Molecular Characterization of a New Virus Species Identified in Yam (Dioscorea spp.) by High-Throughput Sequencing
Abstract
1. Introduction
2. Results
2.1. High Throughput Sequencing (HTS) Analysis
2.2. Genome Organization
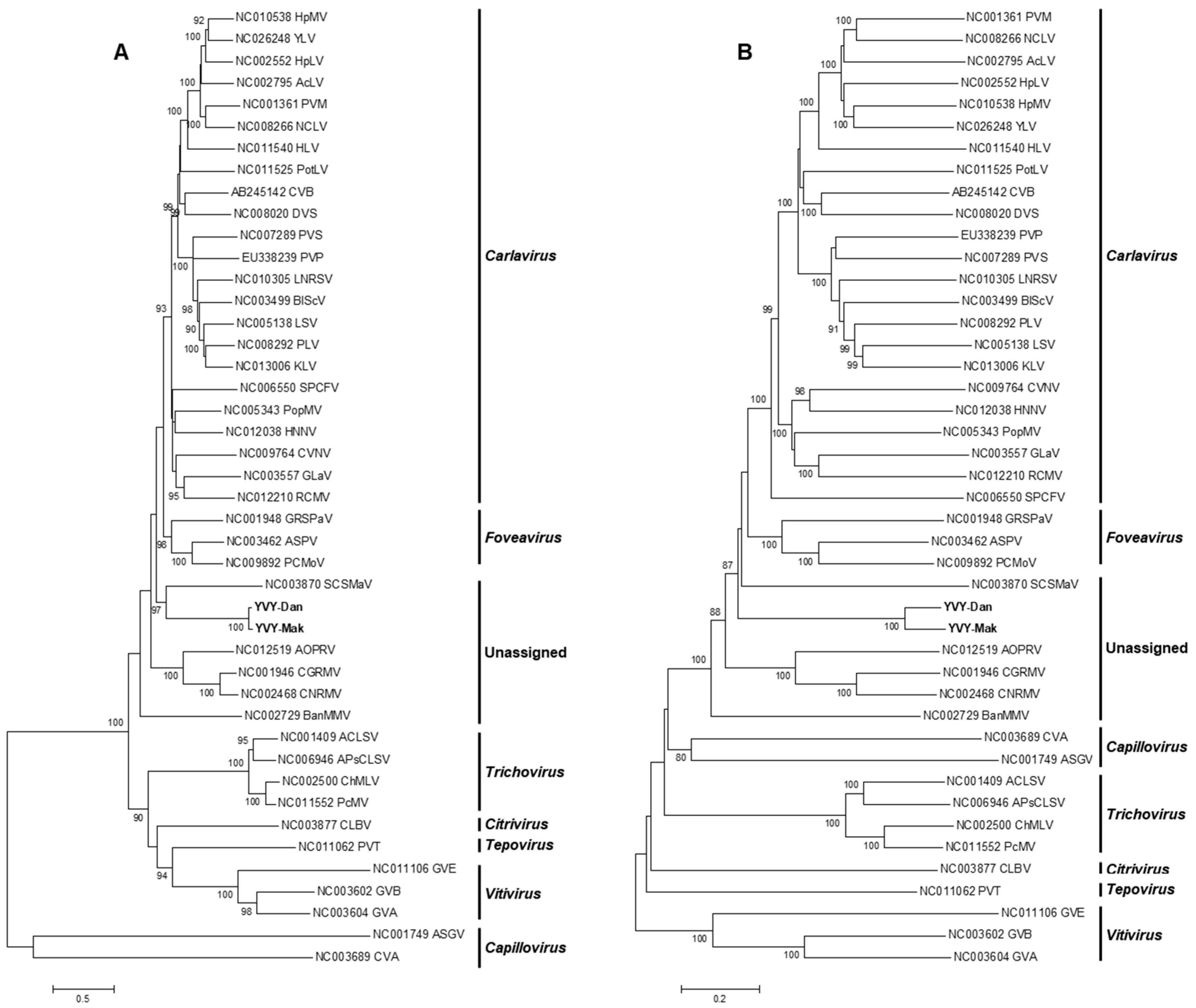
2.3. Phylogenetic Analysis
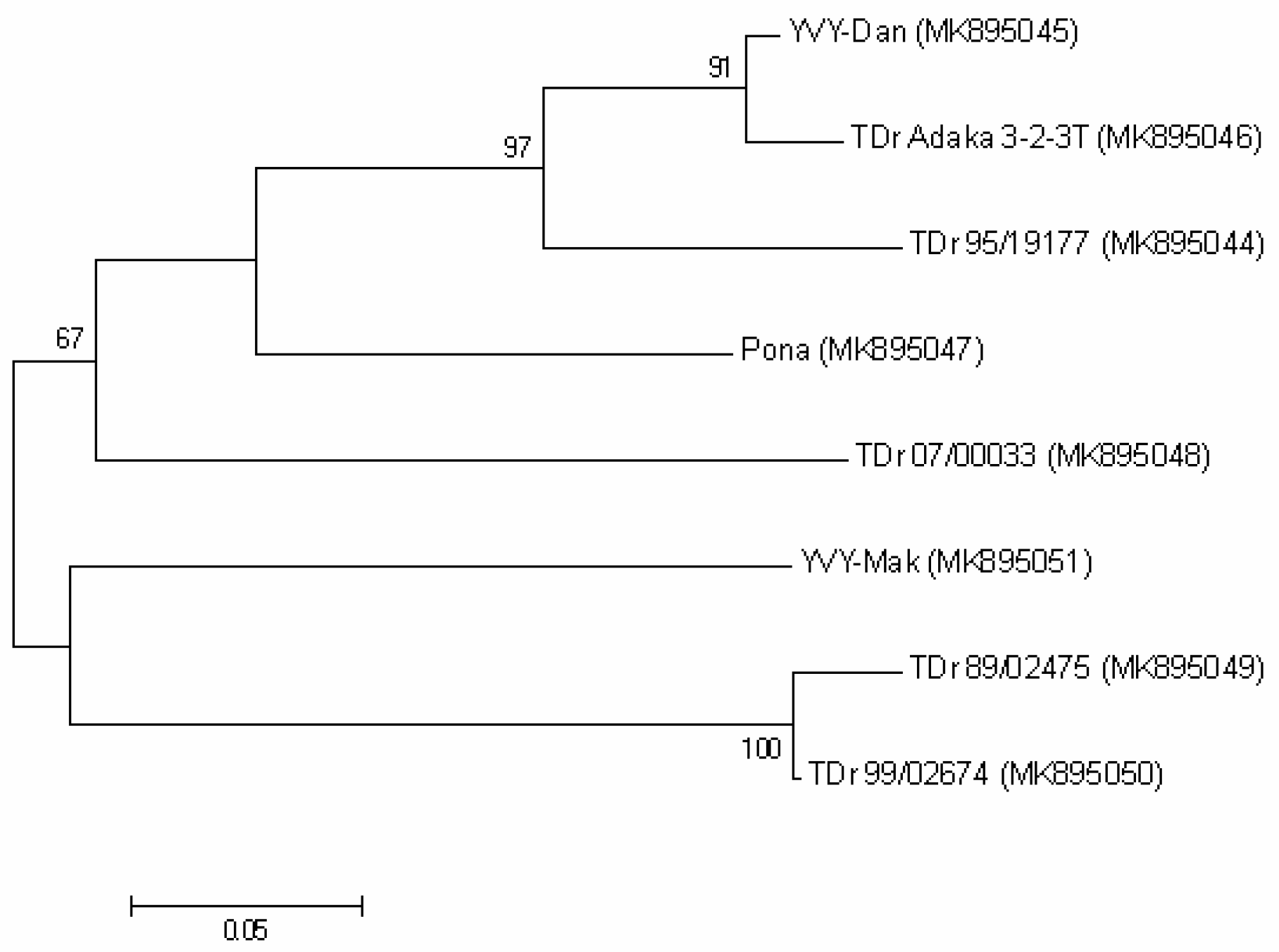
2.4. Prevalence and Diversity of the New Virus
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction and High Throughput Sequencing (HTS)
4.3. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.4. Phylogenetic and Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Asiedu, R.; Sartie, A. Crops that feed the World 1. Yams. Food Secur. 2010, 2, 305–315. [Google Scholar] [CrossRef]
- Balogun, M.O.; Maroya, N.; Asiedu, R. Status and prospects for improving yam seed systems using temporary immersion bioreactors. Afr. J. Biotechnol. 2014, 13, 1614–1622. [Google Scholar]
- Kenyon, L.; Shoyinka, S.A.; Hughes, J.; Odu, B.O. An overview of viruses infecting yams in Sub-Saharan Africa. In Proceedings of the 1st Symposium of Plant Virology for Sub-Saharan Africa (PVSSA), Ibadan, Nigeria, 4–8 June 2001; IITA: Ibadan, Nigeria. [Google Scholar]
- Mambole, I.A.; Bonheur, L.; Dumas, L.S.; Filloux, D.; Gomez, R.M.; Faure, C.; Lange, D.; Anzala, F.; Pavis, C.; Marais, A.; et al. Molecular characterization of yam virus X, a new potexvirus infecting yams (Dioscorea spp.) and evidence for the existence of at least three distinct potexviruses infecting yams. Arch. Virol. 2014, 159, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Menzel, W.; Thottappilly, G.; Winter, S. Characterization of an isometric virus isolated from yam (Dioscorea rotundata) in Nigeria suggests that it belongs to a new species in the genus Aureusvirus. Arch. Virol. 2014, 159, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.; Turaki, A.; Muller, E.; Kumar, P.L.; Kenyon, L.; Filloux, D.; Galzi, S.; Lopez-Montes, A.; Iskra-Caruana, M.L. The prevalence of badnaviruses in West African yams (Dioscorea cayenensis-rotundata) and evidence of endogenous pararetrovirus sequences in their genomes. Virus Res. 2014, 186, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Bömer, M.; Rathnayake, A.I.; Visendi, P.; Sewe, S.O.; Sicat, J.P.A.; Silva, G.; Kumar, P.L.; Seal, S.E. Tissue culture and next-generation sequencing: A combined approach for detecting yam (Dioscorea spp.) viruses. Physiol. Mol. Plant. Pathol. 2019, 105, 54–66. [Google Scholar]
- Bousalem, M.; Douzery, E.J.P.; Fargette, D. High genetic diversity, distant phylogenetic relationships and intraspecies recombination events among natural populations of Yam mosaic virus: A contribution to understanding potyvirus evolution. J. Gen. Virol. 2000, 81, 243–255. [Google Scholar] [CrossRef]
- Amusa, N.; Adigbite, A.; Muhammed, S.; Baiyewu, R. Yam diseases and its management in Nigeria. Afr. J. Biotechnol. 2003, 2, 497–502. [Google Scholar] [CrossRef]
- Silva, G.; Oyekanmi, J.; Nkere, C.K.; Bömer, M.; Kumar, P.L.; Seal, S.E. Rapid detection of potyviruses from crude plant extracts. Anal. Biochem. 2018, 546, 17–22. [Google Scholar] [CrossRef]
- Kenyon, L.; Lebas, B.S.M.; Seal, S.E. Yams (Dioscorea spp.) from the South Pacific Islands contain many novel badnaviruses: Implications for international movement of yam germplasm. Arch. Virol. 2008, 153, 877–889. [Google Scholar] [CrossRef]
- Aighewi, B.A.; Asiedu, R.; Maroya, N.; Balogun, M. Improved propagation methods to raise the productivity of yam (Dioscorea rotundata Poir). Food Secur. 2015, 7, 823–834. [Google Scholar] [CrossRef]
- Filloux, D.; Fernandez, E.; Loire, E.; Claude, L.; Galzi, S.; Candresse, T.; Winter, S.; Jeeva, M.L.; Makeshkumar, T.; Martin, D.P.; et al. Nanopore-based detection and characterization of yam viruses. Sci. Rep. 2018, 8, 17879. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, E.A.I.; Blawid, R.; de Melo, F.L.; Andrade, M.S.; Pio-Ribeiro, G.; de Andrade, G.P.; Nagata, T. Complete genome sequence of a putative new secovirus infecting yam (Dioscorea) plants. Arch. Virol. 2017, 162, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Meng, Y.; Shen, P.; Li, R.; Ma, Y.; Tan, S.; Chen, H.; Cao, M.; Li, F. Complete genome sequence of yam chlorotic necrosis virus, a novel macluravirus infecting yam. Arch. Virol. 2018, 163, 2275–2278. [Google Scholar] [CrossRef]
- Martelli, G.P.; Adams, M.J.; Kreuze, J.F.; Dolja, V.V. Family Flexiviridae: A Case Study in Virion and Genome Plasticity. Annu. Rev. Phytopathol. 2007, 45, 73–100. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; Giampetruzzi, A.; Laghezza, L.; Catalano, L.; Savino, V.N.; Saldarelli, P. Identification and characterization of an isolate of apple green crinkle associated virus involved in a severe disease of quince (Cydonia oblonga, Mill.). Arch. Virol. 2017, 162, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Candresse, T.; Hammond, J.; Kreuze, J.F.; Martelli, G.P.; Namba, S.; Pearson, M.N.; Ryu, K.H.; Saldarelli, P.; Yoshikawa, N. Family Betaflexiviridae. In Virus Taxonomy-Ninth Report on the International Committee on Taxonomy of Viruses; Elsevier Academic Press: Cambridge, MA, USA, 2012; pp. 920–941. [Google Scholar]
- Mumford, R.A.; Seal, S.E. Rapid single-tube immunocapture RT-PCR for the detection of two yam potyviruses. J. Virol. Methods 1997, 69, 73–79. [Google Scholar] [CrossRef]
- Finotello, F.; Lavezzo, E.; Bianco, L.; Barzon, L.; Mazzon, P.; Fontana, P.; Toppo, S.; Di Camillo, B. Reducing bias in RNA sequencing data: A novel approach to compute counts. BMC Bioinform. 2014, 15, S7. [Google Scholar] [CrossRef]
- Hily, J.M.; Candresse, T.; Garcia, S.; Vigne, E.; Tannière, M.; Komar, V.; Barnabé, G.; Alliaume, A.; Gilg, S.; Hommay, G.; et al. High-throughput sequencing and the viromic study of grapevine leaves: From the detection of grapevine-infecting viruses to the description of a new environmental Tymovirales member. Front. Microbiol. 2018, 9, 1782. [Google Scholar] [CrossRef] [PubMed]
- Beuve, M.; Hily, J.M.; Alliaume, A.; Reinbold, C.; Le Maguet, J.; Candresse, T.; Herrbach, E.; Lemaire, O. A complex virome unveiled by deep sequencing analysis of RNAs from a French Pinot Noir grapevine exhibiting strong leafroll symptoms. Arch. Virol. 2018, 163, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Maroya, N.; Asiedu, R. Yam Improvement for Income and Food Security in West Africa: Effectiveness of a Multi-Disciplinary and Multi-Institutional Team-Work. J. Root Crops 2014, 40, 85–92. [Google Scholar]
- Silva, G.; Bömer, M.; Nkere, C.; Kumar, P.L.; Seal, S.E. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J. Virol. Methods 2015, 222, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Nkere, C.K.; Oyekanmi, J.O.; Silva, G.; Bömer, M.; Atiri, G.I.; Onyeka, J.; Maroya, N.G.; Seal, S.E.; Kumar, P.L. Chromogenic detection of yam mosaic virus by closed-tube reverse transcription loop-mediated isothermal amplification (CT-RT-LAMP). Arch. Virol. 2018, 163, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Tamiru, M.; Natsume, S.; Takagi, H.; White, B.; Yaegashi, H.; Shimizu, M.; Yoshida, K.; Uemura, A.; Oikawa, K.; Abe, A.; et al. Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination. BMC Biol. 2017, 15, 1–20. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate DNA and RNA-seq aligner for long and short reads. BioRxiv 2018, 390013. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef]
- Abarshi, M.M.; Mohammed, I.U.; Wasswa, P.; Hillocks, R.J.; Holt, J.; Legg, J.P.; Seal, S.E.; Maruthi, M.N. Optimization of diagnostic RT-PCR protocols and sampling procedures for the reliable and cost-effective detection of Cassava brown streak virus. J. Virol. Methods 2010, 163, 353–359. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]


| ORFs 1 | Gene 2 | Length (nt/aa) | % Identity (nt/aa) YVY-Dan vs. YVY-Mak | |
|---|---|---|---|---|
| YVY-Dan | YVY-Mak | |||
| 1 | Replicase | 5451/1816 | 5454/1817 | 83/93 |
| 2 | TGB1 | 702/233 | 702/233 | 84/91 |
| 3 | TGB2 | 348/115 | 348/115 | 86/92 |
| 4 | TGB3 | 198/65 | 198/65 | 86/89 |
| 5 | CP | 711/236 | 711/236 | 85/94 |
| Virus | YMV + | YMV − | Total |
|---|---|---|---|
| YVY + | 23 (42%) | 8 (15%) | 31 |
| YVY − | 8 (15%) | 16 (29%) | 24 |
| Total | 31 | 24 | 55 |
| Name | Sequence (5′–3′) | Product Size (bp) | Location |
|---|---|---|---|
| YVY-RdRp1-PF | GTAATTGAAAATCACAGTGAGC | 790 | RdRp |
| YVY-RdRp1-PR | CTTCAAGTGCATAATTGTCTAT | ||
| YVY-CP-F | TTGATTAGTTAAGTATTTAGC | 788 | CP-3′UTR |
| YVY-CP-R | CCAGTTTTTCCTGCTGGCAAAC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, G.; Bömer, M.; Rathnayake, A.I.; Sewe, S.O.; Visendi, P.; Oyekanmi, J.O.; Quain, M.D.; Akomeah, B.; Kumar, P.L.; Seal, S.E. Molecular Characterization of a New Virus Species Identified in Yam (Dioscorea spp.) by High-Throughput Sequencing. Plants 2019, 8, 167. https://doi.org/10.3390/plants8060167
Silva G, Bömer M, Rathnayake AI, Sewe SO, Visendi P, Oyekanmi JO, Quain MD, Akomeah B, Kumar PL, Seal SE. Molecular Characterization of a New Virus Species Identified in Yam (Dioscorea spp.) by High-Throughput Sequencing. Plants. 2019; 8(6):167. https://doi.org/10.3390/plants8060167
Chicago/Turabian StyleSilva, Gonçalo, Moritz Bömer, Ajith I. Rathnayake, Steven O. Sewe, Paul Visendi, Joshua O. Oyekanmi, Marian D. Quain, Belinda Akomeah, P. Lava Kumar, and Susan E. Seal. 2019. "Molecular Characterization of a New Virus Species Identified in Yam (Dioscorea spp.) by High-Throughput Sequencing" Plants 8, no. 6: 167. https://doi.org/10.3390/plants8060167
APA StyleSilva, G., Bömer, M., Rathnayake, A. I., Sewe, S. O., Visendi, P., Oyekanmi, J. O., Quain, M. D., Akomeah, B., Kumar, P. L., & Seal, S. E. (2019). Molecular Characterization of a New Virus Species Identified in Yam (Dioscorea spp.) by High-Throughput Sequencing. Plants, 8(6), 167. https://doi.org/10.3390/plants8060167





