The Rooting of Stem Cuttings and the Stability of uidA Gene Expression in Generative and Vegetative Progeny of Transgenic Pear Rootstock in the Field
Abstract
:1. Introduction
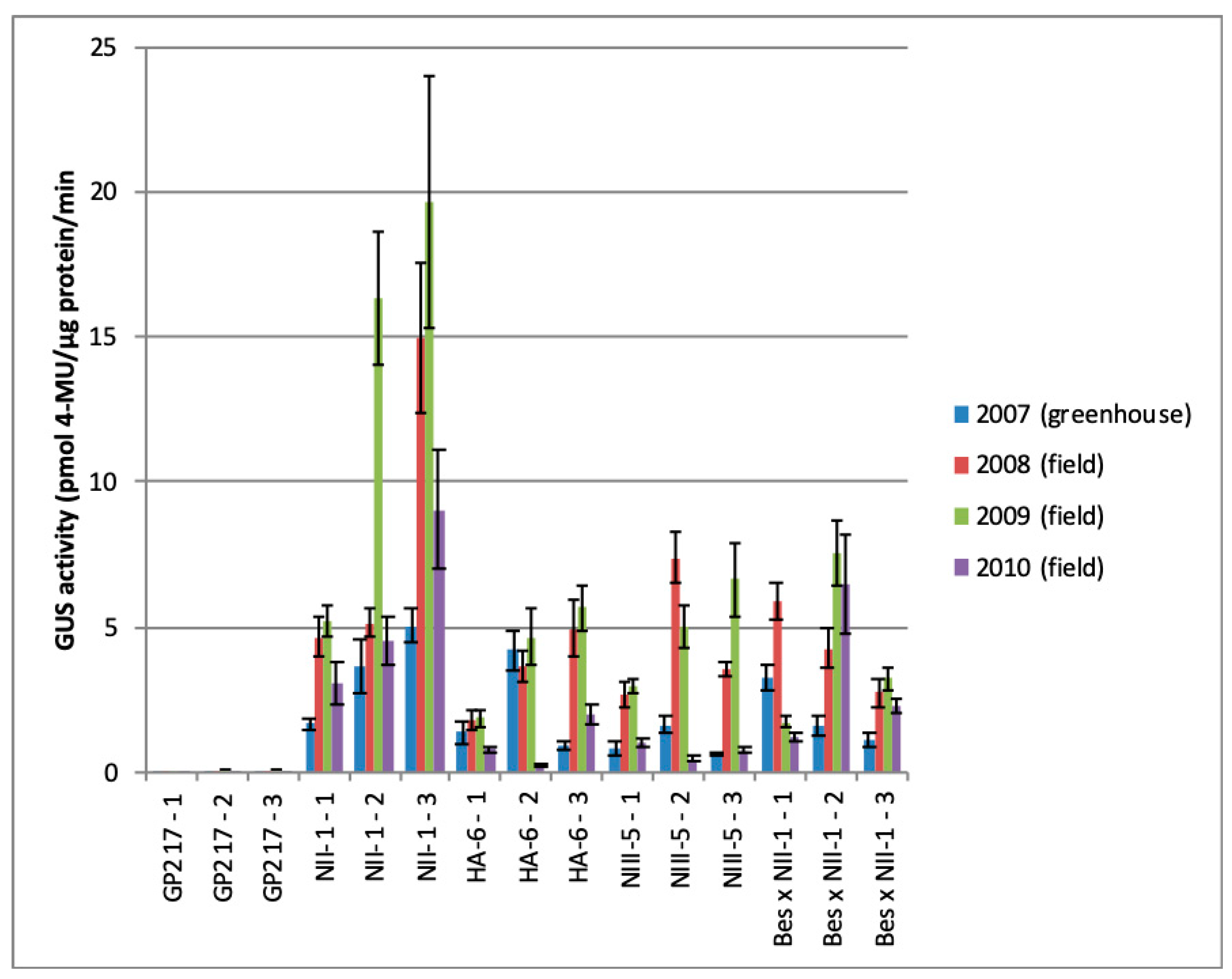
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. In Vitro Rooting
4.3. Cutting Experiments
4.4. Obtaining Generative and Vegetative Progeny of Pear
4.5. GUS Histochemical and Fluorometric Assays
4.6. Statistical Analysis
5. Conclusions
Funding
Conflicts of Interest
References
- FAOSTAT. 2019. Available online: http://www.fao.org/faostat/ (accessed on 14 June 2019).
- Ahloowalia, B.S.; Maluszynski, M.; Nichterlein, K. Global impact of mutation-derived varieties. Euphytica 2004, 135, 187–204. [Google Scholar] [CrossRef]
- Mourgues, F.; Chevreau, E.; Lambert, C.; de Bondt, A. Efficient Agrobacterium-mediated transformation and recovery of transgenic plants from pear (Pyrus communis L.). Plant Cell Rep. 1996, 16, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Cheng, Z.-M.; Wayne, A. Sargent expression of the rolB gene enhances adventitious root formation in hardwood cuttings of aspen. In Vitro Cell. Dev. Biol. Plant 2004, 40, 366–370. [Google Scholar] [CrossRef]
- Cheng, Z.-M.; Dai, W.; Bosela, M.J.; Osburn, L.D. Genetic engineering approach to enhance adventitious root formation of hardwood cuttings. J. Crop Improv. 2006, 17, 211–225. [Google Scholar] [CrossRef]
- Nakhooda, M.; Jain, S.M. A review of Eucalyptus propagation and conservation. Propag. Ornam. Plants 2016, 16, 101–119. [Google Scholar]
- Fischer, C. Shortening of the juvenile period in apple breeding. In Developments in Plant Breeding: Progress in Temperate Fruit Breeding; Schmidt, H., Kellerhals, M., Eds.; Kluwer Academic Publishers: London, UK, 1994; pp. 161–164. [Google Scholar]
- Mudge, K.; Janick, J.; Scofield, S.; Goldschmidt, E.E. A history of grafting. Hortic. Rev. 2009, 35, 437–493. [Google Scholar] [CrossRef]
- Lemgo, G.; Sabbadini, S.; Pandolfini, T.; Mezzetti, B. Biosafety considerations of RNAi-mediated virus resistance in fruit-tree cultivars and in rootstock. Transgenic Res. 2013, 22, 1073–1088. [Google Scholar] [CrossRef]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Igarashi, M.; Ogasawara, H.; Hatsuyama, Y.; Saito, A.; Suzuki, M. Introduction of rolC into Marubakaidou [Malus prunifolia Borkh. var. ringo Asami Mo 84-A] apple rootstock via Agrobacterium tumefaciens. Plant Sci. 2002, 163, 463–473. [Google Scholar] [CrossRef]
- Necas, T.; Kosina, J. Vegetative propagation of pear and quince rootstocks using hardwood cuttings. Acta Hortic. 2008, 800, 701–706. [Google Scholar] [CrossRef]
- Da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef]
- Li, K.; Liang, Y.; Xing, L.; Mao, J.; Liu, Z.; Dong, F.; Meng, Y.; Han, M.; Zhao, C.; Bao, L.; et al. Transcriptome analysis reveals multiple hormones, wounding and sugar signaling pathways mediate adventitious root formation in apple rootstock. Int. J. Mol. Sci. 2018, 19, 2201. [Google Scholar] [CrossRef]
- Bell, R.L.; Scorza, R.; Srinivasan, C.; Webb, K. Transformation of ‘Beurre Bosc’ pear with the rolC gene. J. Am. Soc. Hortic. Sci. 1999, 124, 570–574. [Google Scholar] [CrossRef]
- Sedira, M.; Holefors, A.; Welander, M. Protocol for transformation of the apple rootstock Jork 9 with the rolB gene and its influence on rooting. Plant Cell Rep. 2001, 20, 517–524. [Google Scholar] [CrossRef]
- Vahdati, K.; McKenna, J.R.; Dandekar, A.M.; Leslie, C.A.; Uratsu, S.L.; Hackett, W.P.; Negri, P.; McGranahan, G.H. Rooting and other characteristics of a transgenic walnut hybrid (Juglans hindsii × J. regia) rootstock expressing rolABC. J. Am. Soc. Hortic. Sci. 2002, 127, 724–728. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, A.; Cong, Y.; Sheng, B.; Yao, Q.; Cheng, Z.-M. Agrobacterium-mediated transformation of the apple rootstock Malus micromalus makino with the rolC gene. In Vitro Cell. Dev. Biol. Plant 2006, 42, 491–497. [Google Scholar] [CrossRef]
- Druege, U.; Hilo, A.; Pérez-Pérez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef]
- Ladics, G.S.; Bartholomaeus, A.; Bregitzer, P.; Doerrer, N.G.; Gray, A.; Holzhauser, T.; Jordan, M.; Keese, P.; Kok, E.; Macdonald, P.; et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 2015, 24, 587–603. [Google Scholar] [CrossRef] [Green Version]
- Pons, E.; Peris, J.; Peña, L. Field performance of transgenic citrus trees: Assessment of the long-term expression of uidA and nptII transgenes and its impact on relevant agronomic and phenotypic characteristics. BMC Biotechnol. 2012, 12, 41. [Google Scholar] [CrossRef]
- Hawkins, S.; Leple, J.-C.; Cornu, D.; Jouanin, L.; Pilate, G. Stability of transgene expression in poplar: A model forest tree species. Ann. For. Sci. 2003, 60, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Lebedev, V.G.; Dolgov, S.V. Stability of gus and bar gene expression in transgenic pear clonal rootstock plants during several years. Acta Hortic. 2008, 800, 373–382. [Google Scholar] [CrossRef]
- Borejsza-Wysocka, E.; Norelli, J.L.; Aldwinckle, H.S.; Malnoy, M. Stable expression and phenotypic impact of attacin E transgene in orchard grown apple trees over a 12 year period. BMC Biotechnol. 2010, 10, 41. [Google Scholar] [CrossRef]
- Li, J.; Brunner, A.M.; Meilan, R.; Strauss, S.H. Stability of transgenes in trees: Expression of two reporter genes in poplar over three filed seasons. Tree Physiol. 2009, 29, 299–312. [Google Scholar] [CrossRef]
- Sabbadini, S.; Ricci, A.; Limera, C.; Baldoni, D.; Capriotti, L.; Mezzetti, B. Factors affecting the regeneration, via organogenesis, and the selection of transgenic calli in the peach rootstock Hansen 536 (Prunus persica × Prunus amygdalus) to express an RNAi construct against PPV virus. Plants 2019, 8, 178. [Google Scholar] [CrossRef]
- Freiman, A.; Shlizerman, L.; Golobovitch, S.; Yablovitz, Z.; Korchinsky, R.; Cohen, Y.; Samach, A.; Chevreau, E.; Le Roux, P.-M.; Patocchi, A.; et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 2012, 235, 1239–1251. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Dolgov, S.V. The effect of selective agents and a plant intron on transformation efficiency and expression of heterologous genes in pear Pyrus communis L. Russ. J. Genet. 2000, 36, 650–655. [Google Scholar]
- Lebedev, V.G.; Skryabin, K.G.; Dolgov, S.V. Transgenic pear clonal rootstocks resistant to herbicide “Basta”. Acta Hortic. 2002, 596, 193–197. [Google Scholar] [CrossRef]
- Geiss, G.; Gutierrez, L.; Bellini, C. Adventitious root formation: New insights and perspectives. Ann. Plant Rev. 2009, 37, 127–156. [Google Scholar] [CrossRef]
- Legue, V.; Rigald, A.; Bhalerao, R.P. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant. 2014, 151, 192–198. [Google Scholar] [CrossRef]
- Ribeiro, C.L.; Silva, C.M.; Drost, D.R.; Novaes, E.; Novaes, R.D.B.; Dervinis, C.; Kirst, M. Integration of genetic, genomic and transcriptomic information identifies putative regulators of adventitious root formation in Populus. BMC Plant Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Wang, Z.; Hua, J.; Yin, Y.; Gu, C.; Yu, C.; Shi, Q.; Guo, J.; Xuan, L.; Yu, F. An Integrated transcriptome and proteome analysis reveals putative regulators of adventitious root formation in Taxodium ‘Zhongshanshan’. Int. J. Mol. Sci. 2019, 20, 1225. [Google Scholar] [CrossRef]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 2014, 78, 372–384. [Google Scholar] [CrossRef]
- Filipecki, M.; Malepszy, S. Unintended consequences of plant transformation: A molecular insight. J. Appl. Genet. 2006, 47, 277–286. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Schroder, R.; Hallett, I.C.; Cohen, D.; MacRae, E.A. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef]
- Pasonen, H.L.; Seppänen, S.K.; Degefu, Y.; Rytkönen, A.; von Weissenberg, K.; Pappinen, A. Field performance of chitinase transgenic silver birches (Betula pendula Roth): Resistance to fungal diseases. Theor. Appl. Genet. 2004, 109, 562–570. [Google Scholar] [CrossRef]
- Miki, B.; Abdeen, A.; Manabe, Y.; MacDonald, P. Selectable marker genes and unintended changes to the plant transcriptome. Plant Biotechnol. J. 2009, 7, 211–218. [Google Scholar] [CrossRef]
- Bairu, M.W.; Aremu, A.O.; Staden, J.V. Somaclonal variation in plants: Causes and detection methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Donovan, A.M.; Ridout, M.S.; James, D.J. Assessment of somaclonal variation in apple. II. Rooting ability and shoot proliferation in vitro. J. Hortic. Sci. 1994, 69, 115–122. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Li, X.-Y.; Ahlman, A.; Welander, M. The rooting ability of the dwarfing pear rootstock BP10030 (Pyrus communis) was significantly increased by introduction of the rolB gene. Plant Sci. 2003, 165, 829–835. [Google Scholar] [CrossRef]
- James, D.J.; Passey, A.J.; Barbara, D.J.; Bevan, M. Genetic-transformation of apple (Malus pumila Mill) using a disarmed Ti-binary vector. Plant Cell Rep. 1989, 7, 658–661. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, Y.; Sun, H.; Hammond, R.W.; Davis, R.E.; Xin, L. High-efficiency and stable genetic transformation of pear (Pyrus communis L.) leaf segments and regeneration of transgenic plants. Acta Physiol. Plant. 2011, 33, 383–390. [Google Scholar] [CrossRef]
- Narvaez, I.; Khayreddine, T.; Pliego, C.; Cerezo, S.; Jiménez-Díaz, R.M.; Trapero-Casas, J.L.; López-Herrera, C.; Arjona-Girona, I.; Martín, C.; Mercado, J.A.; et al. Usage of the heterologous expression of the antimicrobial gene afp from Aspergillus giganteus for increasing fungal resistance in olive. Front. Plant Sci. 2018, 9, 680. [Google Scholar] [CrossRef]
- Brodeur-Campbell, S.E.; Vucetich, J.A.; Richter, D.L.; Waite, T.A.; Rosemier, J.; Tsai, C.J. Insect herbivory on low-lignin transgenic aspen. Environ. Entomol. 2006, 35, 1696–1701. [Google Scholar] [CrossRef]
- Axelsson, E.P.; Hjältén, J.; Whitham, T.G.; Julkunen-Tiitto, R.; Pilate, G.; Wennström, A. Leaf ontogeny interacts with Bt-modification to affect innate resistance in GM aspens. Chemoecology 2011, 21, 161–169. [Google Scholar] [CrossRef]
- Trueman, S.J.; McMahon, T.V.; Bristow, M. Production of cuttings in response to stock plant temperature in the subtropical eucalypts, Corymbia citriodora and Eucalyptus dunnii. New For. 2013, 44, 265–279. [Google Scholar] [CrossRef]
- Dole, R.; Hoerling, M.; Perlwitz, J.; Eischeid, J.; Pegion, P.; Zhang, T.; Quan, X.-W.; Xu, T.; Murray, D. Was there a basis for anticipating the 2010 Russian heat wave? Geophys. Res. Lett. 2011, 38, L06702. [Google Scholar] [CrossRef]
- Naidu, R.D.; Jones, N.B. The effect of cutting length on the rooting and growth of subtropical Eucalyptus hybrid clones in South Africa. South. For. 2009, 71, 297–301. [Google Scholar] [CrossRef]
- OuYang, F.; Wang, J.; Li, Y. Effects of cutting size and exogenous hormone treatment on rooting of shoot cuttings in Norway spruce [Picea abies (L.) Karst.]. New For. 2015, 46, 91–105. [Google Scholar] [CrossRef]
- Hartmann, H.T.; Kester, D.E.; Davies, F.T.; Geneve, R.L. Hartmann and Kester’s Plant Propagation: Principles and Practices, 7th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2002. [Google Scholar]
- Exadaktylou, E.; Thomidis, T.; Grout, B.; Zakynthinos, G.; Tsipouridis, C. Methods to improve the rooting of hardwood cuttings of the ‘Gisela 5’ cherry rootstock. HortTechnology 2009, 19, 254–259. [Google Scholar] [CrossRef]
- Smolka, A.; Li, X.-Y.; Heikelt, C.; Welander, M.; Zhu, L.-H. Effects of transgenic rootstocks on growth and development of non-transgenic scion cultivars in apple. Transgenic Res. 2010, 19, 933–948. [Google Scholar] [CrossRef]
- Rugini, E.; Silvestri, C.; Cristofori, V.; Brunori, E.; Biasi, R. Ten years field trial observations of ri-TDNA cherry Colt rootstocks and their effect on grafted sweet cherry cv Lapins. Plant Cell Tissue Organ Cult. 2015, 123, 557–568. [Google Scholar] [CrossRef]
- Fladung, M.; Nowitzki, O.; Ziegenhagen, B.; Kumar, S. Vegetative and generative dispersal capacity of field released transgenic aspen trees. Trees 2003, 17, 412–416. [Google Scholar] [CrossRef]
- Agapito-Tenfen, S.Z.; Vilperte, V.; Benevenuto, R.F.; Rover, C.M.; Traavik, T.I.; Nodari, R.O. Effect of stacking insecticidal cry and herbicide tolerance epsps transgenes on transgenic maize proteome. BMC Plant Biol. 2014, 14, 346–364. [Google Scholar] [CrossRef]
- Cingel, A.; Savic, J.; Cosic, T.; Raspor, M.; Ghalawenji, N.; Smigocki, A.; Ninkovic, S. Phenotypic performance of transgenic potato (Solanum tuberosum L.) plants with pyramided rice cystatin genes (OCI and OCII). Arch. Biol. Sci. 2015, 67, 957–964. [Google Scholar] [CrossRef]
- Maghuly, F.; da Camara Machado, A.; Leopold, S.; Khan, M.A.; Katinger, H.; Laimer, M. Long-term stability of marker gene expression in Prunus subhirtella: A model fruit tree species. J. Biotechnol. 2007, 127, 310–321. [Google Scholar] [CrossRef]
- Lebedev, V.G. Fruiting of transgenic pear trees. In Proceedings of the International Conference Plant Cell Biology and Biotechnology, Minsk, Belarus, 13–15 February 2013; Belarusian State University: Minsk, Belarus, 2013; p. 233. (In Russian). [Google Scholar]
- Laxa, M. Intron-mediated enhancement: A tool for heterologous gene expression in plants? Front. Plant Sci. 2017, 7, 1977. [Google Scholar] [CrossRef]
- Clancy, M.; Hannah, L.C. Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol. 2002, 130, 918–929. [Google Scholar] [CrossRef]
- Rose, A.B.; Carter, A.; Korf, I.; Kojima, N. Intron sequences that stimulate gene expression in Arabidopsis. Plant Mol. Biol. 2016, 92, 337–346. [Google Scholar] [CrossRef]
- Feike, D.; Korolev, A.V.; Soumpourou, E.; Murakami, E.; Reid, D.; Breakspear, A.; Rogers, C.; Radutoiu, S.; Stougaard, J.; Harwood, W.A.; et al. Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Carter, N. Petition for Determination of Nonregulated Status: Arctic™ Apple (Malus × domestica) Events GD743 and GS784; United States Department of Agriculture—Animal and Plant Health Inspection Service: Riverdale, MD, USA, 2012; 163p. [Google Scholar]
- Meilan, R.; Han, K.-H.; Ma, C.; DiFazio, S.P.; Eaton, J.A.; Hoien, E.A.; Stanton, B.J.; Crockett, R.P.; Taylor, M.L.; James, R.R.; et al. The CP4 transgene provides high levels of tolerance to Roundup® herbicide in field-grown hybrid poplars. Can. J. For. Res. 2002, 32, 967–976. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Tränkner, C.; Hanke, M.-V. The MdTFL1 gene of apple (Malus × domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef]
- Flachowsky, H.; Le Roux, P.M.; Peil, A.; Patocchi, A.; Richter, K.; Hanke, M.V. Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol. 2011, 192, 364–377. [Google Scholar] [CrossRef]
- Quoirin, M.; Lepoivre, P. Improved media for in vitro culture of Prunus species. Acta Hortic. 1977, 78, 437–442. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay of tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Scott, R.; Draper, J.; Jefferson, R.; Dury, G.; Jacob, L. Analysis of gene organization and expression in plants. In Plant Genetic Transformation and Gene Expression: A Laboratory Manual; Draper, J., Scott, R., Armitage, P., Walden, R., Eds.; Blackwell Scientific Pubs.: Oxford, UK, 1988; pp. 263–339. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Genotype | Gene | Selective Agent | Rooting Rate, % | |
|---|---|---|---|---|
| sel. Agent − | sel. Agent + | |||
| GP217 | - | - | 47.2 ab | - |
| HB-1 | uidA-intron | Hy | 18.8 c | 26.0 b |
| HB-4 | uidA-intron | Hy | 66.8 a | 52.9 a |
| NII-1 | uidA | Km | 54.8 ab | 48.3 |
| NIII-2 | uidA | Km | 43.3 b | 36.7 |
| GP217 | - | - | 54.0 a | - |
| P-BK | bar | PPT | 17.7 b | 19.9 |
| P-BU | bar | PPT | 36.5 ab | 33.3 |
| T-BO | uidA + bar | PPT | 22.5 b | 23.1 |
| T2-BF | uidA + bar | PPT | 50.9 a | 37.5 |
| T2-ES | uidA + bar | PPT | 44.8 a | 31.9 |
| Genotype | Cutting Length, mm | Cutting Base Diameter, mm | Rooting, % | Number of Roots >2 mm per Cutting | Mean Length of Roots >2 mm, mm | Cuttings with New Shoots, % | New Shoot Height, mm |
|---|---|---|---|---|---|---|---|
| 2009 | |||||||
| GP217 | 132.4 a | 5.3 a | 29.4 c | 2.0 ab | 116.5 | 42.5 b | 45.3 |
| HB-1 | 123.0 ab | 4.7 b | 59.9 ab | 1.8 bc | 134.5 | 49.5 ab | 67.5 |
| HB-4 | 135.6 a | 4.7 b | 71.6 a | 2.2 a | 109.4 | 39.8 b | 56.2 |
| NII-1 | 121.0 ab | 4.4 b | 63.5 ab | 1.5 cd | 97.5 | 51.5 ab | 54.7 |
| NIII-2 | 125.0 a | 4.6 b | 59.7 ab | 2.1 ab | 126.4 | 70.3 a | 60.5 |
| P-BK | 123.0 ab | 4.4 b | 53.8 ab | 1.4 cd | 118.2 | 40.5 b | 61.5 |
| P-BU | 137.6 a | 3.8 d | 25.0 c | 0.9 e | 105.0 | 47.1 b | 58.5 |
| T-BO | 115.1 ab | 3.8 cd | 21.1 c | 1.1 de | 118.8 | 37.5 b | 44.0 |
| T2-BF | 106.2 b | 4.0 bc | 42.4 bc | 1.3 de | 94.0 | 43.6 b | 52.7 |
| T2-ES | 120.5 ab | 4.3 bc | 63.2 ab | 1.1 de | 127.0 | 56.3 ab | 42.5 |
| 2010 | |||||||
| GP217 | 111.2 a | 5.0 a | 34.4 b | 2.7 | 128.7 | 33.3 bc | 51.4 |
| HB-1 | 109.8 a | 4.6 ab | 67.7 a | 1.9 | 118.8 | 32.3 bc | 59.1 |
| HB-4 | 99.6 b | 4.6 ab | 74.0 a | 1.6 | 133.0 | 29.9 c | 49.5 |
| NII-1 | 100.2 b | 4.4 b | 84.4 a | 1.6 | 137.2 | 27.2 c | 36.3 |
| NIII-2 | 96.6 b | 4.3 b | 79.8 a | 1.7 | 116.5 | 39.8 bc | 54.3 |
| P-BK | 94.7 b | 4.2 b | 72.9 a | 1.7 | 144.6 | 51.4 ab | 53.8 |
| P-BU | 92.6 b | 4.3 b | 63.5 a | 1.7 | 139.5 | 59.1 a | 63.9 |
| T-BO | 109.2 a | 4.2 b | 79.8 a | 1.6 | 130.4 | 32.5 bc | 53.2 |
| T2-BF | 107.8 a | 4.6 ab | 81.3 a | 2.3 | 112.6 | 48.7 ab | 52.1 |
| T2-ES | 96.4 b | 4.4 b | 85.2 a | 2.2 | 119.6 | 37.3 bc | 46.7 |
| Genotype | Cutting Length, mm | Cutting Base Diameter, mm | Rooting, % | Number of Roots >2 mm per Cutting | Mean Length of Roots >2 mm, mm | Cuttings with New Shoots, % | New Shoot Height, mm |
|---|---|---|---|---|---|---|---|
| 2010 | |||||||
| GP217 | 120.1 bcd | 4.5 | 31.5 abc | 1.9 ab | 103.4 | 50.8 abc | 89.0 abc |
| HB-1 | 117.7 cd | 4.2 | 15.5 de | 1.2 c | 91.4 | 25.8 de | 72.3 cd |
| HB-4 | 124.7 bc | 4.7 | 36.0 ab | 2.1 a | 107.9 | 54.2 abc | 108.0 ab |
| NII-1 | 119.4 bcd | 4.2 | 19.5 cde | 1.4 bc | 78.9 | 30.8 de | 68.5 cd |
| NIII-2 | 129.7 ab | 4.6 | 44.0 a | 1.9 ab | 102.1 | 39.8 cd | 85.6 abc |
| P-BK | 111.5 d | 4.6 | 27.5 bc | 1.6 abc | 111.9 | 60.0 ab | 96.7 abc |
| P-BU | 127.9 abc | 4.5 | 35.5 ab | 1.6 abc | 105.9 | 64.8 a | 109.7 a |
| T-BO | 121.8 bcd | 4.7 | 24.0 bcd | 1.5 abc | 108.2 | 47.9 bc | 80.1 bcd |
| T2-BF | 135.8 a | 4.5 | 26.0 bcd | 1.8 ab | 105.0 | 63.5 a | 92.3 abc |
| T2-ES | 117.5 cd | 4.0 | 11.5 e | 1.2 c | 80.8 | 21.7 e | 58.0 d |
| 2011 | |||||||
| GP217 | 126.5 cde | 4.7 | 23.0 bc | 1.9 bcd | - | 21.7 | 57.0 bcd |
| HB-1 | 138.8 abc | 4.5 | 26.0 abc | 2.4 ab | - | 19.2 | 84.0 a |
| HB-4 | 143.3 ab | 4.2 | 32.0 ab | 1.6 bcd | - | 18.8 | 53.0 bcde |
| NII-1 | 126.0 de | 4.2 | 22.0 bc | 1.4 d | - | 13.6 | 71.3 ab |
| NIII-2 | 146.3 a | 4.9 | 36.0 a | 2.9 a | - | 16.7 | 59.5 bcd |
| P-BK | 117.0 e | 4.6 | 21.0 bc | 1.8 bcd | - | 33.3 | 53.3 bcde |
| P-BU | 132.6 bcd | 4.7 | 24.0 abc | 2.2 abc | - | 29.2 | 65.6 abc |
| T-BO | 126.7 de | 4.4 | 29.0 ab | 1.5 cd | - | 27.6 | 48.9 cde |
| T2-BF | 124.9 de | 4.3 | 17.0 c | 1.5 cd | - | 11.8 | 38.5 e |
| T2-ES | 119.4 de | 4.4 | 28.0 abc | 1.8 bcd | - | 21.4 | 44.8 de |
| 2012 | |||||||
| GP217 | 130.1 ab | 4.2 | 45.0 bc | 1.4 cd | - | 38.5 | 6.1 |
| HB-1 | 138.4 a | 4.1 | 51.0 ab | 1.2 d | - | 45.2 | 4.5 |
| HB-4 | 142.8 a | 4.2 | 65.0 a | 1.8 ab | - | 58.7 | 7.5 |
| NII-1 | 130.7 ab | 3.9 | 34.0 bc | 1.2 d | - | 28.0 | 6.0 |
| NIII-2 | 135.8 a | 4.2 | 31.0 c | 2.0 a | - | 29.0 | 5.4 |
| P-BK | 131.0 ab | 4.1 | 53.0 ab | 1.5 bcd | - | 26.0 | 5.3 |
| P-BU | 142.5 a | 4.2 | 43.0 bc | 1.7 abc | - | 41.5 | 9.6 |
| T-BO | 122.3 b | 4.0 | 49.0 abc | 1.4 cd | - | 39.8 | 6.4 |
| T2-BF | 132.0 ab | 4.5 | 43.0 bc | 2.0 a | - | 40.0 | 6.6 |
| T2-ES | 134.4 ab | 4.3 | 54.0 ab | 1.4 cd | - | 37.1 | 5.7 |
| Gene | Line | Harvest of 2009 in the Greenhouse (2010) | Harvest of 2007 in the Field (2010) | Harvest of 2008 in the Field (2011) |
|---|---|---|---|---|
| uidA-intron | HB-1 | 12.6 ± 3.1 | 50.1 ± 5.0 | |
| 11.6 ± 3.5 | 100.4 ± 12.0 | |||
| 10.5 ± 1.5 | 68.0 ± 4.1 | |||
| HA-2 | 5.1 ± 0.9 | |||
| 0.6 ± 0.2 | ||||
| 1.9 ± 0.5 | ||||
| HA-3 | 10.7 ± 1.3 | |||
| 16.8 ± 2.2 | ||||
| 1.0 ± 0.2 | ||||
| 13.1 ± 2.6 | ||||
| HA-4 | 6.9 ± 0.6 | 27.6 ± 5.5 | ||
| 7.8 ± 1.2 | 47.4 ± 3.8 | |||
| 10.7 ± 1.8 | 41.6 ± 5.0 | |||
| 8.0 ± 1.0 | ||||
| HA-5 | 12.6 ± 1.1 | |||
| 29.6 ± 4.4 | ||||
| 11.9 ± 1.5 | ||||
| HA-6 | 35.4 ± 5.1 | 2.2 ± 0.1 | ||
| 7.6 ± 1.5 | 2.2 ± 0.4 | |||
| 11.8 ± 2.5 | 36.0 ± 5.0 | |||
| 1.7 ± 0.2 | ||||
| uidA | NII-1 | 25.7 ± 4.2 | 1.3 ± 0.2 | 33.7 ± 7.4 |
| 7.3 ± 1.5 | 14.9 ± 3.3 | 41.0 ± 2.1 | ||
| 10.4 ± 2.5 | 2.4 ± 0.2 | 30.5 ± 2.7 | ||
| 2.0 ± 0.3 | ||||
| NII-2 | 3.9 ± 0.7 | |||
| 3.8 ± 0.3 | ||||
| 7.2 ± 1.7 | ||||
| NII-3 | 5.3 ± 1.1 | 13.5 ± 1.6 | ||
| 3.4 ± 0.8 | 30.6 ± 6.1 | |||
| 2.4 ± 0.4 | 25.0 ± 3.2 | |||
| NIII-2 | 1.6 ± 0.4 | 6.7 ± 1.1 | ||
| 1.3 ± 0.3 | 5.8 ± 1.6 | |||
| 4.7 ± 0.9 | 1.4 ± 0.3 | |||
| 5.5 ± 1.0 | ||||
| NIII-4 | 1.7 ± 0.4 | 5.9 ± 0.6 | ||
| 2.0 ± 0.6 | 1.4 ± 0.4 | |||
| 11.2 ± 2.1 | 21.2 ± 4.0 | |||
| 9.6 ± 1.1 | ||||
| NIII-5 | 0.7 ± 0.1 | 1.1 ± 0.2 | ||
| 7.9 ± 1.2 | 19.4 ± 2.1 | |||
| 2.7 ± 0.5 | 1.3 ± 0.2 | |||
| 0.8 ± 0.1 | ||||
| NIV-2 | 19.2 ± 4.0 | 1.7 ± 0.2 | ||
| 4.4 ± 0.8 | 2.5 ± 0.2 | |||
| 1.1 ± 0.2 | 1.3 ± 0.2 | |||
| 3.2 ± 0.5 |
| Line | Graftings from 2008 in the Field (2009) | Cuttings from 2009 in the Field | |
|---|---|---|---|
| 2010 | 2011 | ||
| HB-1 | 14.3 | 15.3 | 14.5 |
| 16.8 | 17.3 | 25.1 | |
| 20.1 | 13.6 | 46.6 | |
| HB-4 | 8.5 | 2.5 | 5.9 |
| 5.8 | 6.1 | 8.5 | |
| 4.3 | 6.1 | 11.0 | |
| NII-1 | 20.1 | 7.9 | 17.2 |
| 15.3 | 11.2 | 25.8 | |
| 14.5 | 12.6 | 12.8 | |
| NIII-2 | 8.8 | 3.5 | 11.0 |
| 7.6 | 3.1 | 12.4 | |
| 5.7 | 3.3 | 15.1 | |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedev, V. The Rooting of Stem Cuttings and the Stability of uidA Gene Expression in Generative and Vegetative Progeny of Transgenic Pear Rootstock in the Field. Plants 2019, 8, 291. https://doi.org/10.3390/plants8080291
Lebedev V. The Rooting of Stem Cuttings and the Stability of uidA Gene Expression in Generative and Vegetative Progeny of Transgenic Pear Rootstock in the Field. Plants. 2019; 8(8):291. https://doi.org/10.3390/plants8080291
Chicago/Turabian StyleLebedev, Vadim. 2019. "The Rooting of Stem Cuttings and the Stability of uidA Gene Expression in Generative and Vegetative Progeny of Transgenic Pear Rootstock in the Field" Plants 8, no. 8: 291. https://doi.org/10.3390/plants8080291
APA StyleLebedev, V. (2019). The Rooting of Stem Cuttings and the Stability of uidA Gene Expression in Generative and Vegetative Progeny of Transgenic Pear Rootstock in the Field. Plants, 8(8), 291. https://doi.org/10.3390/plants8080291




