The HKT Transporter Gene from Arabidopsis, AtHKT1;1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive
Abstract
1. Introduction
2. Results
2.1. Production of Transgenic Arabidopsis Expressing β-glucuronidase (GUS) Gene Driven by AtHKT1;1 Promoter
2.2. Histochemical Analysis of GUS Activity in Transgenic Lines
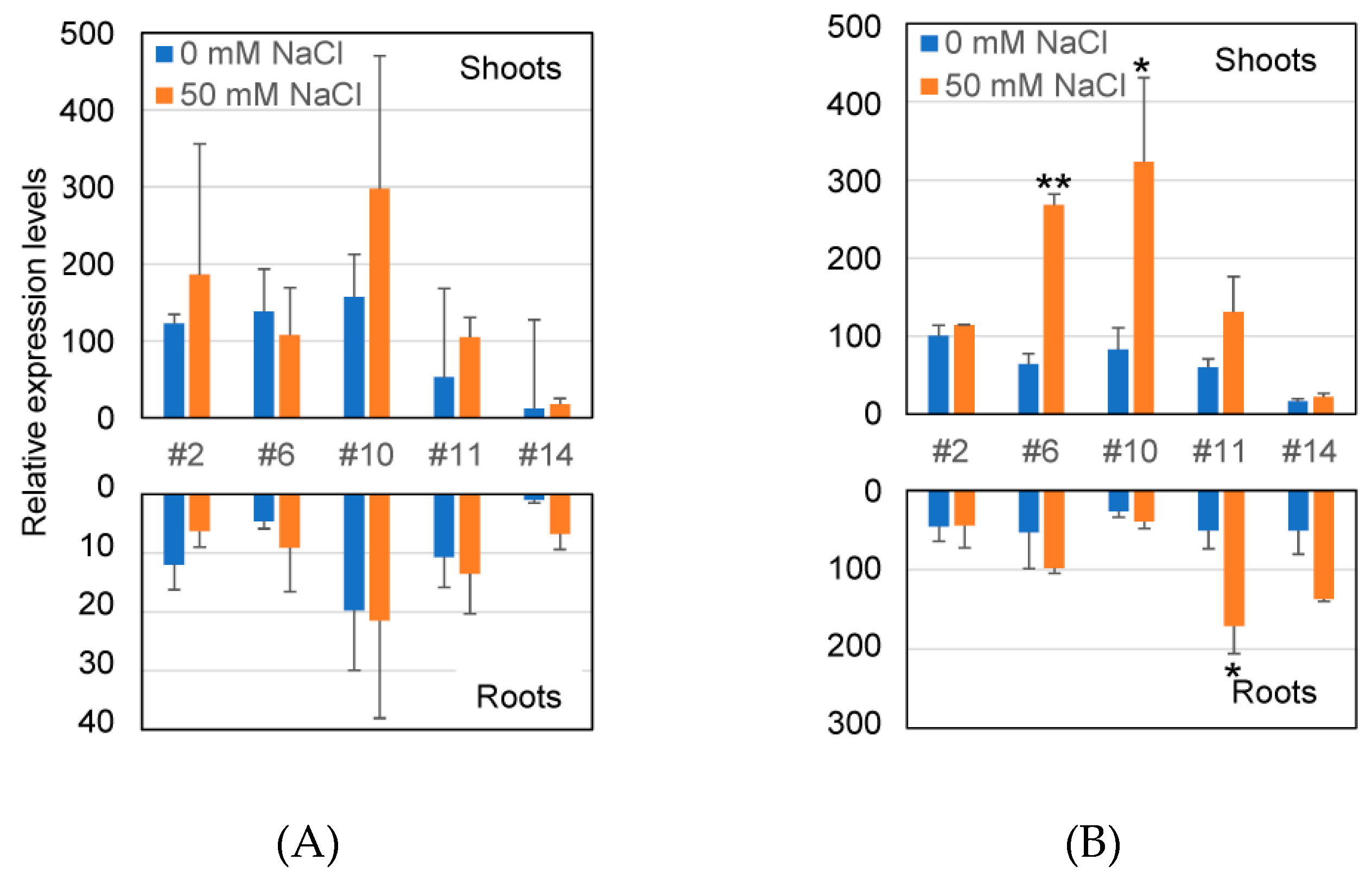
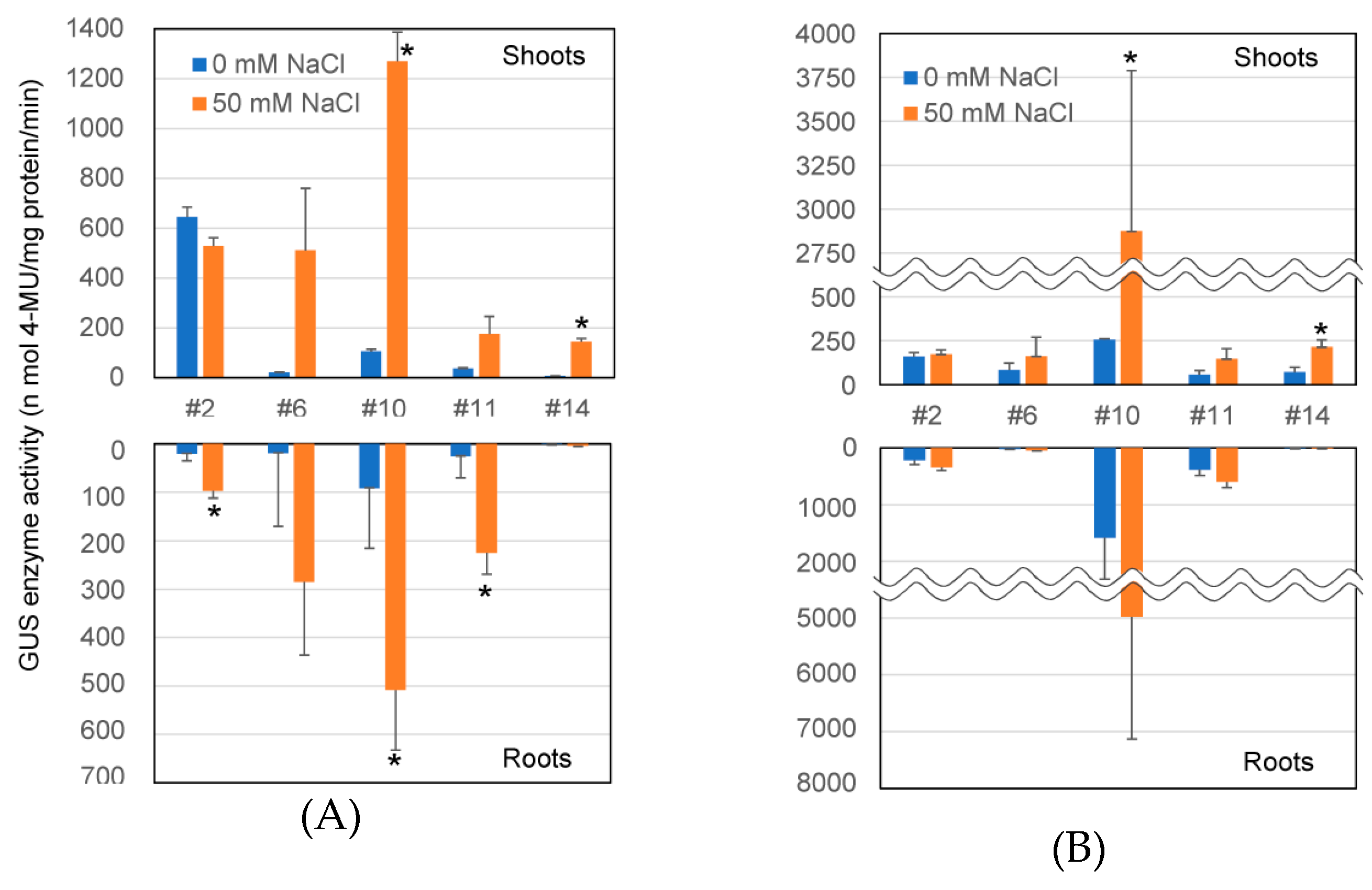
2.3. Transcriptional Level of GUS Gene and GUS Enzyme Activity in Transformants
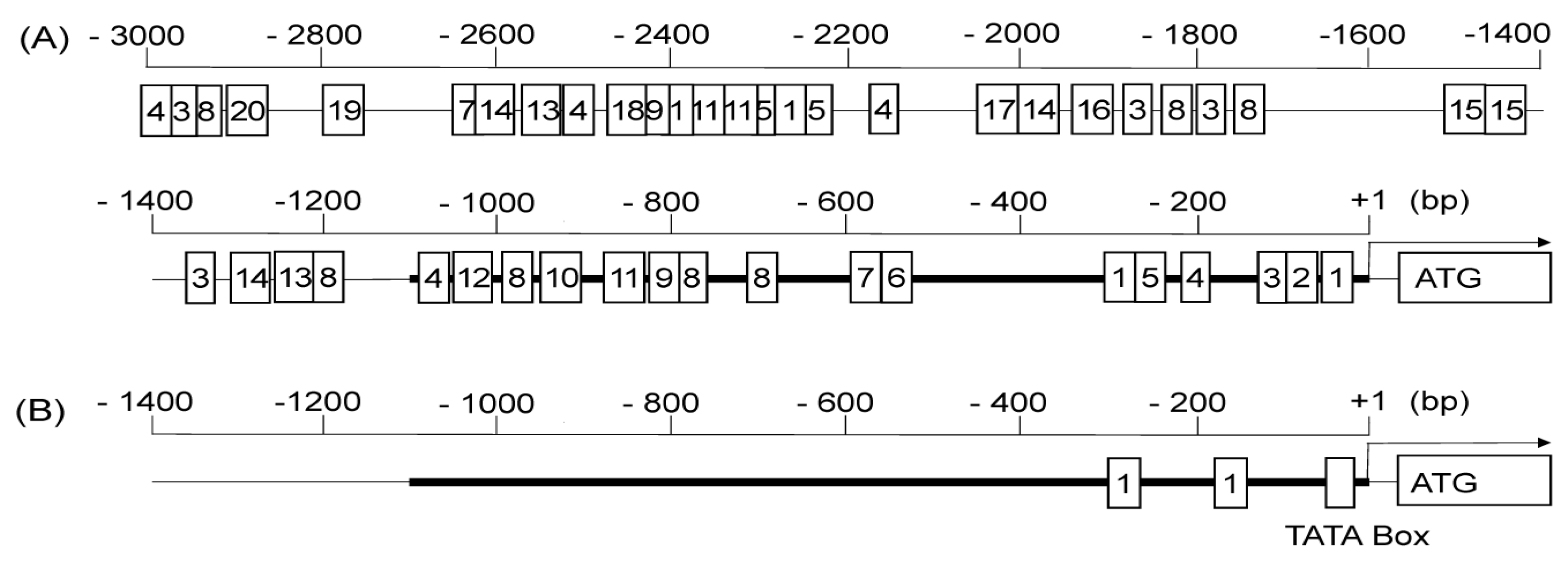
2.4. In Silico Analysis of Regulatory Motifs in the Promoter Sequence
3. Discussion
4. Materials and Methods
4.1. Production of Transgenic Arabidopsis
4.2. Plant-Growth Conditions
4.3. Real-Time qRT-PCR
4.4. GUS Assay
4.5. Promoter and Regulatory Element Resources
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Boyer, J.S. Plant Productivity and Environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Schroeder, J.I. Sodium Transporters in Plants. Diverse Genes and Physiological Functions. Plant Physiol. 2004, 136, 2457–2462. [Google Scholar] [PubMed]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.L.; Conejero, G. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009, 150, 1955–1971. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Davenport, R. Na+ Tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Yao, X.; Luo, W.; Jha, D.; Tester, M.; Horie, T.; Schroeder, J.I. AtHKT1;1 mediates nernstian sodium channel transport properties in Arabidopsis root stelar cells. PloS one 2011, 6, e24725. [Google Scholar] [CrossRef] [PubMed]
- Berthomieu, P.; Conéjéro, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Davenport, R.J.; MuÑOz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.N.A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Sunarpi; Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Chen, J.-G.; Gao, Y.-Q.; Li, X.; Chao, Z.-F.; Chen, Z.-R.; Li, Q.-Q.; Han, M.-L.; Wang, Y.-L.; Wang, Y.-F.; et al. AtHKT1 drives adaptation of Arabidopsis thaliana to salinity by reducing floral sodium content. PLoS Genet. 2017, 13, e1007086. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, C.; Wang, P.; Ma, Q.; Bao, A.-K.; Zhang, J.-L.; Wang, S.-M. The Effect of AtHKT1;1 or AtSOS1 mutation on the expressions of Na+ or K+ transporter genes and ion homeostasis in Arabidopsis thaliana under salt stress. Int. J. Mol. Sci. 2019, 20, 1085. [Google Scholar] [CrossRef] [PubMed]
- Davuluri, R.V.; Sun, H.; Palaniswamy, S.K.; Matthews, N.; Molina, C.; Kurtz, M.; Grotewold, E. AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform. 2003, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Hieno, A.; Naznin, H.A.; Hyakumachi, M.; Sakurai, T.; Tokizawa, M.; Koyama, H.; Sato, N.; Nishiyama, T.; Hasebe, M.; Zimmer, A.D.; et al. ppdb: Plant promoter database version 3.0. Nucleic Acids Res. 2014, 42, D1188–D1192. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Jiang, J.; Chung, J.-S.; Wang, B.; Chen, J.; Xin, Z.; Shi, H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2010, 52, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Endo, C.; Katsuhara, M.; Horie, T.; Shibasaka, M.; Nakahara, Y.; Kurusu, T. High-affinity K+ transporters from a halophyte, Sporobolus virginicus, mediate both K+ and Na+ transport in transgenic Arabidopsis, X. laevis oocytes and yeast. Plant Cell Physiol. 2019, 60, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
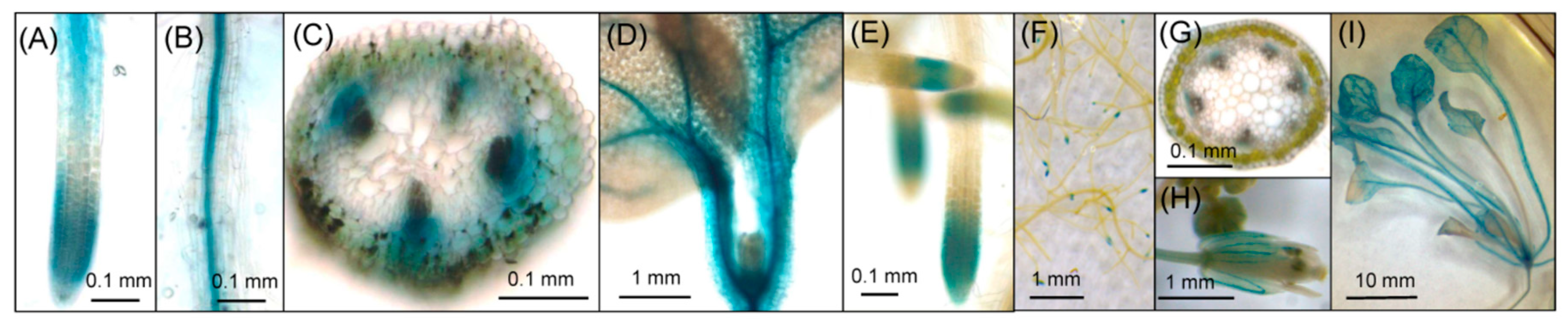
| Reproductive Growth Stage | Seedling Stage | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Plant # | Root Tips | Root Vascular Bundle | Hypocotyl Vascular Bundle | Leaf Vascular Bundle | Leaf Petiole Vascular Bundle | Root Tips | Root Vascular Bundle | Hypocotyl Vascular Bundle | Leaf Vascular Bundle | Leaf Petiole Vascular Bundle |
| A | 2, 3, 10, 13 | ++ | ++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| B | 4, 11 | ++ | − | − | + | + | +++ | +++ | −/+ | +++ | −/+++ |
| C | 1, 6, 7, 8, 9, 12 | + | + | + | +/++ | +++ | +++ | ++/+++ | +++ | +++ | +++ |
| D | 5, 14 | −/+ | −/+ | + | + | +/++ | + | −/+ | +++ | + | +++ |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tada, Y. The HKT Transporter Gene from Arabidopsis, AtHKT1;1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive. Plants 2019, 8, 204. https://doi.org/10.3390/plants8070204
Tada Y. The HKT Transporter Gene from Arabidopsis, AtHKT1;1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive. Plants. 2019; 8(7):204. https://doi.org/10.3390/plants8070204
Chicago/Turabian StyleTada, Yuichi. 2019. "The HKT Transporter Gene from Arabidopsis, AtHKT1;1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive" Plants 8, no. 7: 204. https://doi.org/10.3390/plants8070204
APA StyleTada, Y. (2019). The HKT Transporter Gene from Arabidopsis, AtHKT1;1, Is Dominantly Expressed in Shoot Vascular Tissue and Root Tips and Is Mild Salt Stress-Responsive. Plants, 8(7), 204. https://doi.org/10.3390/plants8070204





