Enzymes Involved in the Biosynthesis of Arginine from Ornithine in Maritime Pine (Pinus pinaster Ait.)
Abstract
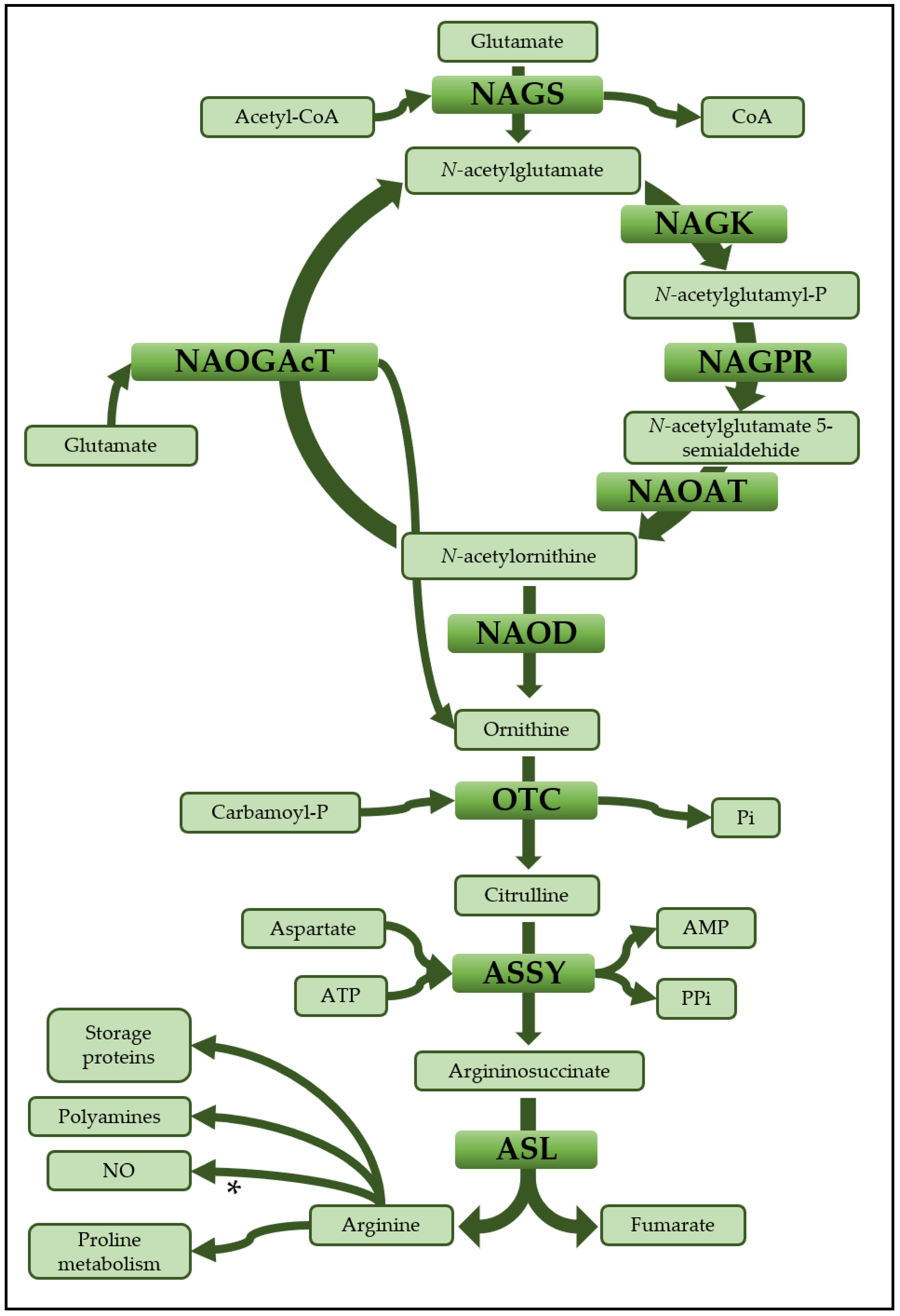
1. Introduction
2. Results
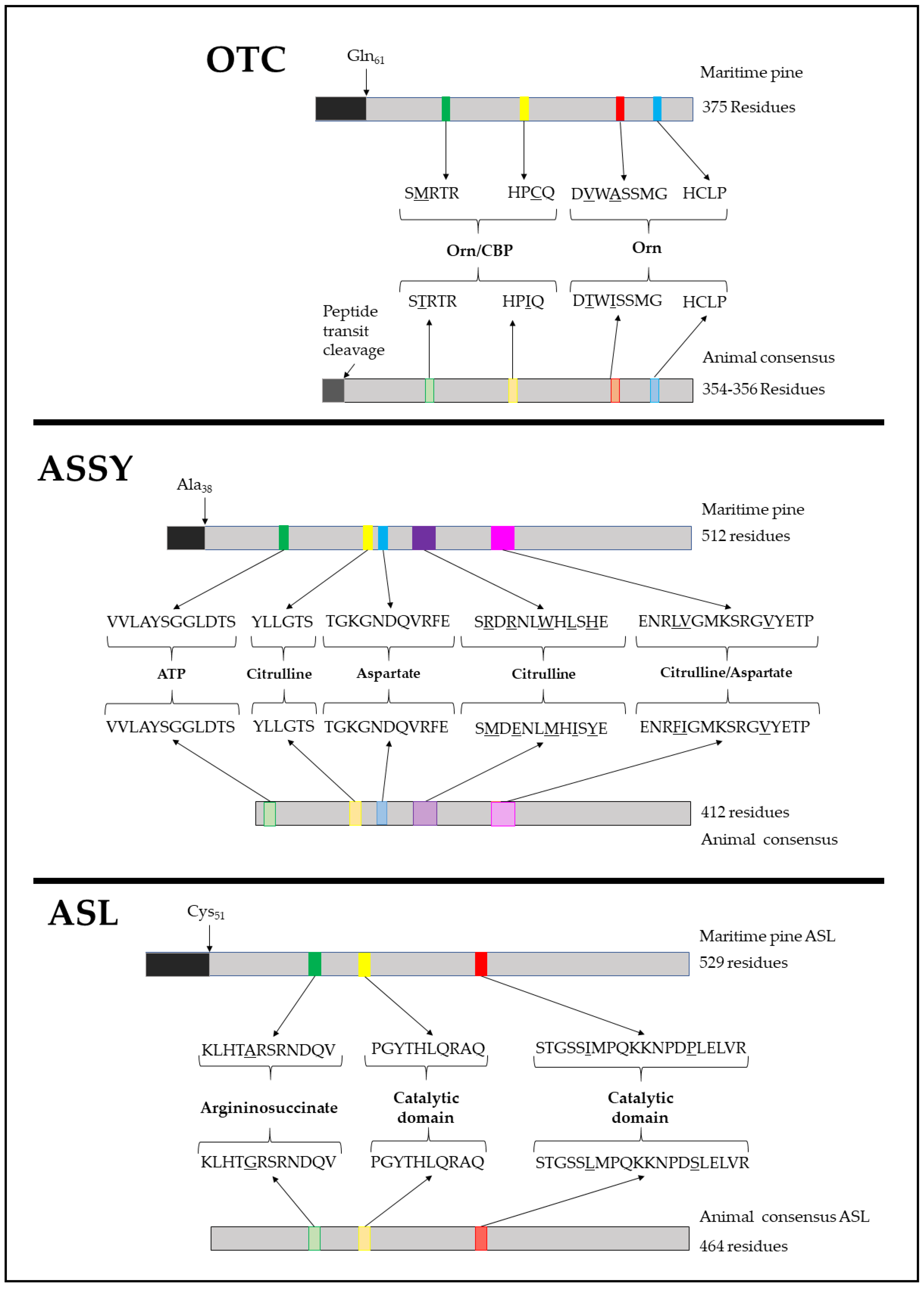
2.1. Identification of Conserved Protein Motifs in the Catalytic Mechanism of PpOTC, PpASSY and PpASL
2.2. Production of Recombinant PpOTC, PpASSY and PpASL Enzymes
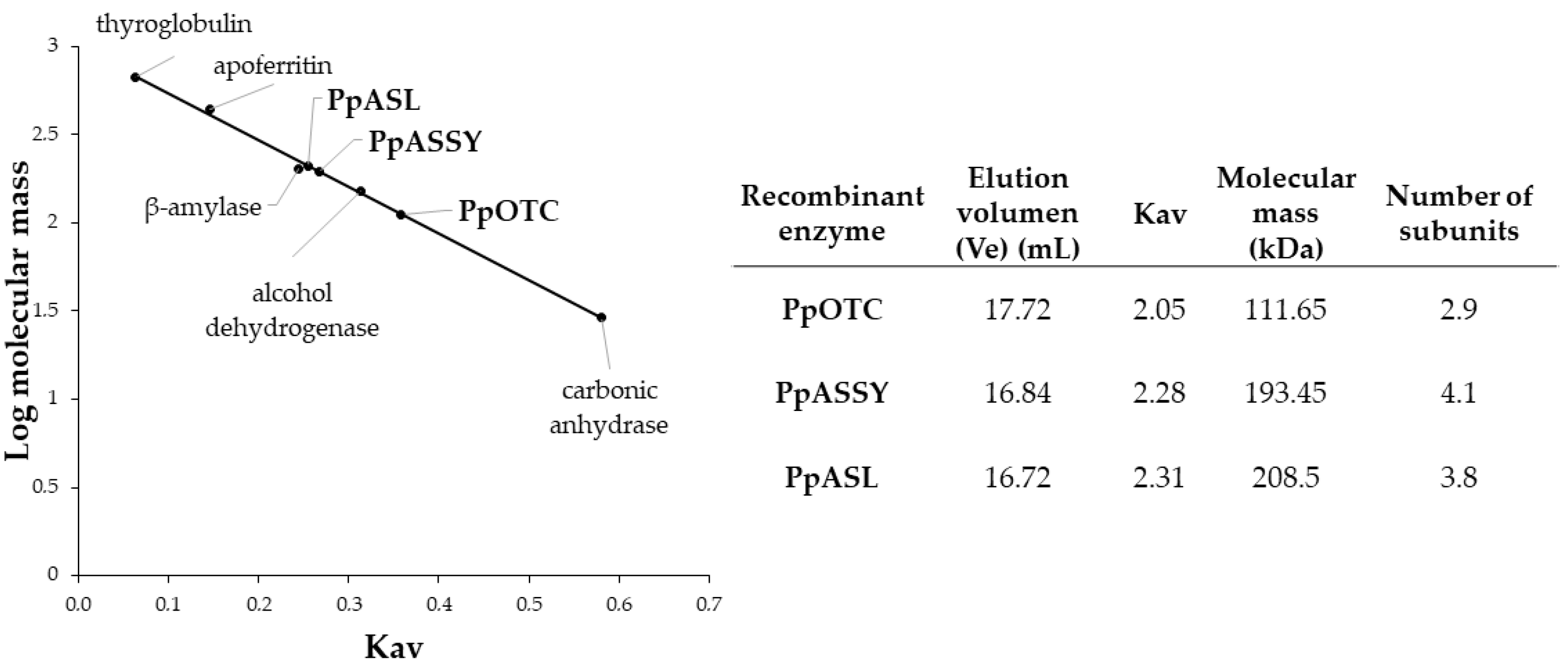
2.3. Molecular Size of Maritime Pine OTC, ASSY and ASL Enzymes
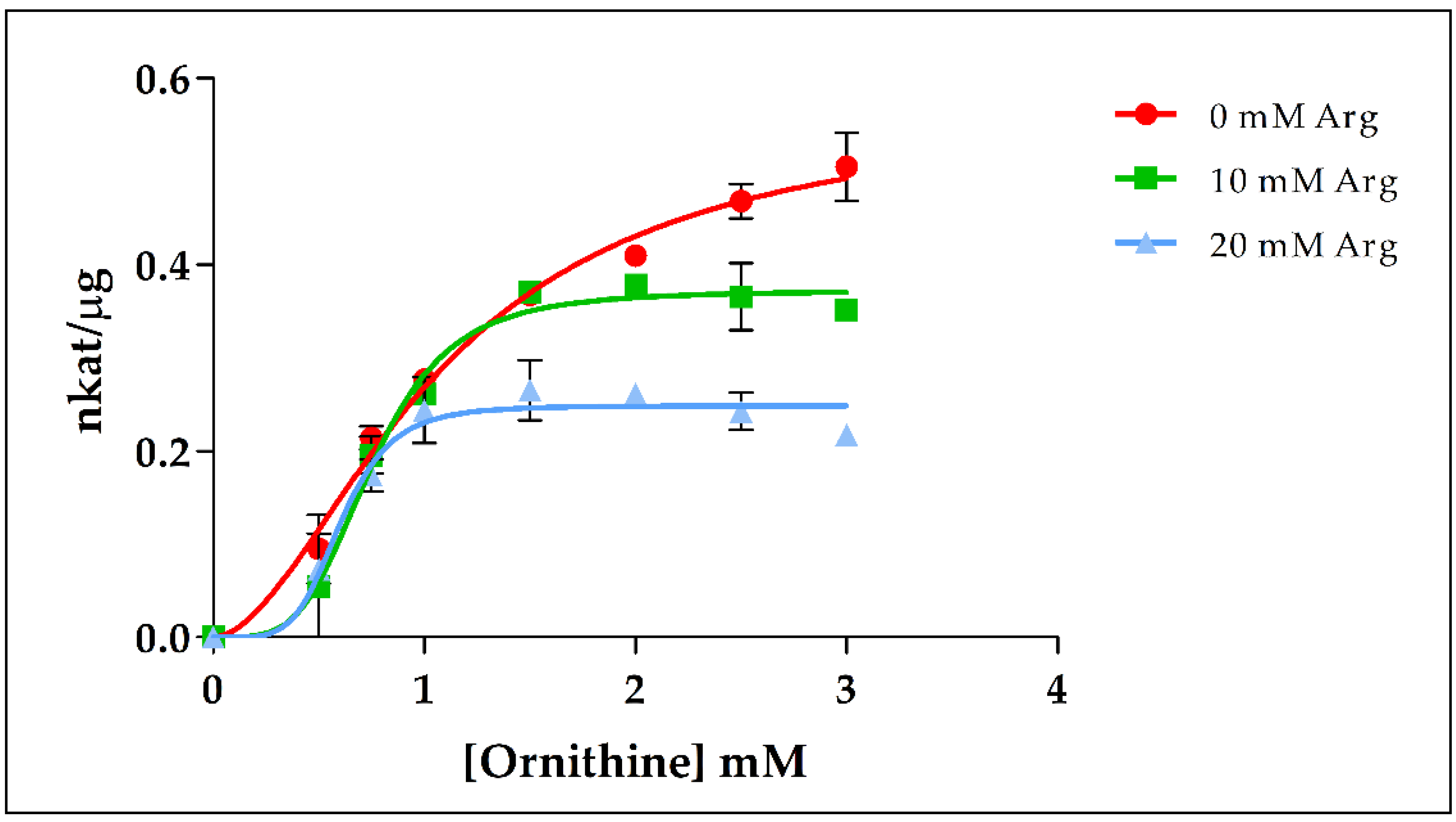
2.4. Catalytic Properties of the Recombinant PpOTC, PpASSY and PpASL Enzymes
2.5. Effect of Several Metabolites on PpOTC, PpASSY and PpASL Activities
3. Discussion
3.1. Enzyme Structure and Recombinant Production
3.2. Enzyme Kinetics
3.3. Relevance of the Ornithine-Arginine Conversion in N Metabolism
3.4. Effector-Mediated Regulation of Ornithine and Arginine Biosynthesis
4. Material and Methods
4.1. RNA Isolation and cDNA Cloning
4.2. Overexpression of Recombinant Enzymes
4.3. Enzyme Assays
4.4. Size-Exclusion Chromatography
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ågren, G.I.; Wetterstedt, J.Å.M.; Billberg, M.F.K. Nutrient limitation on terrestrial plant growth—Modeling the interaction between nitrogen and phosphorus. New Phytol. 2012, 194, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; de la Torre, F.; Pascual, M.B.; Ávila, C.; Cánovas, F.M. Nitrogen economy and nitrogen environmental interactions in conifers. Agronomy 2016, 6, 26. [Google Scholar] [CrossRef]
- Li, G.; Coleman, G.D. Nitrogen storage and cycling in trees. Adv. Bot. Res. 2019, 89, 127–148. [Google Scholar] [CrossRef]
- Llebrés, M.T.; Pascual, M.B.; Debille, S.; Trontin, J.-F.; Harvengt, L.; Ávila, C.; Cánovas, F.M. The role of arginine metabolic pathway during embryogenesis and germination in maritime pine (Pinus pinaster Ait.). Tree Phys. 2018, 38, 471–484. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M.; del Río, L.A.; Barroso, J.B. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009, 184, 9–14. [Google Scholar] [CrossRef]
- Krebs, H.A.; Henseleit, K. Untersuchungen über die Harnstoffbildung im Tierkörper. Klin. Wochenschr. 1932, 11, 757–759. [Google Scholar] [CrossRef]
- Mathews, C.K.; van Holde, K.E.; Appling, D.R.; Anthony-Cahill, S.J. Biochemistry, 4th ed.; Pearson: London, UK, 2013; ISBN 0138004641. [Google Scholar]
- Cohen, S.N.; Kuda, A. Argininosuccinate Synthetase and Argininosuccinate Lyase Are Localized Around Mitochondria: An Immunocytochemical Study. J. Cell Biochem. 1996, 60, 334–340. [Google Scholar] [CrossRef]
- Summar, M.L.; Koelker, S.; Freedenberg, D.; Le Mons, C.; Haberle, J.; Lee, H.-S.; Kirmse, B. The incidence of urea cycle disorders. Mol. Gen. Metab. 2013, 110, 179–180. [Google Scholar] [CrossRef]
- Szefel, J.; Danilelak, A.; Kruszewski, W.J. Metabolic pathways of L-arginine and therapeutic consequences in tumors. Adv. Med. Sci. 2019, 64, 104–110. [Google Scholar] [CrossRef]
- Durzan, D.J. Arginine, scurvy and Cartiers’s “tree of life”. J. Ethnobiol. Ethnomed 2009, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Shargool, P.D.; Jain, J.C.; McKay, G. Ornithine biosynthesis, and arginine biosynthesis and degradation in plant cells. Phytochemistry 1988, 27, 1571–1574. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Phys. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef]
- Fremont, N.; Riefler, M.; Stolz, A.; Schmülling, T. The Arabidopsis TUMOR PRONE5 gene encodes an acetylornithine aminotransferase required for arginine biosynthesis and root meristem maintenance in blue light. Plant Phys. 2013, 161, 1127–1140. [Google Scholar] [CrossRef]
- Xia, J.; Yamaji, N.; Che, J.; Shen, R.F.; Ma, J.F. Normal root elongation requires arginine produced by argininosuccinate lyase in rice. Plant J. 2014, 78, 2. [Google Scholar] [CrossRef]
- Allona, I.; Collada, C.; Casado, R.; Aragoncillo, C. 2S Arginine-rich proteins from Pinus pinaster seeds. Tree Phys. 1994, 14, 211–218. [Google Scholar] [CrossRef]
- Klimaszewska, K.; Morency, F.; Jones-Overton, C.; Cooke, J. Accumulation pattern and identification of seed storage proteins in zygotic embryos of Pinus strobus and in somatic embryos from different maturation treatments. Physiol. Plant 2004, 121, 682–690. [Google Scholar] [CrossRef]
- Trontin, J.F.; Teyssier, C.; Morel, A.; Harvengt, L.; Lelu-Walter, M.-A. Prospects for new variety deployment through somatic embryogenesis in maritime pine. In Vegetative Propagation of Forest Trees; Online ed.; Park, Y.-S., Bonga, J.M., Moon, H.-K., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 653–692. [Google Scholar]
- Cañas, R.A.; Li, Z.; Pascual, M.B.; Castro-Rodríguez, V.; Ávila, C.; Sterck, L. The gene expression landscape of pine seedling tissue. Plant J. 2017, 91, 1064–1087. [Google Scholar] [CrossRef]
- Cañas, R.A.; Pascual, M.B.; de la Torre, F.N.; Ávila, C.; Cánovas, F.M. Resources for conifer functional genomics at the omics era. Adv. Bot. Res. 2019, 89, 39–76. [Google Scholar] [CrossRef]
- Shi, D.; Morizono, H.; Yu, X.; Tong, L.; Allewell, N.M.; Tuchman, M. Human ornithine transcarbamoylase: Crystallographic insights into substrate recognition and conformational changes. Biochem. J. 2001, 354, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, N.; Kobayashi, K.; Terazono, H.; Fukushige, T.; Horiuchi, M.; Takeyori, S. Characterization of Human Wild-Type and Mutant Argininosuccinate Synthetase Proteins Expressed in Bacterial Cells. Enz. Prot. 1994, 48, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lemke, C.T.; Howell, P.L. The 1.6A crystal structure of E. coli argininosuccinate synthetase suggests a conformational change during catalysis. Structure 2001, 9, 1153–1164. [Google Scholar] [CrossRef]
- Karlberg, T.; Collins, R.; van den Berg, S.; Hammarström, M.; Högbom, M.; Schiavone, L.H.; Uppenberg, J. Structure of human argininosuccinate synthetase. Acta Cryst. Sect. D 2008, D64, 279–286. [Google Scholar] [CrossRef]
- Sampaleanu, L.M.; Codding, P.W.; Lobsanov, Y.D.; Tsai, M.; Smith, G.D.; Horvatin, C.; Howell, P.L. Structural studies of duck δ2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis. Biochem. J. 2004, 384, 437–447. [Google Scholar] [CrossRef]
- Sampaleanu, L.M.; Yu, B.; Howell, P.L. Mutational Analysis of Duck δ2 Crystallin and the Structure of an Inactive Mutant with Bound Substrate Provide Insight into the Enzymatic Mechanism of Argininosuccinate Lyase. J. Biol. Chem. 2002, 277, 4166–4175. [Google Scholar] [CrossRef]
- Lee, H.-J.; Chiou, S.-H.; Chang, G.-G. Inactivation of the endogenous argininosuccinate lyase activity of duck δ-crystallin by modification of an essential histidine residue with diethyl pyrocarbonate. Biochem. J. 1993, 293, 537–544. [Google Scholar] [CrossRef]
- Bhaumik, P.; Koski, K.M.; Bergmann, U.; Wierenga, R.K. Structure determination and refinement at 2.44 Å resolution of argininosuccinate lyase from Escherichia coli. Acta Cryst. Sect. D 2004, D60, 1964–1970. [Google Scholar] [CrossRef]
- Almagro-Armenteros, J.J.; Salvatore, M.; Winther, O.; Emanuelsson, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting Sequence Signals in Targeting Peptides Using Deep Learning. Life Sci. All. 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localizator predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; DiPiazza, J.; Joshi, V. Functional Relevance of Citrulline in the Vegetative Tissues of Watermelon During Abiotic Stresses. Front. Plant Sci. 2020, 11, 512. [Google Scholar] [CrossRef]
- Raushel, F.M.; Nygaard, R. Kinetic Mechanism of Bovine Liver Argininosuccinate Lyase. Arch. Biochem. Biophys. 1983, 221, 143–147. [Google Scholar] [CrossRef]
- Slocum, R.D.; Nichols III, H.F.; Williamson, C.L. Purification and characterization of Arabidopsis ornithine transcarbamoylase (OTCase), a member of a distinct and evolutionary-conserved group of plant OTCases. Plant Physiol. Biochem. 2000, 38, 279–288. [Google Scholar] [CrossRef]
- Kimball, M.E.; Jacoby, L.B. Purification and properties of argininosuccinate synthetase from normal and canavanine-resistant human lymphoblasts. Biochemistry 1980, 19, 705–709. [Google Scholar] [CrossRef]
- Tuner, M.A.; Simpson, A.; McInnes, R.R.; Howell, P.L. Human argininosuccinate lyase: A structural basis for intragenic complementation. Proc. Natl. Acad. Sci. USA 1997, 94, 9063–9068. [Google Scholar] [CrossRef]
- Palmieri, L.; Todd, C.D.; Arrigoni, R.; Hoyos, M.E.; Santoro, A.; Polacco, J.C.; Palmieri, F. Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim. Biophys. Acta 2006, 1757, 1277–1283. [Google Scholar] [CrossRef]
- Monné, M.; Miniero, D.V.; Daddabbo, L.; Palmieri, L.; Porcelli, V.; Palmieri, F. Mitochondrial transporters for ornithine and related amino acids: A review. Amino Acids 2015, 47, 1763–1777. [Google Scholar] [CrossRef]
- Ludwig, R.A. Arabidopsis chloroplasts dissimilate L-arginine and L-citrulline for use as N source. Plant Physiol. 1993, 101, 429–434. [Google Scholar] [CrossRef]
- Cánovas, F.M.; Ávila, C.; Cantón, F.R.; Cañas, R.A.; de la Torre, F. Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 2007, 58, 2307–2318. [Google Scholar] [CrossRef]
- Miyazawa, S.I.; Nishiguchi, M.; Futamura, N.; Yukawa, T.; Miyao, M.; Maruyama, T.E.; Kawahara, T. Low assimilation efficiency of photorespiratory ammonia in conifer leaves. J. Plant Res. 2018, 131, 789–902. [Google Scholar] [CrossRef]
- Molesini, B.; Mennella, G.; Martini, F.; Francese, G.; Pandolfini, T. Involvement of the putative N-acetylornithine deacetylase from Arabidopsis thaliana in flowering and fruit development. Plant Cell Physiol. 2015, 56, 1084–1096. [Google Scholar] [CrossRef]
- Kalamaki, S.M.; Merkoiropoulos, G.; Kanellis, A.K. Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis? Plant Sign. Behav. 2009, 4, 1099–1101. [Google Scholar] [CrossRef]
- Joshi, V.; Fernie, A.R. Citrulline metabolism in plants. Amino Acids 2017, 49, 1543–1559. [Google Scholar] [CrossRef]
- Cañas, R.A.; Villalobos, D.P.; Díaz-Moreno, S.M.; Cánovas, F.M.; Cantón, F.R. Molecular and Functional Analyses Support a Role of Ornithine-δ-Aminotransferase in the Provision of Glutamate Biosynthesis during Pine Germination. Plant Phys. 2008, 148, 77–88. [Google Scholar] [CrossRef]
- Ramón-Maiques, S.; Marina, A.; Gil-Ortiz, F.; Fita, F.; Rubio, V. Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, during Catalysis. Structure 2002, 10, 329–342. [Google Scholar] [CrossRef]
- Llácer, J.L.; Contreras, A.; Forchhammer, K.; Marco-Marín, C.; Gil-Ortiz, F.; Maldonado, R.; Fita, I.; Rubio, V. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc. Natl. Acad. Sci. USA 2007, 104, 17644–17649. [Google Scholar] [CrossRef]
- Llácer, J.L.; Fita, I.; Rubio, V. Arginine and nitrogen storage. Curr. Opin. Struct. Biol. 2008, 18, 673–681. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Chellamuthu, V.R.; Ermilova, E.; Lapina, T.; Lüddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A widespread glutamine-sensing mechanism on the plant kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef]
- Llebrés, M.T.; Pascual, M.B.; de la Torre, F.N.; Gómez, L.; Ávila, C.; Cánovas, F.M. Structural and functional characteristics of two molecular variants of the nitrogen sensor PII in maritime pine. Front. Plant Sci. 2020, 11, 823. [Google Scholar] [CrossRef]
- Marshall, M.; Cohen, P.P. Ornithine Transcarbamylase from Streptococcus faecalis and Bovine Liver. (II) Multiple binding sites for carbamoyl-P and L-Norvaline, correlation with steady state kinetics. J. Biol. Chem. 1972, 247, 1654–1668. [Google Scholar]
- Yang, Q.; Zhao, D.; Liu, Q. Connections Between Amino Acid Metabolisms in Plants: Lysine as an Example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Tam, L.Q.; Patil, S.S. Mode of action of the toxin from Pseudomonas phaseolicola. II. Mechanism of inhibition of bean ornithine carbamoyl transferase. Plant Phys. 1972, 49, 808–812. [Google Scholar] [CrossRef][Green Version]
- Arrebola, E.; Cazorla, F.M.; Durán, V.E.; Rivera, E.; Olea, F.; Codina, J.C.; Pérez-García, A.; de Vicente, A. Mangotoxin: A novel antimetabolite toxin produced by Pseudomonas syringae inhibiting ornithine/arginine biosynthesis. Phys. Mol. Plant Path. 2003, 63, 117–127. [Google Scholar] [CrossRef]
- de la Torre, F.; El-Azaz, J.; Ávila, C.; Cánovas, F.M. Deciphering the Role of Aspartate and Prephenate Aminotransferase Activities in Plastid Nitrogen Metabolism. Plant Phys. 2014, 164, 92–104. [Google Scholar] [CrossRef]
- Canales, J.; Rueda-López, M.; Craven-Bartle, B.; Ávila, C.; Cánovas, F.M. Novel insights into regulation of asparagine synthetase in conifers. Front. Plant Sci. 2012, 3, 100. [Google Scholar] [CrossRef]
- Cánovas, F.M.; Cantón, F.R.; Gallardo, F.; García-Gutiérrez, A.; de Vicente, A. Accumulation of glutamine synthetase during early development of maritime pine (Pinus pinaster) seedlings. Planta 1991, 185, 372–378. [Google Scholar] [CrossRef]
- Cantón, F.R.; García-Gutiérrez, A.; Crespillo, R.; Cánovas, F.M. High-level expression of Pinus sylvestris glutamine synthetase in Escherichia coli. Production of polyclonal antibodies against the recombinant protein and expression studies in pine seedlings. FEBS Lett. 1996, 393, 205–210. [Google Scholar] [CrossRef]
- Berhart, D.N.; Wreath, A.R. Colorimetric Determination of Phosphorus by Modified Phosphomolybdate Method. Anal. Chem. 1955, 27, 440–441. [Google Scholar] [CrossRef]
- Katano, H.; Tanaka, R.; Maruyama, C.; Hamano, Y. Assay of enzymes forming AMP + PPi by the pyrophosphate determination based on the formation of 18-molybdopyrophosphate. Anal. Biochem. 2012, 421, 308–312. [Google Scholar] [CrossRef]
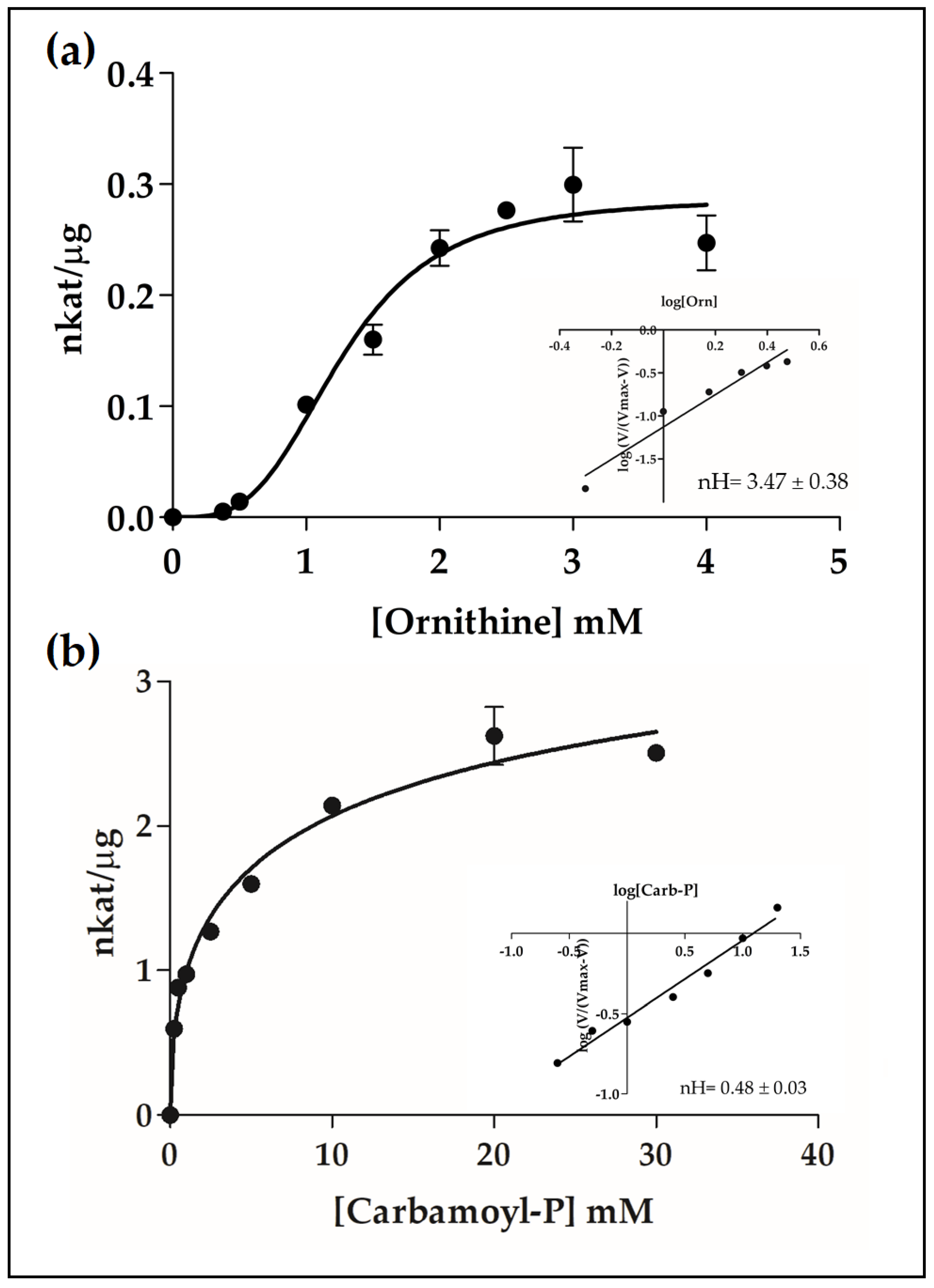

| Enzyme | Substrate | Km (mM) | S0.5 (mM) | Vmax (nkat/μg) | nH | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|---|---|---|---|
| PpOTC | ornithine | 1.28 ± 0.16 | 4.44 ± 1.4 | 3.47 ± 0.38 | 11.03 | ||
| carbamoyl-P | 3.48 ± 1.4 | 0.48 ± 0.03 | |||||
| PpASSY | citrulline | 0.12 ± 0.03 | 0.07 ± 0.01 | 2.82 | 2.46 × 104 | ||
| aspartate | 0.12 ± 0.05 | ||||||
| PpASL | argininosuccinate | 0.22 ± 0.03 | 20.93 ± 0.75 | 1139.24 | 5.18 × 106 |
| Enzyme | Compound | IC50 Value (mM) |
|---|---|---|
| PpOTC | arginine | 10.28 |
| valine | 7.19 | |
| PpASSY | arginine | 1.94 |
| PpASL | spermidine | 4.97 |
| spermine | 19.24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbano-Gámez, J.A.; El-Azaz, J.; Ávila, C.; de la Torre, F.N.; Cánovas, F.M. Enzymes Involved in the Biosynthesis of Arginine from Ornithine in Maritime Pine (Pinus pinaster Ait.). Plants 2020, 9, 1271. https://doi.org/10.3390/plants9101271
Urbano-Gámez JA, El-Azaz J, Ávila C, de la Torre FN, Cánovas FM. Enzymes Involved in the Biosynthesis of Arginine from Ornithine in Maritime Pine (Pinus pinaster Ait.). Plants. 2020; 9(10):1271. https://doi.org/10.3390/plants9101271
Chicago/Turabian StyleUrbano-Gámez, José Alberto, Jorge El-Azaz, Concepción Ávila, Fernando N. de la Torre, and Francisco M. Cánovas. 2020. "Enzymes Involved in the Biosynthesis of Arginine from Ornithine in Maritime Pine (Pinus pinaster Ait.)" Plants 9, no. 10: 1271. https://doi.org/10.3390/plants9101271
APA StyleUrbano-Gámez, J. A., El-Azaz, J., Ávila, C., de la Torre, F. N., & Cánovas, F. M. (2020). Enzymes Involved in the Biosynthesis of Arginine from Ornithine in Maritime Pine (Pinus pinaster Ait.). Plants, 9(10), 1271. https://doi.org/10.3390/plants9101271








