Complete Chloroplast Genome of Paphiopedilum delenatii and Phylogenetic Relationships among Orchidaceae
Abstract
1. Introduction
2. Results and Discussion
2.1. Chloroplast Genome of Paphiopedilum delenatii
2.2. Repeat and Microsatellite Analysis
2.3. Phylogenetic and Species Resolution Analyses
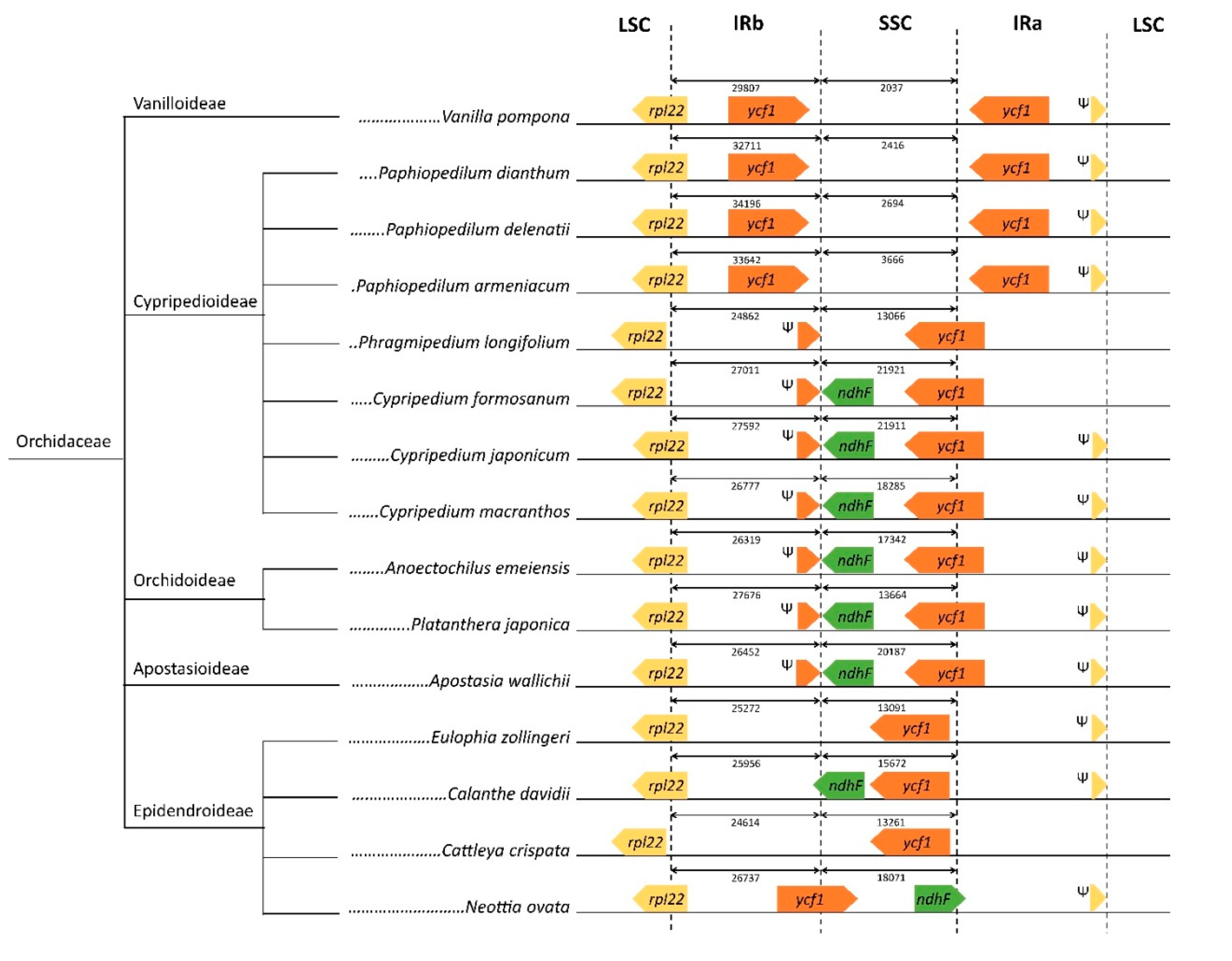
2.4. Divergence of Hotspot Regions
3. Materials and Methods
3.1. Plant Material, DNA Extraction, and Sequencing
3.2. Read Data Processing and Chloroplast Genome Assembly
3.3. Repeat Sequence and Microsatellite Identification
3.4. Examination of IR Junctions
3.5. Phylogenetic Analysis
3.6. Nucleotide Variability Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| IUCN | International Union for Conservation of Nature |
| Cp | chloroplast |
| CR | Critically endangered |
| IR | Inverted repeat region |
| SSC | Small single-copy region |
| LSC | Large single-copy region |
| gBGC | GC-biased gene conversion |
| SSR | Simple sequence repeat |
| SNP | Single nucleotide polymorphism |
References
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Xiang, l.; Su, Y.; Li, X.; Xue, G.; Wang, Q.; Shi, J.; Wang, L.; Chen, S. Identification of Fritillariae bulbus from adulterants using ITS2 regions. Plant Gene 2016, 7, 42–49. [Google Scholar] [CrossRef]
- Yeisoo, Y.; Hyun Oh, L.; Joong Hyoun, C.; Han Yong, P.; Soo-Cheul, Y. The complete chloroplast genome sequence of Oryza sativa aus-type variety Nagina-22 (Poaceae). Mitochondrial DNA Part B 2017, 2, 819–820. [Google Scholar]
- Tian, N.; Han, L.; Chen, C.; Wang, Z. The complete chloroplast genome sequence of Epipremnum aureum and its comparative analysis among eight Araceae species. PLoS ONE 2018, 13, e0192956. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.-L.; Wang, R.-N.; Zhang, N.-Y.; Fan, W.-B.; Fang, M.-F.; Li, Z.-H. Molecular Evolution of Chloroplast Genomes of Orchid Species: Insights into Phylogenetic Relationship and Adaptive Evolution. Int. J. Mol. Sci. 2018, 19, 716. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Wang, H.; Xie, X.; Zhu, S.; Liu, W.; Ding, X. Mutational Biases and GC-Biased Gene Conversion Affect GC Content in the Plastomes of Dendrobium Genus. Int. J. Mol. Sci. 2017, 18, 2307. [Google Scholar]
- Manzanilla, V.; Kool, A.; Nguyen Nhat, L.; Nong Van, H.; Le Thi Thu, H.; de Boer, H.J. Phylogenomics and barcoding of Panax: Toward the identification of ginseng species. BMC Evol. Biol. 2018, 18, 44. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature. The IUCN Red List of Threatened Species; Version 2019-2; International Union for Conservation of Nature: Gland, Switzerland, 2019. [Google Scholar]
- Zhou, Y.; Nie, J.; Xiao, L.; Hu, Z.; Wang, B. Comparative Chloroplast Genome Analysis of Rhubarb Botanical Origins and the Development of Specific Identification Markers. Molecules 2018, 23, 2811. [Google Scholar] [CrossRef]
- Li, X.; Tan, W.; Sun, J.; Du, J.; Zheng, C.; Tian, X.; Zheng, M.; Xiang, B.; Wang, Y. Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination. Sci. Rep. 2019, 9, 10567. [Google Scholar]
- Park, I.; Yang, S.; Kim, W.J.; Song, J.-H.; Lee, H.-S.; Lee, H.O.; Lee, J.-H.; Ahn, S.-N.; Moon, B.C. Sequencing and Comparative Analysis of the Chloroplast Genome of Angelica polymorpha and the Development of a Novel Indel Marker for Species Identification. Molecules 2019, 24, 1038. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, J.; Jia, Y.; Li, W.; Xu, F.; Wang, X. Comparative Chloroplast Genome Analyses of Species in Gentiana section Cruciata (Gentianaceae) and the Development of Authentication Markers. Int. J. Mol. Sci. 2018, 19, 1962. [Google Scholar] [CrossRef] [PubMed]
- Jheng, C.F.; Chen, T.C.; Lin, J.Y.; Chen, T.C.; Wu, W.L.; Chang, C.C. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 2012, 190, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, H.T.; Son, S.-W.; Kim, J.-H. Molecular identification of endangered Korean lady’s slipper orchids (Cypripedium, Orchidaceae) and related taxa. Botany 2015, 93, 603–610. [Google Scholar] [CrossRef]
- Peyachoknagul, S.; Mongkolsiriwatana, C.; Wannapinpong, S.; Huehne, P.S.; Srikulnath, K. Identification of native Dendrobium species in Thailand by PCR-RFLP of rDNA-ITS and chloroplast DNA. Sci. Asia 2014, 40, 113–120. [Google Scholar] [CrossRef]
- Sun, Y.W.; Liao, Y.J.; Hung, Y.S.; Chang, J.C.; Sung, J.M. Development of ITS sequence based SCAR markers for discrimination of Paphiopedilum armeniacum, Paphiopedilum micranthum, Paphiopedilum delenatii and their hybrids. Sci. Hortic. 2011, 127, 405–410. [Google Scholar] [CrossRef]
- Fattmah, D.; Sukma, D. Development of Sequence-Based Microsatellite Marker for Phalaenopsis Orchid. HAYATI J. Biosci. 2011, 18, 71–76. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Lin, B.-Y.; Chang, C.-D.; Liao, S.-C.; Liu, Y.-C.; Wu, W.-L.; Chang, C.-C. Evaluation of chloroplast DNA markers for distinguishing Phalaenopsis species. Sci. Hortic. 2015, 192, 302–310. [Google Scholar] [CrossRef]
- Yu, X.Q.; Drew, B.T.; Yang, J.B.; Gao, L.M.; Li, D.Z. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS ONE 2017, 12, e0178026. [Google Scholar] [CrossRef]
- Sui, C.; Liu, S.; Liu, H.; Liu, H. The complete chloroplast genome of Paphiopedilum tranlimianum (Orchidaceae). Mitochondrial DNA Part B 2018, 3, 820–822. [Google Scholar] [CrossRef]
- Hou, N.; Wang, G.; Zhu, Y.; Wang, L.; Xu, J. The complete chloroplast genome of the rare and endangered herb Paphiopedilum dianthum (Asparagales: Orchidaceae). Conserv. Genet. Resour. 2017, 10, 709–712. [Google Scholar] [CrossRef]
- Lin, C.-S.; Chen, J.J.W.; Huang, Y.-T.; Chan, M.-T.; Daniell, H.; Chang, W.-J.; Hsu, C.-T.; Liao, D.-C.; Wu, F.-H.; Lin, S.-Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.-T.; Nguyen, T.-D.; Ly, L.; Nguyen, T.-A.; Vu, Q.-L.; Nguyen, T.-C.; Tran, H.-D. Construction of complete chloroplast genome of the endemic Paphiopedilum delenatii species of Vietnam. Vietnam J. Biotechnol. 2019. accepted. [Google Scholar]
- Vu, H.-T.; Nguyen, T.-D.; Tran, H.-D.; Vu, Q.-L.; Tran, N.; Nguyen, T.-C.; Luu, P.-N.; Tran, D.-D.; Nguyen, T.-K.; Le, L. Identification of Vietnamese Paphiopedilum species using DNA sequences. Biology 2019. accepted. [Google Scholar] [CrossRef]
- Li, B.; Zheng, Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 2018, 8, 9285. [Google Scholar] [CrossRef]
- Wu, C.S.; Chaw, S.M. Evolutionary Stasis in Cycad Plastomes and the First Case of Plastome GC-Biased Gene Conversion. Genome Biol. Evol. 2015, 7, 2000–2009. [Google Scholar] [CrossRef]
- Duret, L.; Galtier, N. Biased Gene Conversion and the Evolution of Mammalian Genomic Landscapes. Annu. Rev. Genom. Hum. Genet. 2009, 10, 285–311. [Google Scholar] [CrossRef]
- Glemin, S.; Arndt, P.F.; Messer, P.W.; Petrov, D.; Galtier, N.; Duret, L. Quantification of GC-biased gene conversion in the human genome. Genome Res. 2015, 25, 1215–1228. [Google Scholar] [CrossRef]
- Palmer, J.D. Chloroplast DNA exists in two orientations. Nature 1983, 301, 92–93. [Google Scholar] [CrossRef]
- Li, X.-Q.; Du, D. Variation, evolution, and correlation analysis of C+G content and genome or chromosome size in different kingdoms and phyla. PLoS ONE 2014, 9, e88339. [Google Scholar] [CrossRef]
- Singh, R.; Ming, R.R.; Yu, Q. Comparative Analysis of GC Content Variations in Plant Genomes. Trop. Plant Biol. 2016, 9, 136–149. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The Complete Plastome Sequences of Four Orchid Species: Insights into the Evolution of the Orchidaceae and the Utility of Plastomic Mutational Hotspots. Front. Plant Sci. 2017, 8, 715. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Shuying, Z.; Jiajia, P.; Ludan, L.; Jing, S.; Xiaoyu, D. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 2073. [Google Scholar]
- Karimi, K.; Wuitchik, D.M.; Oldach, M.J.; Vize, P.D. Distinguishing Species Using GC Contents in Mixed DNA or RNA Sequences. Evol. Bioinform. Online 2018, 14, 1176934318788866. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Luo, Y.-B.; Liu, Z.-J.; Wang, X.-Q. Evolution and Biogeography of the Slipper Orchids: Eocene Vicariance of the Conduplicate Genera in the Old and New World Tropics. PLoS ONE 2012, 7, e38788. [Google Scholar] [CrossRef]
- Ifuku, K.; Endo, T.; Shikanai, T.; Aro, E.M. Structure of the chloroplast NADH dehydrogenase-like complex: Nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 2011, 52, 1560–1568. [Google Scholar] [CrossRef]
- Nelson, N.F.; Yocum, C. Structure and Function of Photosystem Ι and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef]
- Niu, Z.; Pan, J.; Zhu, S.; Li, L.; Xue, Q.; Liu, W.; Ding, X. Comparative analysis of the complete plastomes of Apostasia wallichii and Neuwiedia singapureana (Apostasioideae) reveals different evolutionary dynamics of IR/SSC boundary among photosynthetic orchids. Front. Plant Sci. 2017, 8, 1713. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S.; Moore, M.J.; Neubig, K.M.; Williams, N.H.; Whitten, W.M.; Kim, J.-H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE 2015, 10, e0142215. [Google Scholar] [CrossRef]
- Luo, J.; Hou, B.-W.; Niu, Z.-T.; Liu, W.; Xue, Q.-Y.; Ding, X.-Y. Comparative Chloroplast Genomes of Photosynthetic Orchids: Insights into Evolution of the Orchidaceae and Development of Molecular Markers for Phylogenetic Applications. PLoS ONE 2014, 9, e99016. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr. Biol. 2002, 12, R62–R64. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Chang, C.H. Common repeat sequences in bacterial genomes. J. Med. Biol. Eng. 2003, 23, 65–72. [Google Scholar]
- Chinnappareddy, L.R.D.; Khandagale, K.; Srinivas Reddy, S.H.; Kanupriya, C.; Chennareddy, A.; Singh, T.H. SSR-Based DNA Barcodes as a Tool for Identification of Eggplant Genotypes. Int. J. Veg. Sci. 2012, 18, 260–271. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Wu, P.-Y.; Kuo, C.-C.; Huang, M.-C.; Yu, S.-K.; Hsu, T.-W.; Chiang, T.-Y.; Chiang, Y.-C. Analysis of microsatellites in the vulnerable orchid Gastrodia flavilabella: The development of microsatellite markers, and cross-species amplification in Gastrodia. Bot. Stud. 2014, 55, 72. [Google Scholar] [CrossRef][Green Version]
- Phuekvilai, P.; Pongtongkam, P.; Peyachoknagul, S. Development of Microsatellite Markers for Vanda Orchid. Kasetsart J. Nat. Sci. 2009, 43, 497–506. [Google Scholar]
- Liu, Y.T.; Chen, R.K.; Lin, S.J.; Chen, Y.C.; Chin, S.W.; Chen, F.C.; Lee, C.Y. Analysis of sequence diversity through internal transcribed spacers and simple sequence repeats to identify Dendrobium species. Genet. Mol. Res. 2014, 13, 2709–2717. [Google Scholar] [CrossRef]
- Jonah, P.M.; Bello, L.L.; Lucky, O.; Midau, A.; Moruppa, S. Review: The importance of molecular markers in plant breeding programs. Glob. J. Sci. Front. Res. 2011, 11, 4–12. [Google Scholar]
- Kalia, R.; Rai, M.; Kalia, S.; Singh, R.; Dhawan, A. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Echt, C.S.; Deverno, L.L.; Anzidei, M.; Vendramin, G.G. Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol. Ecol. 1998, 7, 307–316. [Google Scholar] [CrossRef]
- Huang, J.; Yang, X.; Zhang, C.; Yin, X.; Liu, S.; Li, X. Development of Chloroplast Microsatellite Markers and Analysis of Chloroplast Diversity in Chinese Jujube (Ziziphus jujuba Mill.) and Wild Jujube (Ziziphus acidojujuba Mill.). PLoS ONE 2015, 10, e0134519. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Zhang, H.; He, Y.; Wang, T.; Su, Y. Chloroplast microsatellite markers for Pseudotaxus chienii developed from the whole chloroplast genome of Taxus chinensis var. mairei (Taxaceae). Appl. Plant Sci. 2017, 5, 1600153. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Kim, C.; Lee, Y.-M.; Kim, J.-H. Development of chloroplast microsatellite markers for the endangered Maianthemum bicolor (Asparagaceae s.l.). Appl. Plant Sci. 2016, 4, 1600032. [Google Scholar] [CrossRef]
- Pinheiro, F.; Palma-Silva, C.; Barros, F.; Pessoa Felix, L.; Lexer, C.; Cozzolino, S.; Fay, M. Chloroplast microsatellite markers for the Neotropical orchid genus Epidendrum, and cross-amplification in other Laeliinae species (Orchidaceae). Conserv. Genet. Resour. 2009, 1, 505–511. [Google Scholar] [CrossRef]
- Ebert, D.; Peakall, R.O.D. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009, 9, 673–690. [Google Scholar] [CrossRef]
- Nepal, M.; Piya, S. Characterization of Nuclear and Chloroplast Microsatellite Markers for Falcaria vulgaris (Apiaceae). Am. J. Plant Sci. 2013, 4, 590–595. [Google Scholar]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Ballardini, M.; Mercuri, A.; Littardi, C.; Abbas, S.; Couderc, M.; Ludeña, B.; Pintaud, J.-C. The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix L. (Arecaceae). ZooKeys 2013, 365, 71–82. [Google Scholar] [CrossRef]
- Singh, H.K.; Parveen, I.; Raghuvanshi, S.; Babbar, S.B. The loci recommended as universal barcodes for plants on the basis of floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res. Notes 2012, 5, 42. [Google Scholar] [CrossRef]
- Krawczyk, K.; Nobis, M.; Myszczyński, K.; Klichowska, E.; Sawicki, J. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci. Rep. 2018, 8, 1924. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, J.; Cui, Y.; Wang, Y.; Duan, B.; Yao, H. Identification of Ligularia Herbs Using the Complete Chloroplast Genome as a Super-Barcode. Front. Pharmacol. 2018, 9, 695. [Google Scholar] [CrossRef] [PubMed]
- Nock, C.; Waters, D.; Shepherd, M.; Bundock, P.; Henry, R. Plant DNA Barcoding using chloroplast genome sequences. In Proceedings of the IBC XVII International Botanical Congress, Melbourne, Australia, 23–30 July 2011. [Google Scholar]
- Vu, H.-T.; Huynh, P.; Tran, H.-D.; Le, L. In Silico Study on Molecular Sequences for Identification of Paphiopedilum Species. Evol. Bioinform. 2018, 14, 117693431877454. [Google Scholar] [CrossRef] [PubMed]
- Hosein, F.N.; Austin, N.; Maharaj, S.; Johnson, W.; Rostant, L.; Ramdass, A.C.; Rampersad, S.N. Utility of DNA barcoding to identify rare endemic vascular plant species in Trinidad. Ecol. Evol. 2017, 7, 7311–7333. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, M.-F.; Xue, J.; Dong, R.; Du, Y.-P.; Zhang, X.-H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef] [PubMed]
- Somaratne, Y.; Guan, D.-L.; Abbood, N.N.; Zhao, L.; Wang, W.-Q.; Xu, S.-Q. Comparison of the Complete Eragrostis pilosa Chloroplast Genome with Its Relatives in Eragrostideae (Chloridoideae; Poaceae). Plants 2019, 8, 485. [Google Scholar] [CrossRef]
- Cui, Y.; Nie, L.; Sun, W.; Xu, Z.; Wang, Y.; Yu, J.; Song, J.; Yao, H. Comparative and Phylogenetic Analyses of Ginger (Zingiber officinale) in the Family Zingiberaceae Based on the Complete Chloroplast Genome. Plants 2019, 8, 283. [Google Scholar] [CrossRef]
- Averyanov, L.; Cribb, P.; Phan, K.L.; Nguyen, T.H. Slipper Orchids of Vietnam; (Vietnamese edition); Giao Thong Van Tai Publishing House: Ho Chi Minh City, Vietnam, 2004; p. 308. [Google Scholar]
- Kress, W.J.; Erickson, D.L. DNA barcodes: Genes, genomics, and bioinformatics. Proc. Natl. Acad. Sci. USA 2008, 105, 2761–2762. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple Multilocus DNA Barcodes from the Plastid Genome Discriminate Plant Species Equally Well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Dong, W.; Liu, H.; Xu, C.; Zuo, Y.; Chen, Z.; Zhou, S. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genet. 2014, 15, 138. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Choi, B.S.; Lee, S.C.; Kim, N.H.; Park, J.Y.; Jang, W.; Lakshmanan, M.; Mohan, S.V.G.; Lee, D.Y.; Yang, T.J. Ginseng Genome Database: An open-access platform for genomics of Panax ginseng. BMC Plant Biol. 2018, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Scarcelli, N.; Barnaud, A.; Eiserhardt, W.; Treier, U.A.; Seveno, M.; d’Anfray, A.; Vigouroux, Y.; Pintaud, J.-C. A Set of 100 Chloroplast DNA Primer Pairs to Study Population Genetics and Phylogeny in Monocotyledons. PLoS ONE 2011, 6, e19954. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, X.; Yu, Y.; Yuan, S.; Jiang, D.; Zhang, Y.; Zhang, T.; Zhong, W.; Yuan, Q.; Huang, L. Complete chloroplast genome sequences of Dioscorea: Characterization, genomic resources, and phylogenetic analyses. PeerJ 2018, 6, e6032. [Google Scholar] [CrossRef]
- Yaradua, S.S.; Alzahrani, D.A.; Albokhary, E.J.; Abba, A.; Bello, A. Complete Chloroplast Genome Sequence of Justicia flava: Genome Comparative Analysis and Phylogenetic Relationships among Acanthaceae. BioMed Res. Int. 2019, 2019, 4370258. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Yasui, Y.; Ohnishi, O. Intraspecific cpDNA variations of diploid and tetraploid perennial buckwheat, Fagopyrum cymosum (Polygonaceae). Am. J. Bot. 2003, 90, 339–346. [Google Scholar] [CrossRef]
- Cho, K.S.; Yun, B.K.; Yoon, Y.H.; Hong, S.Y.; Mekapogu, M.; Kim, K.H.; Yang, T.J. Complete Chloroplast Genome Sequence of Tartary Buckwheat (Fagopyrum tataricum) and Comparative Analysis with Common Buckwheat (F. esculentum). PLoS ONE 2015, 10, e0125332. [Google Scholar] [CrossRef]
- Song, Y.; Chen, Y.; Lv, J.; Xu, J.; Zhu, S.; Li, M. Comparative Chloroplast Genomes of Sorghum Species: Sequence Divergence and Phylogenetic Relationships. BioMed Res. Int. 2019, 2019, 5046958. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2010, 27, 592–593. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree: Tree Figure Drawing Tool Version 1.2.2. 2009. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 11 January 2019).
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
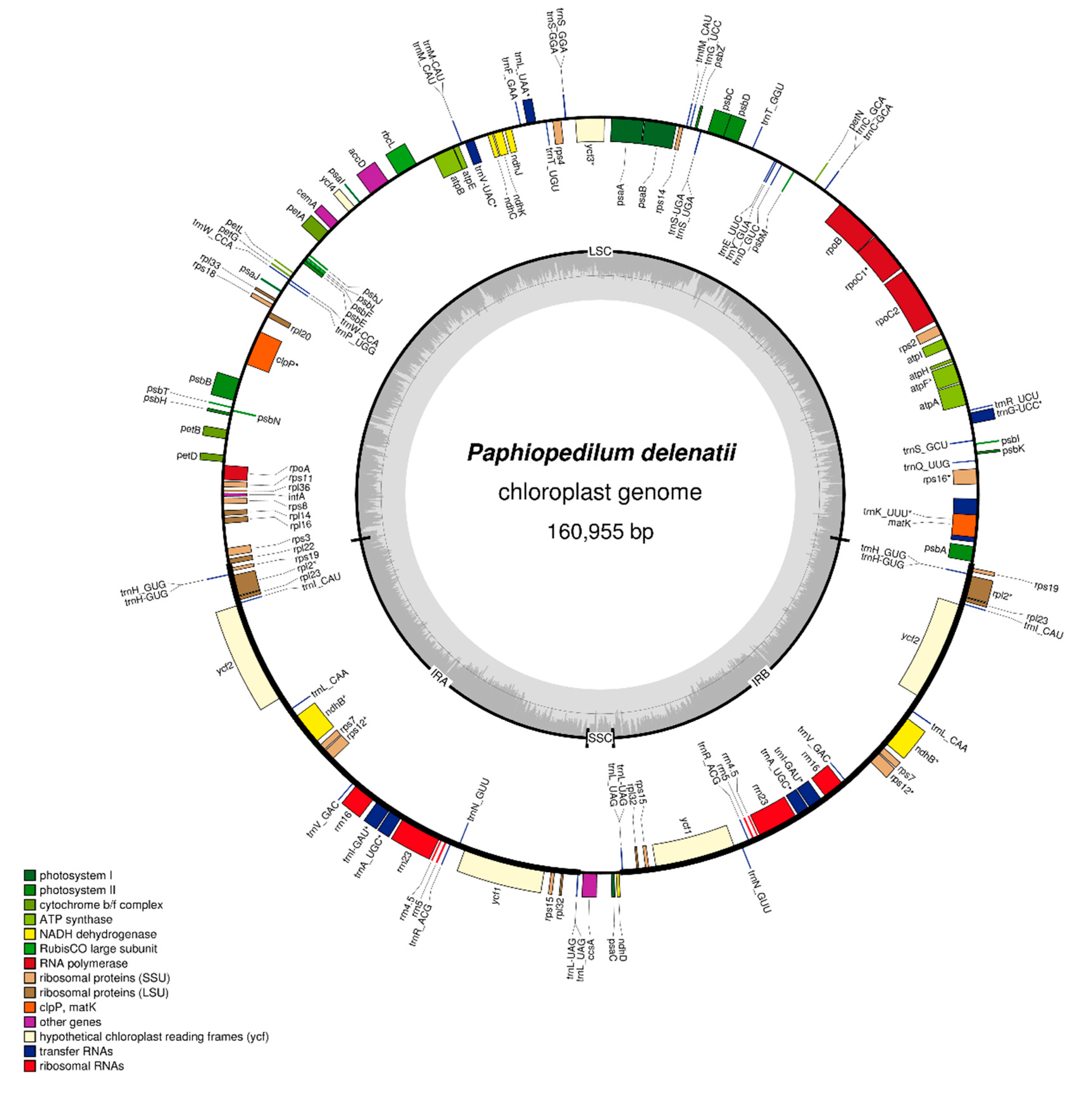



| Classification of Genes | Name of Genes | Number | |
|---|---|---|---|
| RNA genes | Ribosomal RNAs | rrn4.5(× 2), rrn5(× 2), rrn16(× 2), rrn23(× 2) | 8 |
| Transfer RNAs | trnA_UGC(× 2), trnC_GCA, trnD_GUC, trnE_UUC, trnF_GAA, trnfM_CAU, trnG_GCC, trnG_UCC, trnH_GUG(× 2), trnI_CAU(× 2), trnI_GAU(× 2), trnK_UUU, trnL_CAA(× 2), trnL_UAA, trnL_UAG(× 2), trnM_CAU, trnN_GUU(× 2), trnP_UGG, trnQ_UUG, trnR_ACG(× 2), trnR_UCU, trnS_GCU, trnS_GGA, trnS_UGA, trnT_GGU, trnT_UGU, trnV_GAC(× 2), trnV_UAC, trnW_CCA, trnY_GUA | 39 | |
| Protein-coding genes | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| Cytochrome | petA, petB, petD, petG, petL, petN | 6 | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | 6 | |
| Rubisco | rbcL | 1 | |
| Ribosomal proteins-small units | rps11, rps12(× 2), rps14, rps15(× 2), rps16, rps18, rps19(× 2), rps2, rps3, rps4, rps7(× 2), rps8 | 16 | |
| Ribosomal proteins-large units | rpl14, rpl16, rpl2(× 2), rpl20, rpl22, rpl23(× 2), rpl32(× 2), rpl33, rpl36 | 12 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | 4 | |
| Miscellaneous | accD, ccsA, cemA, clpP, infA, matK | 6 | |
| Hypothetical chloroplast reading frames (ycf) | ycf1(× 2), ycf2(× 2), ycf3, ycf4 | 6 | |
| Pseudogenes | NADH dehydrogenase | ndhB(× 2), ndhC, ndhD, ndhJ, ndhK | 6 |
| Total | 130 | ||
| Species | Paphiopedilum delenatii | Paphiopedilumarmeniacum | Paphiopedilumniveum | Paphiopedilumdianthum |
|---|---|---|---|---|
| Total length (bp) | 160,955 | 162,682 | 159,108 | 154,699 |
| IR length (bp) | 34,196 | 33,641 | 31,978 | 32,711 |
| LSC length (bp) | 89,869 | 91,734 | 89,958 | 86,861 |
| SSC length (bp) | 2694 | 3666 | 5194 | 2416 |
| Total gene number | 130 | 129 | 126 | 130 |
| Coding sequence (CDS) number | 77 | 77 | 74 | 79 |
| rRNA number | 8 | 8 | 8 | 8 |
| tRNA number | 39 | 38 | 38 | 38 |
| Pseudogene number | 6 | 6 | 6 | 5 |
| Overall GC content (%) | 35.6 | 35.4 | 35.0 | 35.0 |
| GC content of IR (%) | 39.3 | 39.0 | 40.0 | 39.0 |
| GC content of LSC (%) | 33 | 32.6 | 32.0 | 33.0 |
| GC content of SSC (%) | 28.5 | 31.0 | 29.0 | 29.0 |
| GenBank accession | MK463585 | NC_026779.1 | NC_026776.1 | NC_036958.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, H.-T.; Tran, N.; Nguyen, T.-D.; Vu, Q.-L.; Bui, M.-H.; Le, M.-T.; Le, L. Complete Chloroplast Genome of Paphiopedilum delenatii and Phylogenetic Relationships among Orchidaceae. Plants 2020, 9, 61. https://doi.org/10.3390/plants9010061
Vu H-T, Tran N, Nguyen T-D, Vu Q-L, Bui M-H, Le M-T, Le L. Complete Chloroplast Genome of Paphiopedilum delenatii and Phylogenetic Relationships among Orchidaceae. Plants. 2020; 9(1):61. https://doi.org/10.3390/plants9010061
Chicago/Turabian StyleVu, Huyen-Trang, Ngan Tran, Thanh-Diem Nguyen, Quoc-Luan Vu, My-Huyen Bui, Minh-Tri Le, and Ly Le. 2020. "Complete Chloroplast Genome of Paphiopedilum delenatii and Phylogenetic Relationships among Orchidaceae" Plants 9, no. 1: 61. https://doi.org/10.3390/plants9010061
APA StyleVu, H.-T., Tran, N., Nguyen, T.-D., Vu, Q.-L., Bui, M.-H., Le, M.-T., & Le, L. (2020). Complete Chloroplast Genome of Paphiopedilum delenatii and Phylogenetic Relationships among Orchidaceae. Plants, 9(1), 61. https://doi.org/10.3390/plants9010061





