Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants
Abstract
1. Introduction
2. LSD1, EDS1, and PAD4 Play a Crucial Role in the Response to Both Biotic and Abiotic Stresses
3. LSD1, EDS1 and PAD4 Molecular Properties
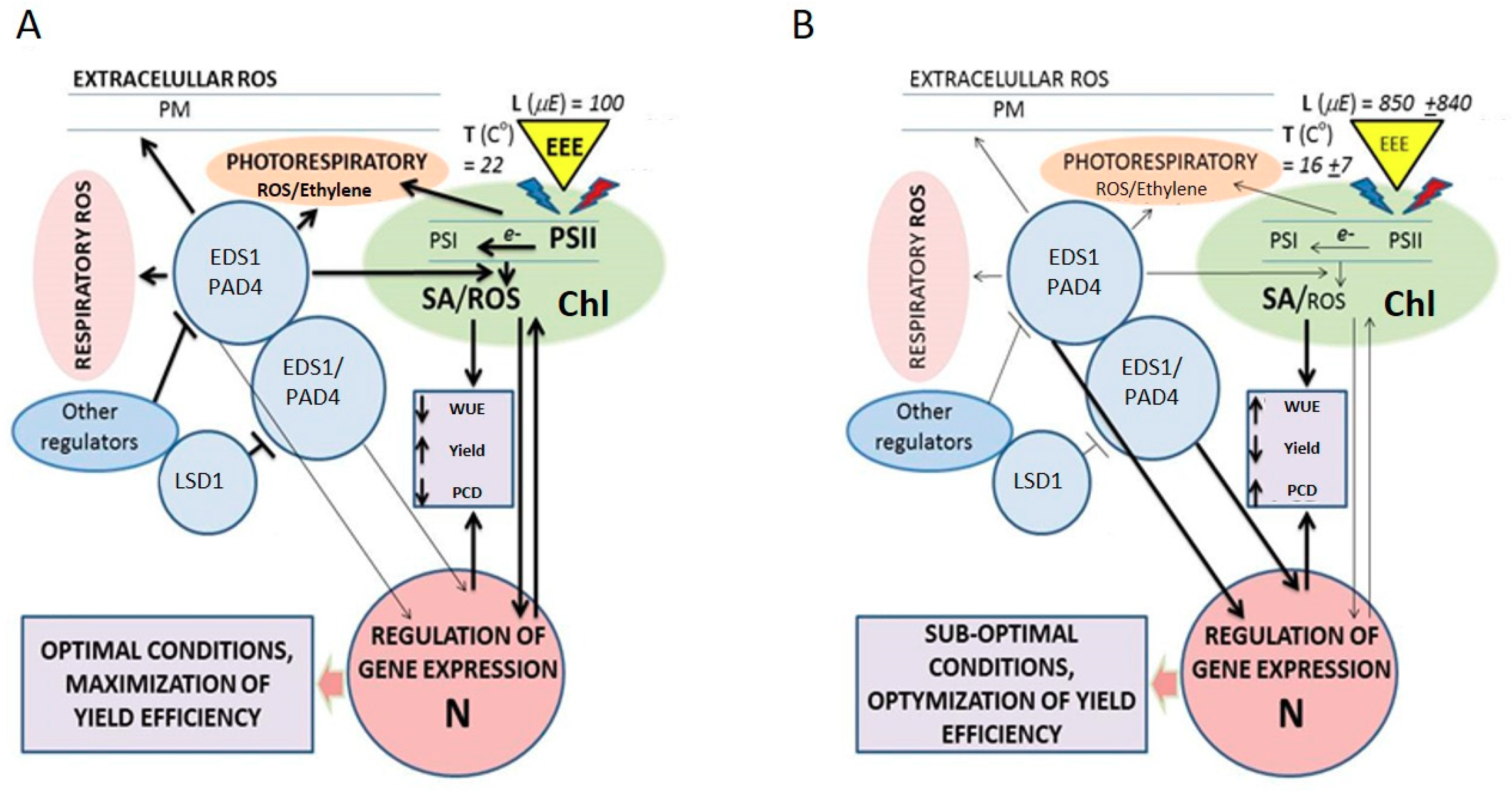
4. The LSD1, EDS1, and PAD4 Regulatory Hub Links Plastoquinone, Salicylic acid, Ethylene, and ROS Signaling in Arabidopsis thaliana
5. Involvement of Salicylic Acid, Ethylene, and ROS in Plant Productivity
6. LSD1, EDS1, and PAD4 are Involved in Biomass Production, Seed Yield Regulation, and Water Use Efficiency in Arabidopsis thaliana
7. LSD1, EDS1, and PAD4 Regulate Morphology, Photosynthetic Efficiency, and Wood Properties in Populus tremula L. × P. tremuloides
8. Orthologs of LSD1, EDS1, and PAD4 in Crops
9. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Atafar, Z.; Mesdaghinia, A.; Nouri, J.; Homaee, M.; Yunesian, M.; Ahmadimoghaddam, M.; Mahvi, A.H. Effect of fertilizer application on soil heavy metal concentration. Environ. Monit. Assess. 2010, 160, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M. Environmental toxins and health: The health impact of pesticides. Aust. Fam. Phys. Melb. 2007, 36, 1002–1004. [Google Scholar]
- Geiger, F.; Bengtsson, J.; Berendse, F.; Weisser, W.W.; Emmerson, M.; Morales, M.B.; Ceryngier, P.; Liira, J.; Tscharntke, T.; Winqvist, C.; et al. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl. Ecol. 2010, 11, 97–105. [Google Scholar] [CrossRef]
- Van der Werf, H.M.G. Assessing the impact of pesticides on the environment. Agric. Ecosyst. Environ. 1996, 60, 81–96. [Google Scholar] [CrossRef]
- Wilson, C.; Tisdell, C. Why farmers continue to use pesticides despite environmental, health and sustainability costs. Ecol. Econ. 2001, 39, 449–462. [Google Scholar] [CrossRef]
- Bernacki, Z. Biomass production of maize (Zea mays L.) cropping in exceptionally advantageous conditions in central Wielkopolska (Poland). Biomass Bioenergy 2018, 110, 25–32. [Google Scholar] [CrossRef]
- Kundzewicz, Z.W.; Krysanova, V.; Benestad, R.E.; Hov, Ø.; Piniewski, M.; Otto, I.M. Uncertainty in climate change impacts on water resources. Environ. Sci. Policy 2018, 79, 1–8. [Google Scholar] [CrossRef]
- Polley, H.W. Implications of atmospheric and climatic change for crop yield and water use efficiency. Crop Sci. 2002, 42, 131–140. [Google Scholar] [CrossRef]
- Tambussi, E.A.; Bort, J.; Araus, J. Water use efficiency in C3 cereals under Mediterranean conditions: A review of physiological aspects. Ann. Appl. Biol. 2007, 150, 307–321. [Google Scholar] [CrossRef]
- Kausch, A.; Rhodes, R. Research and Technology Development for Genetic Improvement of Switchgrass; Univ. of Rhode Island: Kingston, RI, USA, 2014. Available online: https://www.osti.gov/servlets/purl/1357908 (accessed on 27 May 2019).
- Padgette, S.R.; Kolacz, K.H.; Delannay, X.; Re, D.B.; LaVallee, B.J.; Tinius, C.N.; Rhodes, W.K.; Otero, Y.I.; Barry, G.F.; Eichholtz, D.A.; et al. Development, identification, and characterization of a glyphosate-tolerant soybean line. Crop Sci. 1995, 35, 1451–1461. [Google Scholar] [CrossRef]
- Huang, J.; Rozelle, S.; Pray, C.; Wang, Q. Plant biotechnology in China. Science 2002, 295, 674–676. [Google Scholar] [CrossRef] [PubMed]
- Sasson, A.; Malpica, C. Bioeconomy in Latin America. New Biotechnol. 2018, 40, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Kerr, P.S. Genetically-engineered crops: Their potential use for improvement of human nutrition. Nutr. Rev. 2002, 60, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Brookes, G.; Barfoot, P. The global income and production effects of genetically-modified (GM) crops 1996–2011. GM Crops Food 2013, 4, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Benbrook, C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Jones, H.D.; Halford, N.G. Plant Biotechnology: Transgenic Crops. In Food Biotechnology; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 149–186. ISBN 978-3-540-70535-2. [Google Scholar]
- Szechyńska-Hebda, M.; Wędzony, M.; Tyrka, M.; Gołębiowska, G.; Chrupek, M.; Czyczyło-Mysza, I.; Dubas, E.; Żur, I.; Golemiec, E. Identifying QTLs for cold-induced resistance to Microdochium nivale in winter triticale. Plant Genet. Resour. 2011, 9, 296–299. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Wąsek, I.; Gołębiowska-Pikania, G.; Dubas, E.; Żur, I.; Wędzony, M. Photosynthesis-dependent physiological and genetic crosstalk between cold acclimation and cold-induced resistance to fungal pathogens in triticale (Triticosecale Wittm.). J. Plant Physiol. 2015, 177, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F.; Gao, C. Precision genome engineering and agriculture: Opportunities and regulatory challenges. PLoS Biol. 2014, 12, e1001877. [Google Scholar] [CrossRef]
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S.P. Feedstocks for lignocellulosic biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef]
- Ke, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host-pathogen interactions. Plant J. 2014, 78, 619–631. [Google Scholar] [CrossRef]
- Wang, L.; Pei, Z.; Tian, Y.; He, C. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant. Microbe Interact. 2005, 18, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wei, B.; Li, G.; Gong, C.; Fan, R.; Zhang, X. TaEDS1 genes positively regulate resistance to powdery mildew in wheat. Plant Mol. Biol. 2018, 96, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bai, P.; Yang, Q.; Liu, F.; Wang, X.; Huang, L.; Kang, Z. Wheat zinc finger protein TaLSD1, a negative regulator of programmed cell death, is involved in wheat resistance against stripe rust fungus. Plant Physiol. Biochem. 2013, 71, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Qi, X.; Cheng, H. Molecular cloning and characterization of enhanced disease susceptibility 1 (EDS1) from Gossypium barbadense. Mol. Biol. Rep. 2014, 41, 3821–3828. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Xingfen, W.; Wei, R.; Jun, Y.; Zhiying, M. Island cotton enhanced disease susceptibility 1 gene encoding a lipase-like protein plays a crucial role in response to Verticillium dahliae by regulating the SA level and H2O2 accumulation. Front. Plant Sci. 2016, 7, 1830. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Shu, X.; Ali, M.B.; Howard, S.; Li, N.; Winterhagen, P.; Qiu, W.; Gassmann, W. A functional EDS1 ortholog is differentially regulated in powdery mildew resistant and susceptible grapevines and complements an Arabidopsis eds1 mutant. Planta 2010, 231, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Dai, R.; Pike, S.M.; Qiu, W.; Gassmann, W. Functions of EDS1-like and PAD4 genes in grapevine defenses against powdery mildew. Plant Mol. Biol. 2014, 86, 381–393. [Google Scholar] [CrossRef]
- Hu, G.; deHart, A.K.A.; Li, Y.; Ustach, C.; Handley, V.; Navarre, R.; Hwang, C.-F.; Aegerter, B.J.; Williamson, V.M.; Baker, B. EDS1 in tomato is required for resistance mediated by TIR-class R genes and the receptor-like R gene Ve. Plant J. Cell Mol. Biol. 2005, 42, 376–391. [Google Scholar] [CrossRef]
- Schornack, S.; Ballvora, A.; Gürlebeck, D.; Peart, J.; Baulcombe, D.; Ganal, M.; Baker, B.; Bonas, U.; Lahaye, T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. Cell Mol. Biol. 2004, 37, 46–60. [Google Scholar] [CrossRef]
- He, S.; Huang, K.; Zhang, X.; Yu, X.; Huang, P.; An, C. The LSD1-type zinc finger motifs of Pisum sativa LSD1 are a novel nuclear localization signal and interact with importin alpha. PLoS ONE 2011, 6, e22131. [Google Scholar] [CrossRef]
- Lorrain, S.; Vailleau, F.; Balagué, C.; Roby, D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003, 8, 263–271. [Google Scholar] [CrossRef]
- Salmeron, J.M.; Vernooij, B. Transgenic approaches to microbial disease resistance in crop plants. Curr. Opin. Plant Biol. 1998, 1, 347–352. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Ślesak, I.; Vanderauwera, S.; Szechyńska-Hebda, M.; Kornaś, A.; Kelen, K.V.D.; Mühlenbock, P.; Karpińska, B.; Maćkowski, S.; Breusegem, F.V.; et al. LESION SIMULATING DISEASE 1, ENHANCED DISEASE SUSCEPTIBILITY 1, and PHYTOALEXIN DEFICIENT 4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 2013, 161, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.A.; Delaney, T.P.; Uknes, S.J.; Ward, E.R.; Ryals, J.A.; Dangl, J.L. Arabidopsis mutants simulating disease resistance response. Cell 1994, 77, 565–577. [Google Scholar] [CrossRef]
- Jabs, T.; Dietrich, R.A.; Dangl, J.L. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 1996, 273, 1853–1856. [Google Scholar] [CrossRef]
- Bernacki, M.J.; Czarnocka, W.; Witoń, D.; Rusaczonek, A.; Szechyńska-Hebda, M.; Ślesak, I.; Dąbrowska-Bronk, J.; Karpiński, S. ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) affects development, photosynthesis, and hormonal homeostasis in hybrid aspen (Populus tremula L. × P. tremuloides). J. Plant Physiol. 2018, 226, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Kuo, W.Y.; Weiss, C.; Jinn, T.L. Copper chaperone-dependent and -Independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 2012, 158, 737–746. [Google Scholar] [CrossRef]
- Mateo, A.; Funck, D.; Mühlenbock, P.; Kular, B.; Mullineaux, P.M.; Karpinski, S. Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 2006, 57, 1795–1807. [Google Scholar] [CrossRef]
- Mateo, A.; Mühlenbock, P.; Rustérucci, C.; Chang, C.C.C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef]
- Mühlenbock, P.; Plaszczyca, M.; Plaszczyca, M.; Mellerowicz, E.; Karpinski, S. Lysigenous aerenchyma formation in Arabidopsis is Controlled by LESION SIMULATING DISEASE1. Plant Cell 2007, 19, 3819–3830. [Google Scholar] [CrossRef]
- Rustérucci, C.; Aviv, D.H.; Holt, B.F.; Dangl, J.L.; Parker, J.E. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 2001, 13, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Wituszyńska, W.; Szechyńska-Hebda, M.; Sobczak, M.; Rusaczonek, A.; Kozłowska-Makulska, A.; Witoń, D.; Karpiński, S. Lesion simulating disease 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Bernacki, M.J.; Czarnocka, W.; Rusaczonek, A.; Witoń, D.; Kęska, S.; Czyż, J.; Szechyńska-Hebda, M.; Karpiński, S. LSD1, EDS1 and PAD4-dependent conditional correlation among salicylic acid, hydrogen peroxide, water use efficiency, and seed yield in Arabidopsis thaliana. Physiol. Plant. 2018, 165, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Aviv, D.H.; Rustérucci, C.; Iii, B.F.H.; Dietrich, R.A.; Parker, J.E.; Dangl, J.L. Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 2002, 29, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Szechynska-Hebda, M.; Plaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpinska, B.; Karpinski, S. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [PubMed]
- Szechyńska-Hebda, M.; Czarnocka, W.; Hebda, M.; Karpiński, S. PAD4, LSD1 and EDS1 regulate drought tolerance, plant biomass production, and cell wall properties. Plant Cell Rep. 2016, 35, 527–539. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Zhang, X.; Zuo, J.; Yang, S. The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol. 2010, 187, 301–312. [Google Scholar] [CrossRef]
- Czarnocka, W.; Van Der Kelen, K.; Willems, P.; Szechyńska-Hebda, M.; Shahnejat-Bushehri, S.; Balazadeh, S.; Rusaczonek, A.; Mueller-Roeber, B.; Van Breusegem, F.; Karpiński, S. The dual role of LESION SIMULATING DISEASE 1 as a condition-dependent scaffold protein and transcription regulator: Insight into the LSD1 molecular function. Plant Cell Environ. 2017, 40, 2644–2662. [Google Scholar] [CrossRef]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef]
- Rietz, S.; Stamm, A.; Malonek, S.; Wagner, S.; Becker, D.; Medina-Escobar, N.; Vlot, A.C.; Feys, B.J.; Niefind, K.; Parker, J.E. Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011, 191, 107–119. [Google Scholar] [CrossRef]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.G.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 13583–13588. [Google Scholar] [CrossRef] [PubMed]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.E.; Holub, E.B.; Frost, L.N.; Falk, A.; Gunn, N.D.; Daniels, M.J. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 1996, 8, 2033–2046. [Google Scholar]
- Wiermer, M.; Feys, B.J.; Parker, J.E. Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 2005, 8, 383–389. [Google Scholar] [CrossRef]
- Drew, M.C.; He, C.J.; Morgan, P.W. Programmed cell death and aerenchyma formation in roots. Trends Plant Sci. 2000, 5, 123–127. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Dietrich, R.A.; Richberg, M.H.; Schmidt, R.; Dean, C.; Dangl, J.L. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 1997, 88, 685–694. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. CMLS 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Chai, T.; Zhou, J.; Liu, J.; Xing, D. LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front. Plant Sci. 2015, 6, 292. [Google Scholar] [CrossRef] [PubMed]
- Kaminaka, H.; Näke, C.; Epple, P.; Dittgen, J.; Schütze, K.; Chaban, C.; Holt, B.F.; Merkle, T.; Schäfer, E.; Harter, K.; et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006, 25, 4400–4411. [Google Scholar] [CrossRef] [PubMed]
- Klopffleisch, K.; Phan, N.; Augustin, K.; Bayne, R.S.; Booker, K.S.; Botella, J.R.; Carpita, N.C.; Carr, T.; Chen, J.-G.; Cooke, T.R.; et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 2011, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Trigg, S.A.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.-S.C.; et al. CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 2017, 14, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Yoon, G.M.; Kieber, J.J. 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 2013, 25, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- García, A.V.; Blanvillain-Baufumé, S.; Huibers, R.P.; Wiermer, M.; Li, G.; Gobbato, E.; Rietz, S.; Parker, J.E. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLOS Pathog. 2010, 6, e1000970. [Google Scholar] [CrossRef]
- Zhu, S.; Jeong, R.D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLOS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Altmann, M.; Alkofer, A.; Epple, P.M.; Dangl, J.L.; Falter-Braun, P. LSU network hubs integrate abiotic and biotic stress responses via interaction with the superoxide dismutase FSD2. J. Exp. Bot. 2017, 68, 1185–1197. [Google Scholar] [CrossRef]
- Mohr, T.J.; Mammarella, N.D.; Hoff, T.; Woffenden, B.J.; Jelesko, J.G.; McDowell, J.M. The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant-Microbe Interact. 2010, 23, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Gassmann, W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007, 145, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, S.; Shin, Y.; Hur, Y.S.; Kim, W.Y.; Lee, M.S.; Cheon, C.I.; Verma, D.P.S. Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J. Biol. Chem. 2014, 289, 3901–3912. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Liang, C.; Wang, S.; Hou, Y.; Gao, L.; Liu, L.; He, W.; Ma, W.; Mo, B.; Chen, X. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018, 9, 5080. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.D.T.; Pike, S.; Wang, J.; Nepal Poudel, A.; Heinz, R.; Schultz, J.C.; Koo, A.J.; Mitchum, M.G.; Appel, H.M.; Gassmann, W. The Arabidopsis immune regulator SRFR1 dampens defences against herbivory by Spodoptera exigua and parasitism by Heterodera schachtii. Mol. Plant Pathol. 2016, 17, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Karpiński, S.; Szechyńska-Hebda, M. Secret life of plants: from memory to intelligence. Plant Signal. Behav. 2010, 5, 1391–1394. [Google Scholar] [CrossRef] [PubMed]
- Szechyńska-Hebda, M.; Kruk, J.; Górecka, M.; Karpińska, B.; Karpiński, S. Evidence for light wavelength-specific photoelectrophysiological signaling and memory of excess light episodes in Arabidopsis. Plant Cell 2010, 22, 2201–2218. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. REACTIVE OXYGEN SPECIES: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Mittler, R. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006, 141, 311. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.A.; Klessig, D.F. Salicylic acid, active oxygen species and systemic acquired resistance in plants. Trends Cell Biol. 1994, 4, 334–338. [Google Scholar] [CrossRef]
- Lu, H.; Greenberg, J.T.; Holuigue, L. Editorial: Salicylic acid signaling networks. Front. Plant Sci. 2016, 7, 238. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef]
- Jordan, B.R. Review: Molecular response of plant cells to UV-B stress. Funct. Plant Biol. 2002, 29, 909–916. [Google Scholar] [CrossRef]
- Khudyakova, A.Y.; Kreslavski, V.D.; Shirshikova, G.N.; Zharmukhamedov, S.K.; Kosobryukhov, A.A.; Allakhverdiev, S.I. Resistance of Arabidopsis thaliana L. photosynthetic apparatus to UV-B is reduced by deficit of phytochromes B and A. J. Photochem. Photobiol. B 2017, 169, 41–46. [Google Scholar] [CrossRef]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant 2018, 11, 1053–1066. [Google Scholar] [CrossRef]
- Khokon, M.A.R.; Okuma, E.; Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2011, 34, 434–443. [Google Scholar] [CrossRef]
- Lawson, T.; Blatt, M.R. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 2014, 164, 1556–1570. [Google Scholar] [CrossRef]
- Raven, J.A. Speedy small stomata? J. Exp. Bot. 2014, 65, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Devireddy, A.R.; Zandalinas, S.I.; Gómez-Cadenas, A.; Blumwald, E.; Mittler, R. Coordinating the overall stomatal response of plants: Rapid leaf-to-leaf communication during light stress. Sci Signal 2018, 11, eaam9514. [Google Scholar] [CrossRef] [PubMed]
- Dat, J.; Vandenabeele, S.; Vranová, E.; Montagu, M.V.; Inzé, D.; Breusegem, F.V. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Breusegem, F.V.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1767, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2007, 104, 10270–10275. [Google Scholar] [CrossRef]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakière, B.; Vanacker, H.; Miginiac-Maslow, M.; Breusegem, F.V.; Noctor, G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007, 52, 640–657. [Google Scholar]
- Vandenabeele, S.; Vanderauwera, S.; Vuylsteke, M.; Rombauts, S.; Langebartels, C.; Seidlitz, H.K.; Zabeau, M.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004, 39, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Catinot, J.; Buchala, A.; Abou-Mansour, E.; Métraux, J.-P. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 2008, 582, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.A.; Friedrich, L.; Hunt, M.; Weymann, K.; Delaney, T.; Kessmann, H.; Staub, T.; Ryals, J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. Cell Mol. Biol. 1996, 10, 71–82. [Google Scholar] [CrossRef]
- Abreu, M.E.; Munné-Bosch, S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Witoń, D.; Gawroński, P.; Czarnocka, W.; Ślesak, I.; Rusaczonek, A.; Sujkowska-Rybkowska, M.; Bernacki, M.J.; Dąbrowska-Bronk, J.; Tomsia, N.; Szechyńska-Hebda, M.; et al. Mitogen activated protein kinase 4 (MPK4) influences growth in Populus tremula L. × tremuloides. Environ. Exp. Bot. 2016, 130, 189–205. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Casalongué, C.A. Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal. Behav. 2011, 6, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant. Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef]
- Jirage, D.; Zhou, N.; Cooper, B.; Clarke, J.D.; Dong, X.; Glazebrook, J. Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 2001, 26, 395–407. [Google Scholar] [CrossRef]
- Rate, D.N.; Cuenca, J.V.; Bowman, G.R.; Guttman, D.S.; Greenberg, J.T. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 1999, 11, 1695–1708. [Google Scholar] [CrossRef]
- Makandar, R.; Nalam, V.J.; Chowdhury, Z.; Sarowar, S.; Klossner, G.; Lee, H.; Burdan, D.; Trick, H.N.; Gobbato, E.; Parker, J.E.; et al. The combined action of ENHANCED DISEASE SUSCEPTIBILITY1, PHYTOALEXIN DEFICIENT4, and SENESCENCE-ASSOCIATED101 promotes salicylic acid-mediated defenses to limit Fusarium graminearum infection in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2015, 28, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Coronado, M.A.; Trejo-López, C.; Larqué-Saavedra, A. Effects of salicylic acid on the growth of roots and shoots in soybean. Plant Physiol. Biochem. 1998, 36, 563–565. [Google Scholar] [CrossRef]
- Shakirova, F.M.; Sakhabutdinova, A.R.; Bezrukova, M.V.; Fatkhutdinova, R.A.; Fatkhutdinova, D.R. Changes in the hormonal status of wheat seedlings induced by salicylic acid and salinity. Plant Sci. 2003, 164, 317–322. [Google Scholar] [CrossRef]
- Singh, B.; Usha, K. Salicylic acid induced physiological and biochemical changes in wheat seedlings under water stress. Plant Growth Regul. 2003, 39, 137–141. [Google Scholar] [CrossRef]
- Gunes, A.; Inal, A.; Alpaslan, M.; Eraslan, F.; Bagci, E.G.; Cicek, N. Salicylic acid induced changes on some physiological parameters symptomatic for oxidative stress and mineral nutrition in maize (Zea mays L.) grown under salinity. J. Plant Physiol. 2007, 164, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Kovácik, J.; Grúz, J.; Backor, M.; Strnad, M.; Repcák, M. Salicylic acid-induced changes to growth and phenolic metabolism in Matricaria chamomilla plants. Plant Cell Rep. 2009, 28, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.M.; Clarke, S.M.; Wood, J.E.; Mur, L.A. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 2004, 135, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Gawroński, P.; Witoń, D.; Vashutina, K.; Bederska, M.; Betliński, B.; Rusaczonek, A.; Karpiński, S. Mitogen-activated protein kinase 4 is a salicylic acid-independent regulator of growth but not of photosynthesis in Arabidopsis. Mol. Plant 2014, 7, 1151–1166. [Google Scholar] [CrossRef]
- Ślesak, I.; Szechyńska-Hebda, M.; Fedak, H.; Sidoruk, N.; Dąbrowska-Bronk, J.; Witoń, D.; Rusaczonek, A.; Antczak, A.; Drożdżek, M.; Karpińska, B.; et al. PHYTOALEXIN DEFICIENT 4 affects reactive oxygen species metabolism, cell wall and wood properties in hybrid aspen (Populus tremula L. × tremuloides). Plant Cell Environ. 2015, 38, 1275–1284. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Kangasjärvi, J. ROS signaling loops - production, perception, regulation. Curr. Opin. Plant Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef]
- Ecker, J.R. The ethylene signal transduction pathway in plants. Science 1995, 268, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, I.I.; Qudeimat, E.; Potuschak, T.; Du, Y.; Genschik, P.; Vandenbussche, F.; Straeten, D.V.D. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. 2018, 115, E4130–E4139. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Rodrigues-Pousada, R.A.; De Rycke, R.; Dedonder, A.; Van Caeneghem, W.; Engler, G.; Van Montagu, M.; Van Der Straeten, D. The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell 1993, 5, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Farwell, A.J.; Vesely, S.; Nero, V.; Rodriguez, H.; Shah, S.; Dixon, D.G.; Glick, B.R. The use of transgenic canola (Brassica napus) and plant growth-promoting bacteria to enhance plant biomass at a nickel-contaminated field site. Plant Soil 2006, 288, 309–318. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Karpiński, S. Programmed cell death as a response to high light, UV and drought stress in plants. In Abiotic Stress—Plant Responses and Applications in Agriculture; InTech: Rijeka, Croatia, 2013; pp. 207–246. ISBN 978-953-51-1024-8. [Google Scholar]
- Mullineaux, P.; Karpinski, S. Signal transduction in response to excess light: getting out of the chloroplast. Curr. Opin. Plant Biol. 2002, 5, 43–48. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Lewandowska, M.; Karpiński, S. Electrical signaling, photosynthesis and systemic acquired acclimation. Front. Physiol. 2017, 14, 684. [Google Scholar] [CrossRef]
- Karpiński, S.; Szechyńska-Hebda, M. Cellular light memory, photo-electrochemical and redox retrograde signaling in plants. BioTechnologia 2014, 93, 27–39. [Google Scholar] [CrossRef]
- Wituszynska, W.; Karpinski, S. Determination of Water Use Efficiency for Arabidopsis thaliana. BIO-Protoc. 2014, 4. [Google Scholar] [CrossRef]
- Foyer, C.H.; Neukermans, J.; Queval, G.; Noctor, G.; Harbinson, J. Photosynthetic control of electron transport and the regulation of gene expression. J. Exp. Bot. 2012, 63, 1637–1661. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, S.; Escobar, C.; Karpinska, B.; Creissen, G.; Mullineaux, P.M. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 1997, 9, 627–640. [Google Scholar] [PubMed]
- Lee, S.; Choi, H.; Suh, S.; Doo, I.S.; Oh, K.Y.; Choi, E.J.; Taylor, A.T.S.; Low, P.S.; Lee, Y. Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and commelina communis. Plant Physiol. 1999, 121, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Okamoto, H.; Okuma, E.; Shiba, H.; Kamada, H.; Hasegawa, P.M.; Murata, Y. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. Cell Mol. Biol. 2013, 73, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 2002, 90, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Christersson, L. Poplar plantations for paper and energy in the south of Sweden. Biomass Bioenergy 2008, 32, 997–1000. [Google Scholar] [CrossRef]
- Devappa, R.K.; Rakshit, S.K.; Dekker, R.F.H. Potential of poplar bark phytochemicals as value-added co-products from the wood and cellulosic bioethanol industry. BioEnergy Res. 2015, 8, 1235–1251. [Google Scholar] [CrossRef]
- Barigah, T.S.; Saugier, B.; Mousseau, M.; Guittet, J.; Ceulemans, R. Photosynthesis, leaf area and productivity of 5 poplar clones during their establishment year. Ann. Sci. For. 1994, 51, 613–625. [Google Scholar] [CrossRef]
- Koch, J.R.; Creelman, R.A.; Eshita, S.M.; Seskar, M.; Mullet, J.E.; Davis, K.R. Ozone Sensitivity in Hybrid Poplar Correlates with Insensitivity to Both Salicylic Acid and Jasmonic Acid. The Role of Programmed Cell Death in Lesion Formation. Plant Physiol. 2000, 123, 487–496. [Google Scholar] [CrossRef]
- Matte Risopatron, J.P.; Sun, Y.; Jones, B.J. The vascular cambium: molecular control of cellular structure. Protoplasma 2010, 247, 145–161. [Google Scholar] [CrossRef]
- Gonzalez, N.; Beemster, G.T.; Inzé, D. David and Goliath: What can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr. Opin. Plant Biol. 2009, 12, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Wang, J.; Sandberg, G. Methods of Increasing Plant Growth 2008. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2008125983 (accessed on 20 April 2019).
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. Cell Mol. Biol. 2003, 36, 94–104. [Google Scholar] [CrossRef]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. 2000, 97, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Beemster, G.T.S.; Fiorani, F.; Inzé, D. Cell cycle: the key to plant growth control? Trends Plant Sci. 2003, 8, 154–158. [Google Scholar] [CrossRef]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [PubMed]
- Abramson, M.; Shoseyov, O.; Shani, Z. Plant cell wall reconstruction toward improved lignocellulosic production and processability. Plant Sci. 2010, 178, 61–72. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat Commun. 2019, 30, 1994. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Daudi, A.; Finch, P.; Butt, V.S.; Whitelegge, J.P.; Souda, P.; Ausubel, F.M.; Bolwell, G.P. A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 2012, 158, 2013–2027. [Google Scholar] [CrossRef]
- Potikha, T.S.; Collins, C.C.; Johnson, D.I.; Delmer, D.P.; Levine, A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 1999, 119, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Ros Barceló, A. Xylem parenchyma cells deliver the H2O2 necessary for lignification in differentiating xylem vessels. Planta 2005, 220, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Giraldo, L.; Escamilla-Trevino, L.; Jackson, L.A.; Dixon, R.A. Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20814–20819. [Google Scholar] [CrossRef] [PubMed]
- Madadi, M.; Penga, C.; and Abbas, A. Advances in genetic manipulation of lignocellulose to reduce biomass recalcitrance and enhance biofuel production in bioenergy crops. Plant Biochem. Physiol. 2017, 5, 2. [Google Scholar]
- Montgomery, R. Relative importance of photosynthetic physiology and biomass allocation for tree seedling growth across a broad light gradient. Tree Physiol. 2004, 24, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Humphry, M.; Bednarek, P.; Kemmerling, B.; Koh, S.; Stein, M.; Göbel, U.; Stüber, K.; Piślewska-Bednarek, M.; Loraine, A.; Schulze-Lefert, P.; et al. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 21896–21901. [Google Scholar] [CrossRef] [PubMed]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 05–517. [Google Scholar] [CrossRef]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- Jackson, S.A. Rice: The first crop genome. Rice 2016, 9, 14. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, H.; Hong, Y.; Liu, S.; Li, D.; Song, F. Stress-responsive expression, subcellular localization and protein–protein interactions of the rice metacaspase family. Int. J. Mol. Sci. 2015, 16, 16216–16241. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Lavoie, M.; Jin, Y.; Xu, J.; Fu, Z.; Qian, H. Diclofop-methyl affects microbial rhizosphere community and induces systemic acquired resistance in rice. J. Environ. Sci. 2017, 51, 352–360. [Google Scholar] [CrossRef]
- Duan, C.; Yu, J.; Bai, J.; Zhu, Z.; Wang, X. Induced defense responses in rice plants against small brown planthopper infestation. Crop J. 2014, 2, 55–62. [Google Scholar] [CrossRef]
- Feys, B.J.; Wiermer, M.; Bhat, R.A.; Moisan, L.J.; Medina-Escobar, N.; Neu, C.; Cabral, A.; Parker, J.E. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 2005, 17, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Alagumani, T.; Anjugam, M. Economic analysis of grape production in Tamil Nadu. J. Innov. Agric. 2018, 5, 13–18. [Google Scholar]
- Jiao, L.; Zhang, Y.; Lu, J. Overexpression of a stress-responsive U-box protein gene VaPUB affects the accumulation of resistance related proteins in Vitis vinifera ‘Thompson Seedless’. Plant Physiol. Biochem. 2017, 112, 53–63. [Google Scholar] [CrossRef]
- Mutawila, C.; Stander, C.; Halleen, F.; Vivier, M.A.; Mostert, L. Response of Vitis vinifera cell cultures to Eutypa lata and Trichoderma atroviride culture filtrates: Expression of defence-related genes and phenotypes. Protoplasma 2017, 254, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Stempien, E.; Goddard, M.L.; Leva, Y.; Bénard-Gellon, M.; Laloue, H.; Farine, S.; Kieffer-Mazet, F.; Tarnus, C.; Bertsch, C.; Chong, J. Secreted proteins produced by fungi associated with Botryosphaeria dieback trigger distinct defense responses in Vitis vinifera and Vitis rupestris cells. Protoplasma 2018, 255, 613–628. [Google Scholar] [CrossRef]
- Zaini, P.A.; Nascimento, R.; Gouran, H.; Cantu, D.; Chakraborty, S.; Phu, M.; Goulart, L.R.; Dandekar, A.M. Molecular profiling of pierce’s disease outlines the response circuitry of Vitis vinifera to Xylella fastidiosa infection. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Zhang, N.; Li, R.; Shen, W.; Jiao, S.; Zhang, J.; Xu, W. Genome-wide evolutionary characterization and expression analyses of major latex protein (MLP) family genes in Vitis vinifera. Mol. Genet. Genomics 2018, 293, 1061–1075. [Google Scholar] [CrossRef]
- Islam, M.Z.; Yun, H.K. Three transcripts of EDS1-like genes respond differently to Vitis flexuosa infection. J. Plant Biotechnol. 2017, 44, 125–134. [Google Scholar] [CrossRef]
- Tandon, G.; Jaiswal, S.; Iquebal, M.A.; Kumar, S.; Kaur, S.; Rai, A.; Kumar, D. Evidence of salicylic acid pathway with EDS1 and PAD4 proteins by molecular dynamics simulation for grape improvement. J. Biomol. Struct. Dyn. 2015, 33, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Sunilkumar, G.; Campbell, L.M.; Puckhaber, L.; Stipanovic, R.D.; Rathore, K.S. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. 2006, 103, 18054–18059. [Google Scholar] [CrossRef] [PubMed]
- Bejarano-Alcázar, J.; Blanco-López, M.A.; Melero-Vara, J.M.; Jiménez-Díaz, R.M. The influence of verticillium wilt epidemics on cotton yield in southern Spain. Plant Pathol. 1997, 46, 168–178. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Chen, T.; Yu, W.; Liu, T.; Li, H.; Fan, X.; Ren, Y.; Shen, D.; Liu, L.; et al. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PloS ONE 2012, 7, e51091. [Google Scholar] [CrossRef] [PubMed]
- Greenhalf, W.; Stephan, C.; Chaudhuri, B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996, 380, 169–175. [Google Scholar] [CrossRef]
- Kawai-Yamada, M.; Jin, L.; Yoshinaga, K.; Hirata, A.; Uchimiya, H. Mammalian bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. USA 2001, 98, 12295–12300. [Google Scholar] [CrossRef] [PubMed]
- Lacomme, C.; Cruz, S.S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. 1999, 96, 7956–7961. [Google Scholar] [CrossRef]
- Pajerowska, K.M.; Parker, J.E.; Gebhardt, C. Potato homologs of Arabidopsis thaliana genes functional in defense signaling—identification, genetic mapping, and molecular cloning. Mol. Plant. Microbe Interact. 2005, 18, 1107–1119. [Google Scholar] [CrossRef]
- Pant, S.R.; Krishnavajhala, A.; McNeece, B.T.; Lawrence, G.W.; Klink, V.P. The syntaxin 31-induced gene, LESION SIMULATING DISEASE1 (LSD1), functions in Glycine max defense to the root parasite Heterodera glycines. Plant Signal. Behav. 2015, 10, e977737. [Google Scholar] [CrossRef]
- Sharma, S.; Jaiswal, S.; Archak, S. Annotation of gene sequence and protein structure of brinjal EDS1. Bioinformation 2017, 13, 54–59. [Google Scholar] [CrossRef][Green Version]
- Heidrich, K.; Wirthmueller, L.; Tasset, C.; Pouzet, C.; Deslandes, L.; Parker, J.E. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment–specific immune responses. Science 2011, 334, 1401–1404. [Google Scholar] [CrossRef] [PubMed]
- Veluchamy, S.; Panthee, D.R. Differential expression analysis of a select list of genes in susceptible and resistant heirloom tomatoes with respect. Eur. J. Plant Pathol. 2015, 142, 653–663. [Google Scholar] [CrossRef]
- Singh, V.; Shah, J. Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signal. Behav. 2012, 7, 605–607. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Shine, M.B.; Gao, Q.M.; Navarre, D.; Jiang, W.; Liu, C.; Chen, Q.; Hu, G.; Kachroo, A. Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 2014, 165, 1269–1284. [Google Scholar] [CrossRef] [PubMed]

| Organism | Mutation, Transgene or Gene Silencing | Effect on SA Level | Growth Phenotype | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | Bacterial NahG expression | Lower level of SA in transgenic plants | Higher biomass, higher seed yield | [106] |
| Arabidopsis thaliana | Mutation in ICS1 | Lower level of SA in the mutant | Higher biomass, higher seed yield | [106] |
| Arabidopsis thaliana | Mutation in CPR1 | A significantly higher level of SA in the mutant | Dwarf phenotype | [119] |
| Arabidopsis thaliana | Mutation in LSD1 | A significantly higher level of SA in the mutant | Lower seed yield | [35] |
| Arabidopsis thaliana | Mutation in MPK4 | A significantly higher level of SA in the mutant | Dwarf phenotype | [120] |
| Populus tremula x tremuloides | Lower expression of PAD4 | Lower level of SA in transgenic lines | Higher stem diameter, higher % of dry weight | [48,121] |
| Populus tremula x tremuloides | Lower expression of EDS1 | Lower level of SA in transgenic lines | Higher CO2 assimilation, changed plant morphology | [38] |
| Populus tremula x tremuloides | Lower expression of MPK4 | Two times higher level of SA in transgenic lines | Lower perimeter of main stem | [107] |
| Ortholog of: | Species | Reference |
|---|---|---|
| AtLSD1 | Oryza sativa Triticum aestivum Pisum sativum | [23] [25] [32] |
| AtEDS1 | Oryza sativa, Gossypium barbadense Vitis vinifera Lycopersicon esculentum Triticum aestivum | [166] [26,27] [28,176] [30,186,188] [24] |
| AtPAD4 | Oryza sativa Vitis vinifera, Gossypium barbadense | [22] [176] [179] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernacki, M.J.; Czarnocka, W.; Szechyńska-Hebda, M.; Mittler, R.; Karpiński, S. Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants. Plants 2019, 8, 290. https://doi.org/10.3390/plants8080290
Bernacki MJ, Czarnocka W, Szechyńska-Hebda M, Mittler R, Karpiński S. Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants. Plants. 2019; 8(8):290. https://doi.org/10.3390/plants8080290
Chicago/Turabian StyleBernacki, Maciej Jerzy, Weronika Czarnocka, Magdalena Szechyńska-Hebda, Ron Mittler, and Stanisław Karpiński. 2019. "Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants" Plants 8, no. 8: 290. https://doi.org/10.3390/plants8080290
APA StyleBernacki, M. J., Czarnocka, W., Szechyńska-Hebda, M., Mittler, R., & Karpiński, S. (2019). Biotechnological Potential of LSD1, EDS1, and PAD4 in the Improvement of Crops and Industrial Plants. Plants, 8(8), 290. https://doi.org/10.3390/plants8080290








