymus species. BMC Plant Biol. 2019, 19, 235. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, G.; Shi, B.; Wang, X.; Qiang, H.; Gao, H. Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 2014, 9, e92029. [Google Scholar] [CrossRef]
- Zhang, M.A.; Mao, W.H.; Zhang, G.P.; Wu, F.B. Development and characterization of polymorphic EST-SSR and genomic SSR markers for Tibetan annual wild barley. PLoS ONE 2014, 9, e94881. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Liu, P.; Luo, D.; Liu, W.X.; Wang, Y.R. Exploiting illumina sequencing for the development of 95 novel polymorphic EST-SSR markers in common vetch (Vicia sativa subsp sativa). Molecules 2014, 19, 5777–5789. [Google Scholar] [CrossRef] [PubMed]
- Hajkova, L.; Koznarova, V.; Mozny, M.; Bartosova, L. Influence of climate change on flowering season of birch in the Czech Republic. Int. J. Biometeorol. 2020, 64, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Grenier, C.; Bramel-Cox, P.J.; Hamon, P. Core collection of sorghum: I. Stratification based on eco-geographical data. Crop. Sci. 2001, 41, 234–240. [Google Scholar] [CrossRef]
- Stuerz, S.; Shrestha, S.P.; Schmierer, M.; Vu, D.H.; Hartmann, J.; Sow, A.; Razafindrazaka, A.; Abera, B.B.; Chuma, B.A.; Asch, F. Climatic determinants of lowland rice development. J. Agron. Crop. Sci. 2020, 206, 466–477. [Google Scholar] [CrossRef]
- Burgarella, C.; Chantret, N.; Gay, L.; Prosperi, J.M.; Bonhomme, M.; Tiffin, P.; Young, N.D.; Ronfort, J. Adaptation to climate through flowering phenology: A case study in Medicago truncatula. Mol. Ecol. 2016, 25, 3397–3415. [Google Scholar] [CrossRef]
- Wang, Q.B.; Wang, Y.P.; Zhang, L. Inheritance and molecular marker for flowering time in radish (Raphanus sativus L.). Plant Mol. Biol. Rep. 2018, 36, 878–887. [Google Scholar] [CrossRef]
- Xie, W.G.; Robins, J.G.; Bushman, B.S. A genetic linkage map of tetraploid orchardgrass (Dactylis glomerata L.) and quantitative trait loci for heading date. Genome 2012, 55, 360–369. [Google Scholar] [CrossRef]
- Wang, N. Evaluation of Flowering Time and Mining of Candidate Genes in Elymus Sibiricus. Master’s Thesis, Lanzhou University, Lanzhou, China, 1 May 2020. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Guerra, V.; Beule, L.; Lehtsaar, E.; Liao, H.L.; Karlovsky, P. Improved protocol for DNA extraction from subsoils using phosphate lysis buffer. Microorganisms 2020, 8, 532. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Paul, J. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar] [CrossRef]
- Hudson, R.R. Gennetic data-analysis-methods for discrete population genetic data-Weir, BS. Science 1990, 250, 575. [Google Scholar] [CrossRef]

| Searching Item | Number |
|---|---|
| Total number of sequences examined | 40,639 |
| Total size of examined sequences (bp) | 78,157,064 |
| Total number of identified SSRs | 13,052 |
| Number of SSR containing sequences | 10,286 |
| Number of sequences containing more than 1 SSR | 2176 |
| Number of SSRs present in compound formation | 742 |
| Mono-nucleotide | 5471 |
| Di-nucleotide | 2642 |
| Tri-nucleotide | 4551 |
| Tetra-nucleotide | 314 |
| Penta-nucleotide | 48 |
| Hexa-nucleotide | 26 |
| Primer Code | Flowering Genes | Flowering Pathways/Functions | He | Ho | PIC |
|---|---|---|---|---|---|
| Primer 46865 | FPF1 | Flowering | 0.6592 | 0.9231 | 0.4767 |
| Primer 33680 | FPF1 | Flowering | 0.2268 | 0.1500 | 0.1275 |
| Primer 43088 | ELF3 | Flowering | 0.7966 | 0.9412 | 0.4210 |
| Primer 41496 | GID1 | Gibberellin | 0.4911 | 0.5000 | 0.2367 |
| Primer 40505 | GA2OX6 | Gibberellin | 0.6655 | 0.7000 | 0.2513 |
| Primer 37852 | CIGR | Gibberellin | 0.1975 | 0.0588 | 0.2500 |
| Primer 34975 | XCT | Circadian Clock | 0.5170 | 0.8000 | 0.1567 |
| Primer 48198 | GIGANTEA | Circadian Clock | 0.6584 | 1.0000 | 0.1400 |
| Primer 34500 | MBD9 | CONSTANS-Like | 0.8731 | 1.0000 | 0.2606 |
| Primer 31670 | NFYC4 | CONSTANS-Like | 0.3200 | 0.5946 | 0.1875 |
| Primer 43287 | COL4 | CONSTANS-Like | 0.7138 | 0.9730 | 0.3325 |
| Primer 36067 | COL13 | CONSTANS-Like | 0.8117 | 0.9730 | 0.3025 |
| Primer 36927 | FLC | Central Integrator | 0.8147 | 1.0000 | 0.1500 |
| Primer 34261 | HD3 | Central Integrator | 0.7794 | 1.0000 | 0.2590 |
| Primer 28366 | HD3a | Central Integrator | 0.6509 | 1.0000 | 0.1600 |
| Mean | 0.6117 | 0.7742 | 0.2475 |
| Code | Accession Number | Sample ID | Origins | Longitude/Latitude | Altitude (m) |
|---|---|---|---|---|---|
| 1 | PI598781 | Z1 | Baikal, Buryat, Russia | - | - |
| 2 | PI610876 | Z2 | - | - | - |
| 3 | PI610994 | Z3 | Siberia, Russia | - | 1250 |
| 4 | PI636676 | Z4 | Xiahe, Gansu, China | 35.19 N, 102.65 E | 2720 |
| 5 | PI655199 | Z5 | Hongyuan, Sichuan, China | 32.08 N, 102.57 E | 3280 |
| 6 | W610305 | Z6 | Siberia, Russia | - | - |
| 7 | W630476 | Z7 | Kyrgyzstan | - | - |
| 8 | HZ03 | Z8 | Hezuo, Gansu, China | - | - |
| 9 | LT05 | Z9 | Lintan, Gansu, China | - | - |
| 10 | LQ02 | Z10 | Luqu, Gansu, China | - | - |
| 11 | PI435088 | W1 | Ninshan, Xinjiang, China | - | - |
| 12 | PI504462 | W2 | Xining, Qinghai, China | - | - |
| 13 | PI531665 | W3 | Beijing, China | - | - |
| 14 | PI531669 | W4 | Xining, Qinghai, China | - | 2400 |
| 15 | PI595149 | W5 | Xinjiang, China | 44.17 N, 84.57 E | 1500 |
| 16 | PI595156 | W6 | Xinjiang, China | 45.03 N, 81.12 E | 1300 |
| 17 | PI595162 | W7 | Xinjiang, China | 44.12 N, 87.97 E | 1680 |
| 18 | PI595169 | W8 | Xinjiang, China | 43.77 N, 89.45 E | 1300 |
| 19 | PI595174 | W9 | Xinjiang, China | 43.68 N, 89.30 E | 1870 |
| 20 | PI655092 | W10 | Mongolia | 49.80 N, 94.96 E | 1370 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Zhang, Z.; Wan, Y.; Tian, J.; Xie, W. Development of EST-SSR Markers Linked to Flowering Candidate Genes in Elymus sibiricus L. Based on RNA Sequencing. Plants 2020, 9, 1371. https://doi.org/10.3390/plants9101371
Zheng Y, Zhang Z, Wan Y, Tian J, Xie W. Development of EST-SSR Markers Linked to Flowering Candidate Genes in Elymus sibiricus L. Based on RNA Sequencing. Plants. 2020; 9(10):1371. https://doi.org/10.3390/plants9101371
Chicago/Turabian StyleZheng, Yuying, Zongyu Zhang, Yiyang Wan, Jiaoyang Tian, and Wengang Xie. 2020. "Development of EST-SSR Markers Linked to Flowering Candidate Genes in Elymus sibiricus L. Based on RNA Sequencing" Plants 9, no. 10: 1371. https://doi.org/10.3390/plants9101371
APA StyleZheng, Y., Zhang, Z., Wan, Y., Tian, J., & Xie, W. (2020). Development of EST-SSR Markers Linked to Flowering Candidate Genes in Elymus sibiricus L. Based on RNA Sequencing. Plants, 9(10), 1371. https://doi.org/10.3390/plants9101371




