Molecular Characterization of the Complete Coding Sequence of Olive Leaf Yellowing-Associated Virus
Abstract
1. Introduction
2. Results
2.1. Genome Organization of Olive Yellowing Leaf-Associated Virus
2.1.1. Bioinformatic Analysis
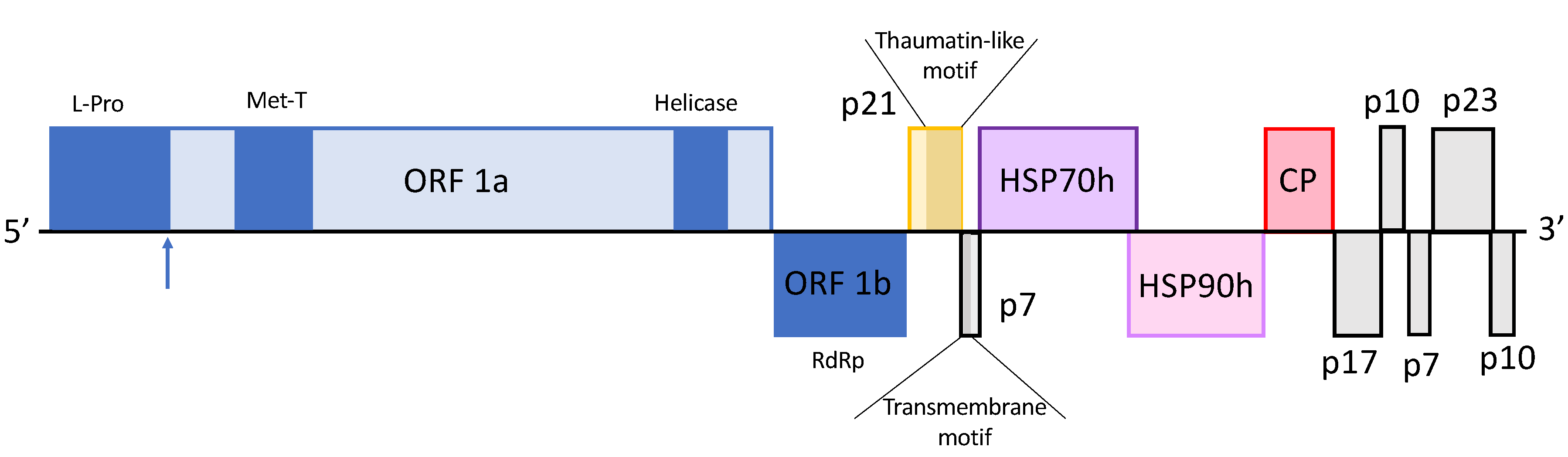
2.1.2. Genomic Structure of OLYaV-CS1
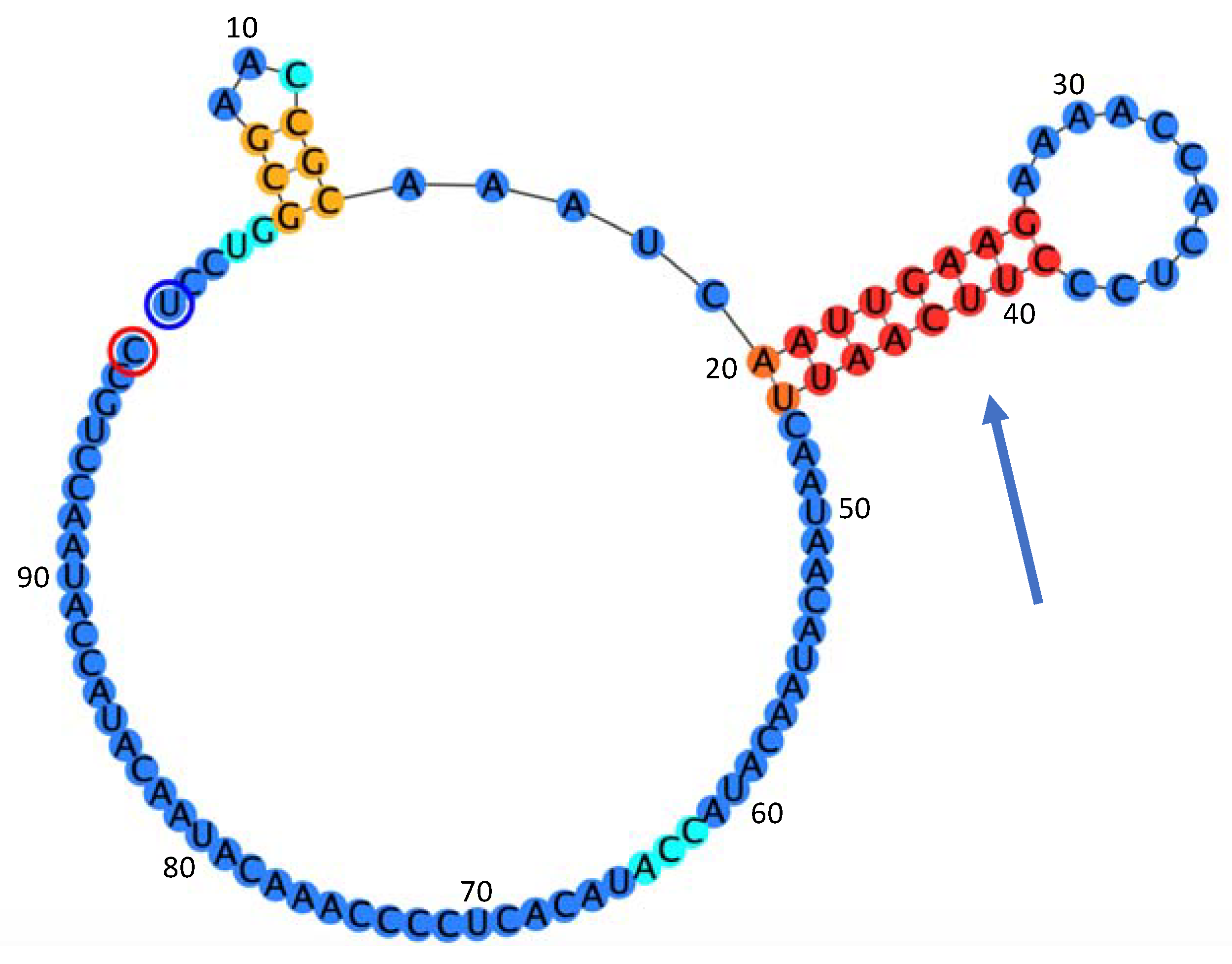
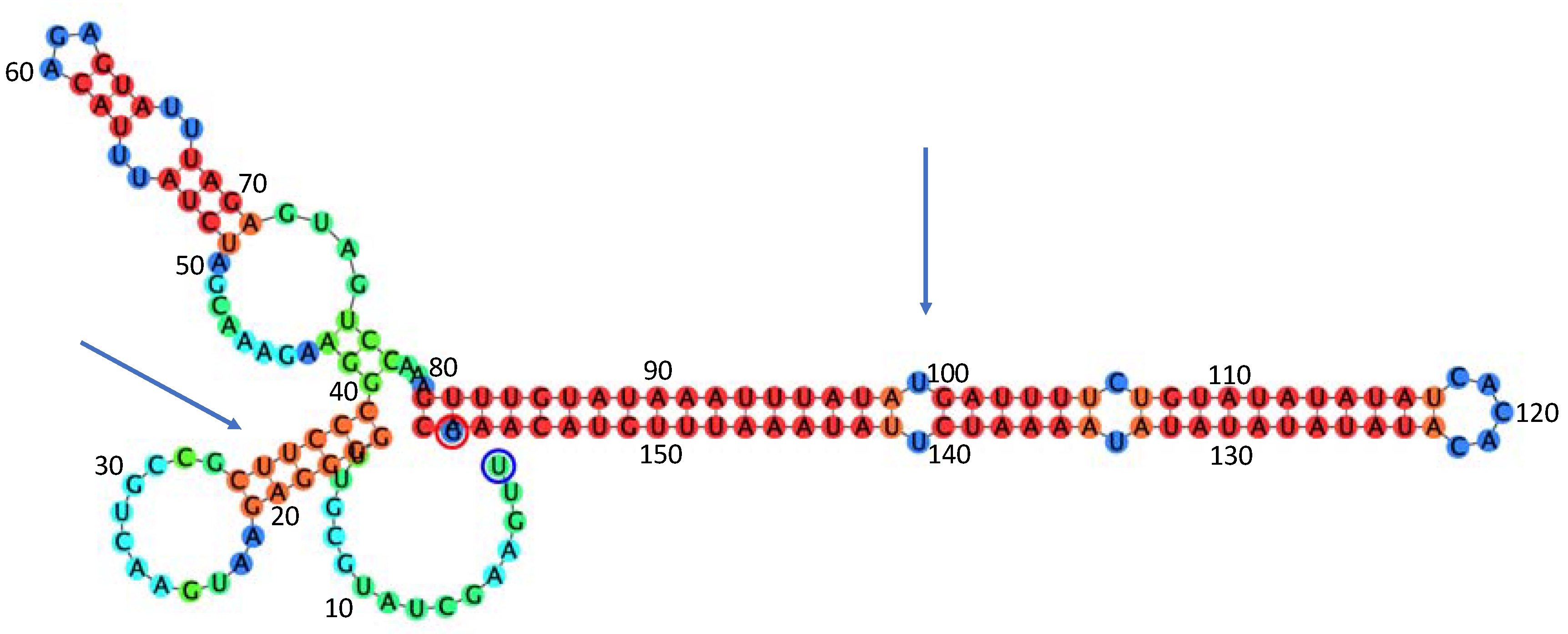
- 5′UTR (1–98 nt)
- ORF1ab (nt 99–9792)
- ORF1a (nt 99–8285)
- 2.
- ORF1b (8287–9793 nt)
- ORF2 (nt 9861–10,430)
- ORF3 (nt 10,430–10,618)
- ORF4 (nt 10,615–12,390)
- ORF5 (nt 12,290–13,837)
- ORF6 (nt 13,904–14,605)
- ORF7 (nt 14,606–15,085), ORF8 (nt 15,087–15,365), ORF9 (nt 15,375–15,569), ORF10 (nt 15,592–16,266) and ORF11 (nt 16,266–16,541).
- 3′UTR (16,542–16,700 nt)
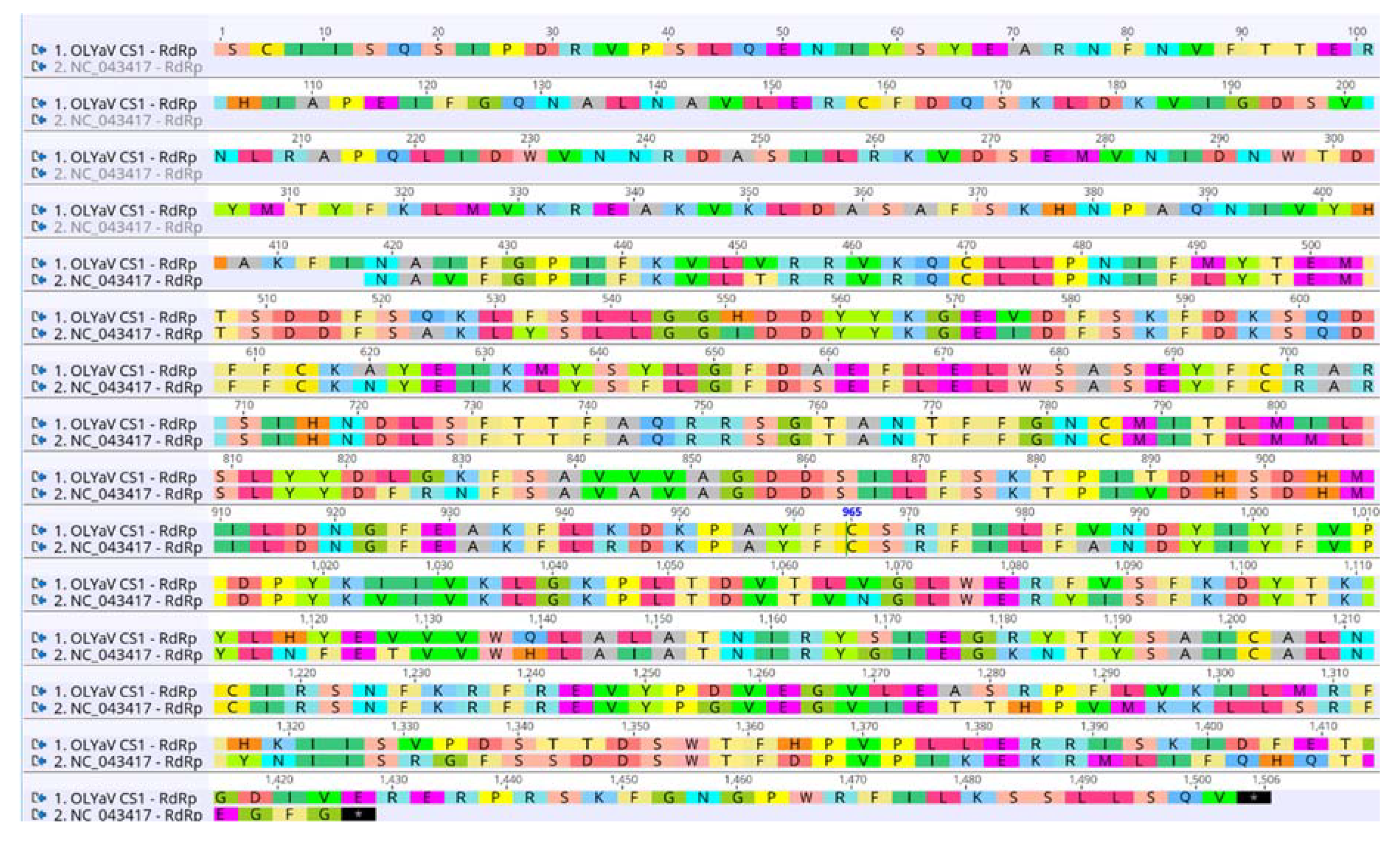
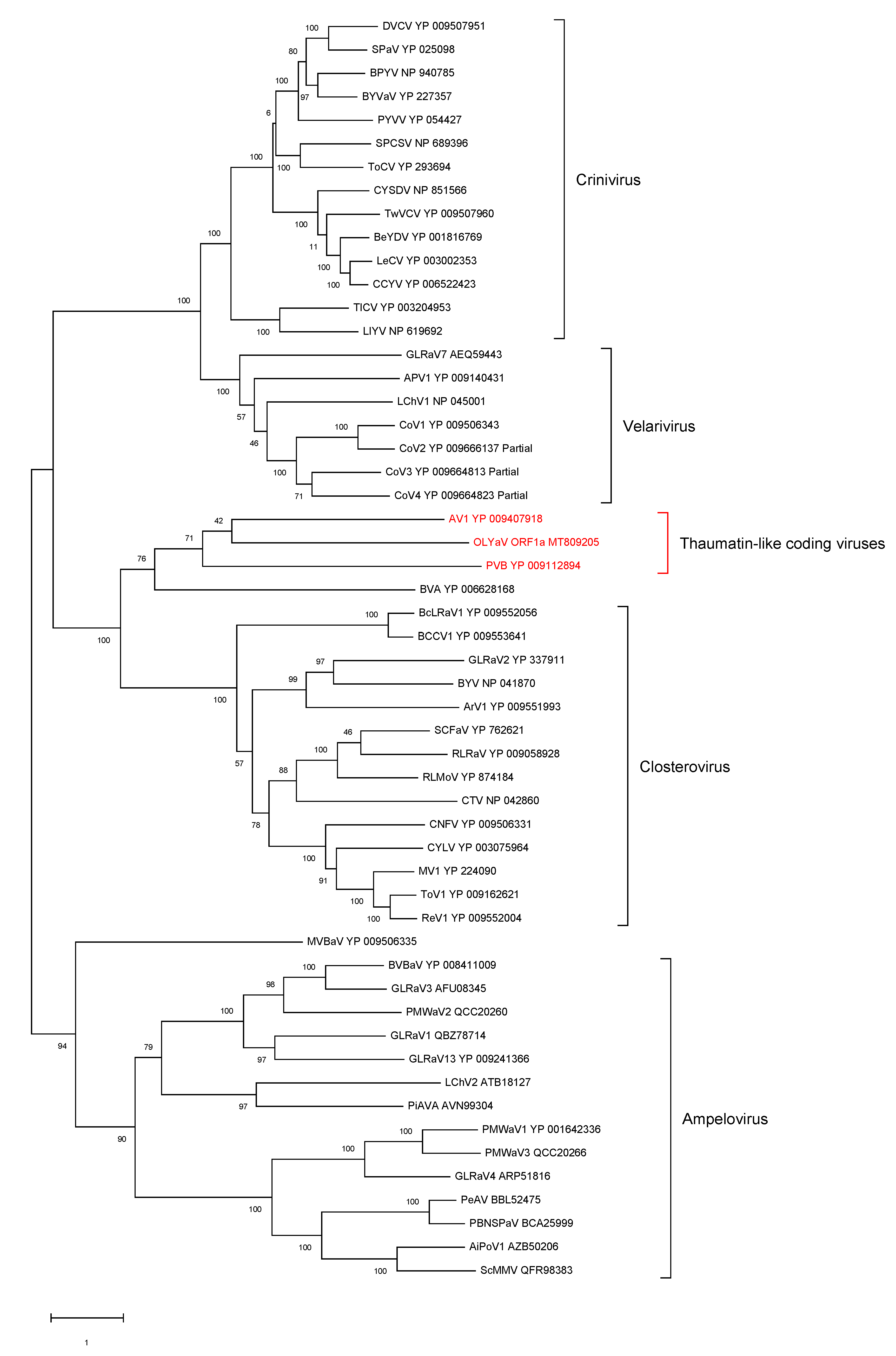
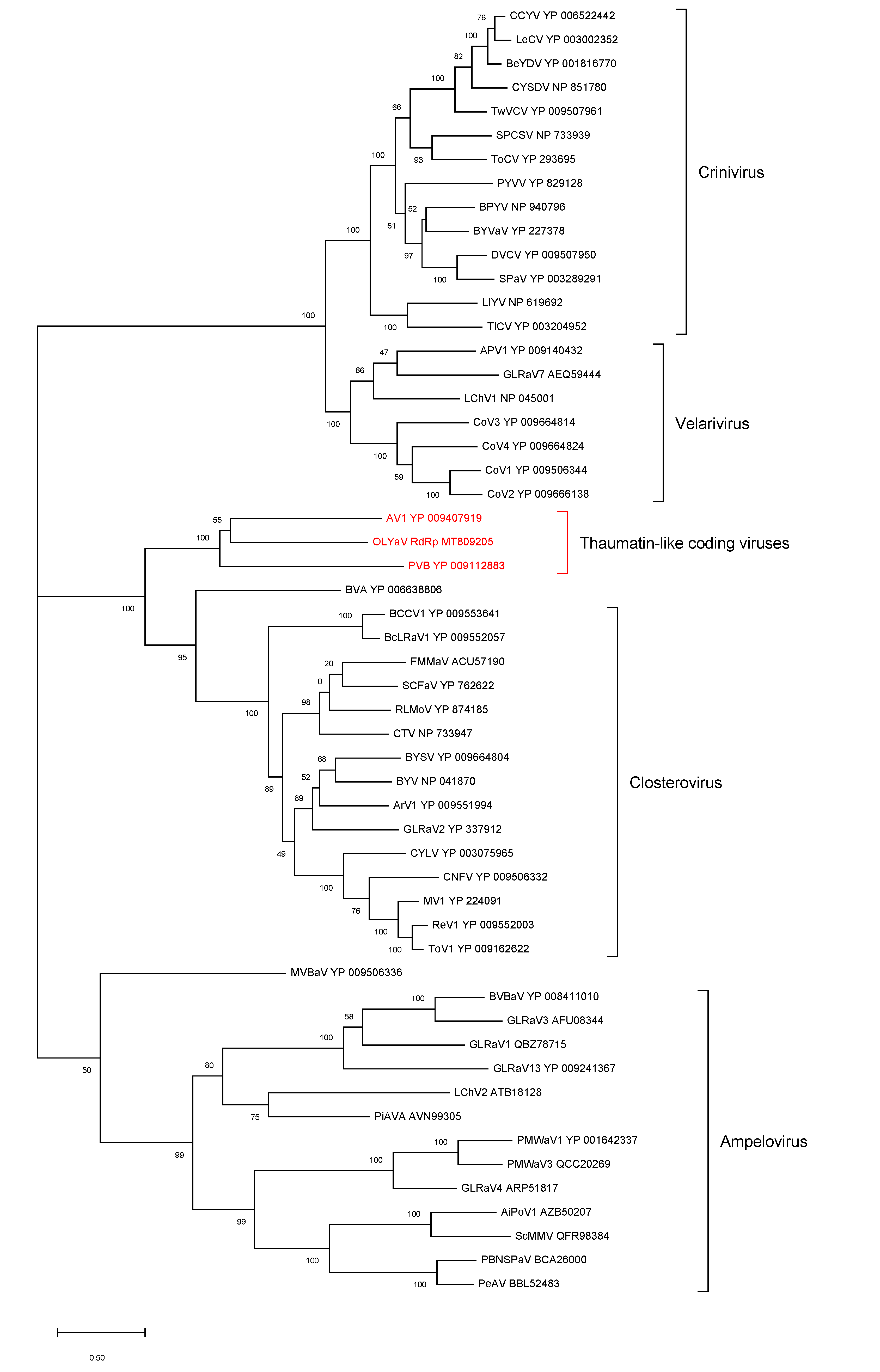
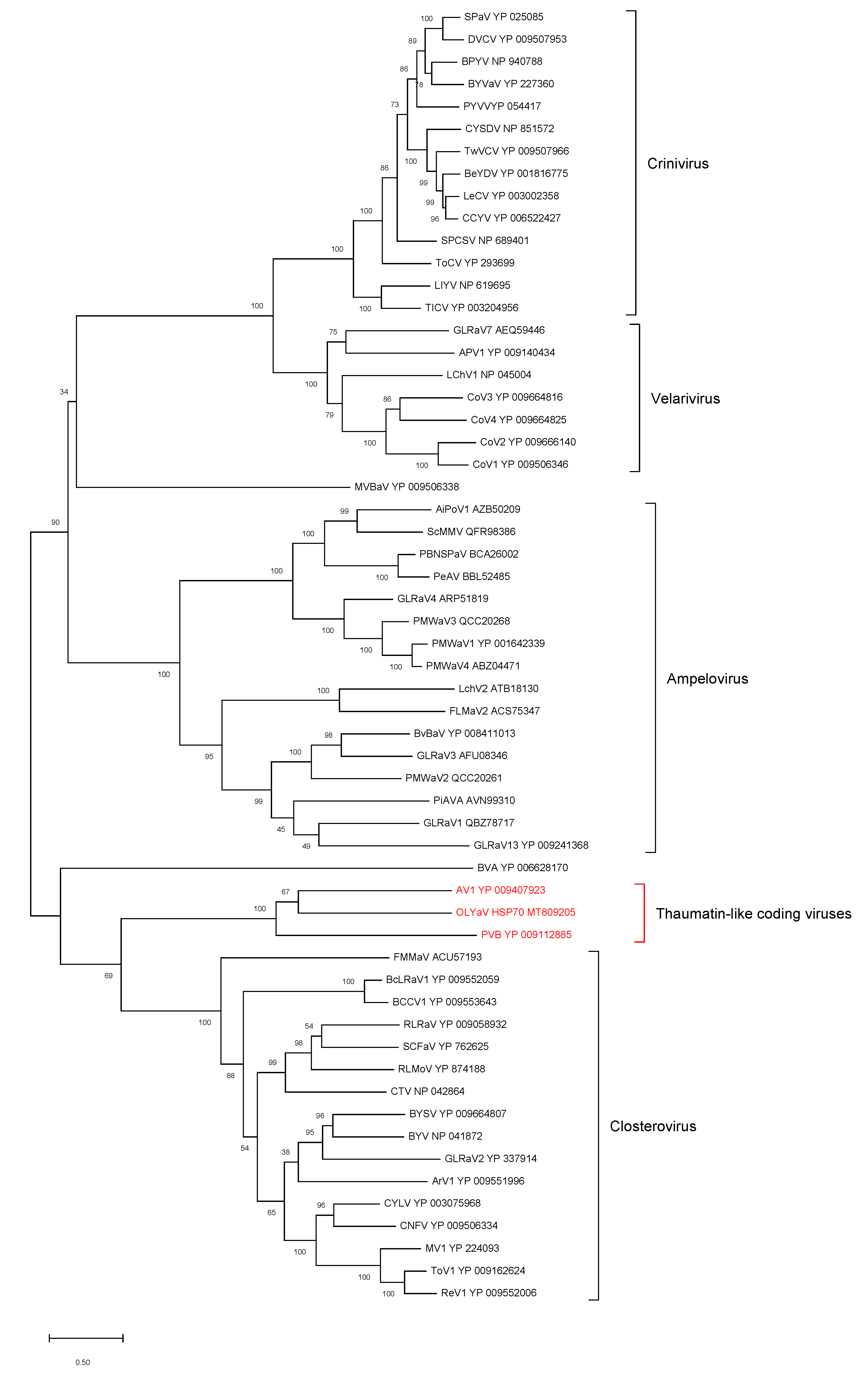
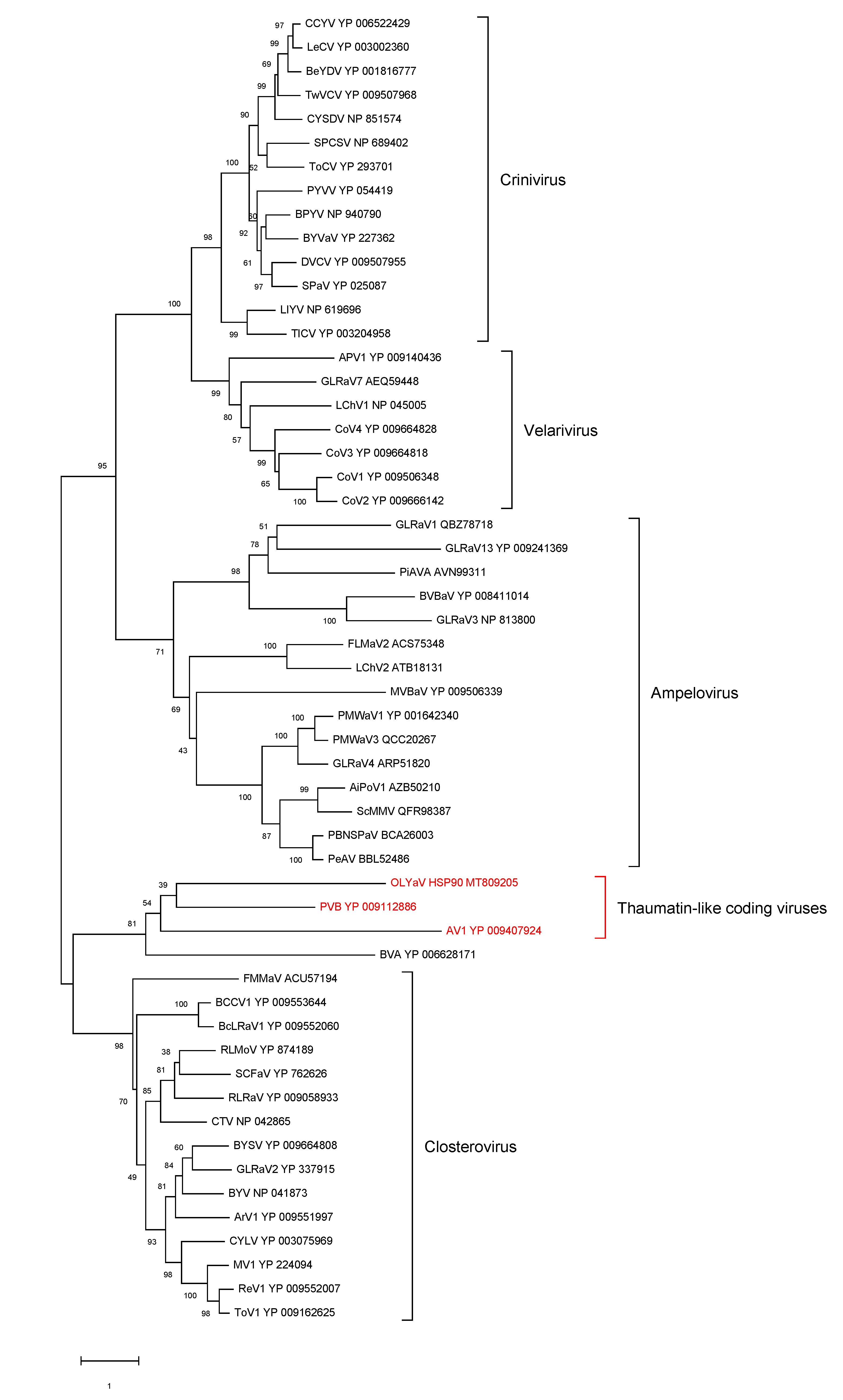
2.2. Phylogenetic Analysis of Conserved Closteroviral Proteins (P1a, P1b, HSP70h, HSP90h, CP)
- ORF1a
- ORF1b (RdRp)
- HSP70h
- HSP90h
- CP
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Sample Preparation and HTS Analysis
4.3. Bioinformatic Analysis of HTS Data
4.4. Analysis of the Strucure of the Genome of OLYaV
4.5. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sabanadzovic, S.; Abou-Ghanem, N.; La Notte, P.; Savino, V.; Scarito, G.; Martelli, G.P. Partial molecular characterization and RT-PCR detection of a putative closterovirus associated with olive leaf yellowing. J. Plant Pathol. 1999, 81, 37–45. [Google Scholar]
- Saponari, M.; Castellano, M.A.; Grieco, F.; Savino, V.; Martelli, G.P. Further studies on olive leaf yellowing- associated virus. J. Plant Pathol. 2004, 86, 33. [Google Scholar]
- El Beaino, T.; Saponari, M.; Minafra, A.; Castellano, M.A.; Savino, V.; Martelli, G. Further characterization of olive leaf yellowing-associated virus. J. Plant Pathol. 2005, 87, 223–228. [Google Scholar]
- Loconsole, G.; Saponari, M.; Faggioli, F.; Albanese, G.; Bouyahia, H.; Elbeaino, T.; Materazzi, A.; Nuzzaci, M.; Prota, V.; Romanazzi, G.; et al. Inter-laboratory validation of PCR-based protocol for detection of olive viruses. EPPO Bull. 2010, 40, 423–428. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Guo, Y.; Daubert, S.; Golino, D.; Rowhani, A. Characterization of latent viral infection of olive trees in the national clonal germplasm repository in California. J. Plant Pathol. 2011, 93, 227–231. [Google Scholar]
- Saponari, M.; Alkowni, R.; Grieco, F.; Driouech, N.; Hassan, M.; Di Terlizzi, B. Detection of Olive-infecting virus in the Mediterranean basin. Acta Hortic. 2002, 586, 787–790. [Google Scholar] [CrossRef]
- Albanese, G.; Faggioli, F.; Ferretti, L.; Sciarroni, R.; La Rosa, R.; Barba, M. Sanitary status evaluation of olive cultivars in Calabria and Sicily. J. Plant Pathol. 2003, 85, 304. [Google Scholar]
- Faggioli, F.; Ferretti, L.; Albanese, G.; Sciarroni, R.; Pasquini, G.; Lumia, V.; Barba, M. Distribution of olive tree viruses in Italy as revealed by one-step RT-PCR. J. Plant Pathol. 2005, 87, 45–51. [Google Scholar]
- El Air, M.; Mahfoudi, N.; Digiaro, M.; Najjar, A.; Elbeaino, T. Detection of olive-infecting viruses in Tunisia. J. Phytopathol. 2011, 159, 283–286. [Google Scholar] [CrossRef]
- Fadel, C.; Digiaro, M.; Choueiri, E.; Elbeaino, T.; Saponari, M.; Savino, V.; Martelli, G.P. On the presence and distribution of olive viruses in Lebanon. EPPO Bull. 2005, 35, 33–36. [Google Scholar] [CrossRef]
- Al Abdullah, A.; Elbeaino, T.; Saponari, M.; Hallakand, M. Preliminary evaluation of the status of oliveinfecting viruses in Syria. EPPO Bull. 2005, 35, 249–252. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Saponari, M.; Hasiów-Jaroszewska, B.; Elbeaino, T.; Koubouris, G. Detection of viruses in olive cultivars in Greece, using a rapid and effective RNA extraction method, for certification of virus-tested propagation material. Phytopathol. Mediterr. 2020, 59, 203–211. [Google Scholar]
- Luigi, M.; Manglli, A.; Thomaj, F.; Buonaurio, R.; Barba, M.; Faggioli, F. Phytosanitary evaluation of olive germplasm in Albania. Phytopathol. Mediterr. 2009, 48, 80–284. [Google Scholar]
- Fuchs, M.; Bar-Joseph, M.; Candresse, T.; Maree, H.J.; Martelli, G.P.; Melzer, M.J.; Menzel, W.; Minafra, A.; Sabanadzovic, S. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Closteroviridae. J. Gen. Virol. 2020, 101, 364–365. [Google Scholar] [PubMed]
- Candresse, T.; Fuchs, M. Closteroviridae; Encyclopedia of Life Sciences; Wiley: Chichester, UK, 2020. [Google Scholar] [CrossRef]
- Bar-Joseph, M.; Hull, R. Purification and partial characterization of sugar beet yellows virus. Virology 1974, 62, 552–562. [Google Scholar] [CrossRef]
- Bar-Joseph, M.; Garnsey, S.M.; Gonsalves, D. The closteroviruses: A distinct group of elongated plant viruses. Adv. Virus Res. 1979, 25, 93–168. [Google Scholar] [PubMed]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Karasev, A.V. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 2000, 38, 293–324. [Google Scholar] [CrossRef]
- Martelli, G.P.; Abou Ghanem-Sabanadzovic, N.; Agranovsky, A.A.; Rwahnih, M.A.; Dolja, V.V.; Dovas, C.I.; Fuchs, M.; Gugerli, P.; Hu, J.S.; Jelkmann, W.; et al. Taxonomic revision of the family Closteroviridae with special reference to the grapevine leafroll-associated members of the genus Ampelovirus and the putative species unassigned to the family. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar]
- Agranovsky, A.A. Closteroviruses: Molecular Biology, Evolution and Interactions with Cells. In Plant Viruses: Evolution and Management; Gaur, R., Petrov, N., Patil, B., Stoyanova, M., Eds.; Springer: Singapore, 2016. [Google Scholar] [CrossRef]
- Ito, T.; Sato, A.; Suzaki, K. An assemblage of divergent variants of a novel putative closterovirus from American persimmon. Virus Genes 2015, 51, 105–111. [Google Scholar] [CrossRef]
- Blouin, A.G.; Biccheri, R.; Khalifa, M.E.; Pearson, M.N.; Poggi Pollini, C.; Hamiaux, C.; Cohen, D.; Ratti, C. Characterization of Actinidia virus 1, a new member of the family Closteroviridae encoding a thaumatin-like protein. Arch. Virol. 2018, 163, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome sequence of the olive tree, Olea europaea. Gigascience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [PubMed]
- Andronescu, M.; Condon, A.; Hoos, H.H.; Mathews, D.H.; Murphy, K.P. Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics 2007, 23, i19–i28. [Google Scholar] [CrossRef]
- Peng, C.W.; Peremyslov, V.V.; Mushegian, A.R.; Dawson, W.O.; Dolja, V.V. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 2001, 75, 12153–12160. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Min, K.; Ha, S.C.; Hasegawa, P.M.; Bressan, R.A.; Yun, D.J.; Kim, K.K. Crystal structure of osmotin, a plant antifungal protein. Proteins Struct. Funct. Bioinform. 2004, 54, 170–173. [Google Scholar] [CrossRef]
- Kim, M.J.; Ham, B.K.; Kim, H.R.; Lee, I.J.; Kim, Y.J.; Ryu, K.H.; Park, Y.I.; Paek, K.H. In vitro and in planta Interaction Evidence between nicotiana tabacum thaumatin-like protein 1 (TLP1) and cucumber mosaic virus proteins. Plant Mol. Biol. 2005, 59, 981–994. [Google Scholar] [CrossRef]
- Liu, J.J.; Sturrock, R.; Ekramoddoullah, A.K. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-García, A.B.; Candresse, T.; Canales, C.; Morán, F.; Machado de Oliveira, C.; Bertolini, E.; Olmos, A. Molecular Characterization of the Complete Coding Sequence of Olive Leaf Yellowing-Associated Virus. Plants 2020, 9, 1272. https://doi.org/10.3390/plants9101272
Ruiz-García AB, Candresse T, Canales C, Morán F, Machado de Oliveira C, Bertolini E, Olmos A. Molecular Characterization of the Complete Coding Sequence of Olive Leaf Yellowing-Associated Virus. Plants. 2020; 9(10):1272. https://doi.org/10.3390/plants9101272
Chicago/Turabian StyleRuiz-García, Ana Belén, Thierry Candresse, Celia Canales, Félix Morán, Carlos Machado de Oliveira, Edson Bertolini, and Antonio Olmos. 2020. "Molecular Characterization of the Complete Coding Sequence of Olive Leaf Yellowing-Associated Virus" Plants 9, no. 10: 1272. https://doi.org/10.3390/plants9101272
APA StyleRuiz-García, A. B., Candresse, T., Canales, C., Morán, F., Machado de Oliveira, C., Bertolini, E., & Olmos, A. (2020). Molecular Characterization of the Complete Coding Sequence of Olive Leaf Yellowing-Associated Virus. Plants, 9(10), 1272. https://doi.org/10.3390/plants9101272







