Ethylene Response of Plum ACC Synthase 1 (ACS1) Promoter is Mediated through the Binding Site of Abscisic Acid Insensitive 5 (ABI5)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
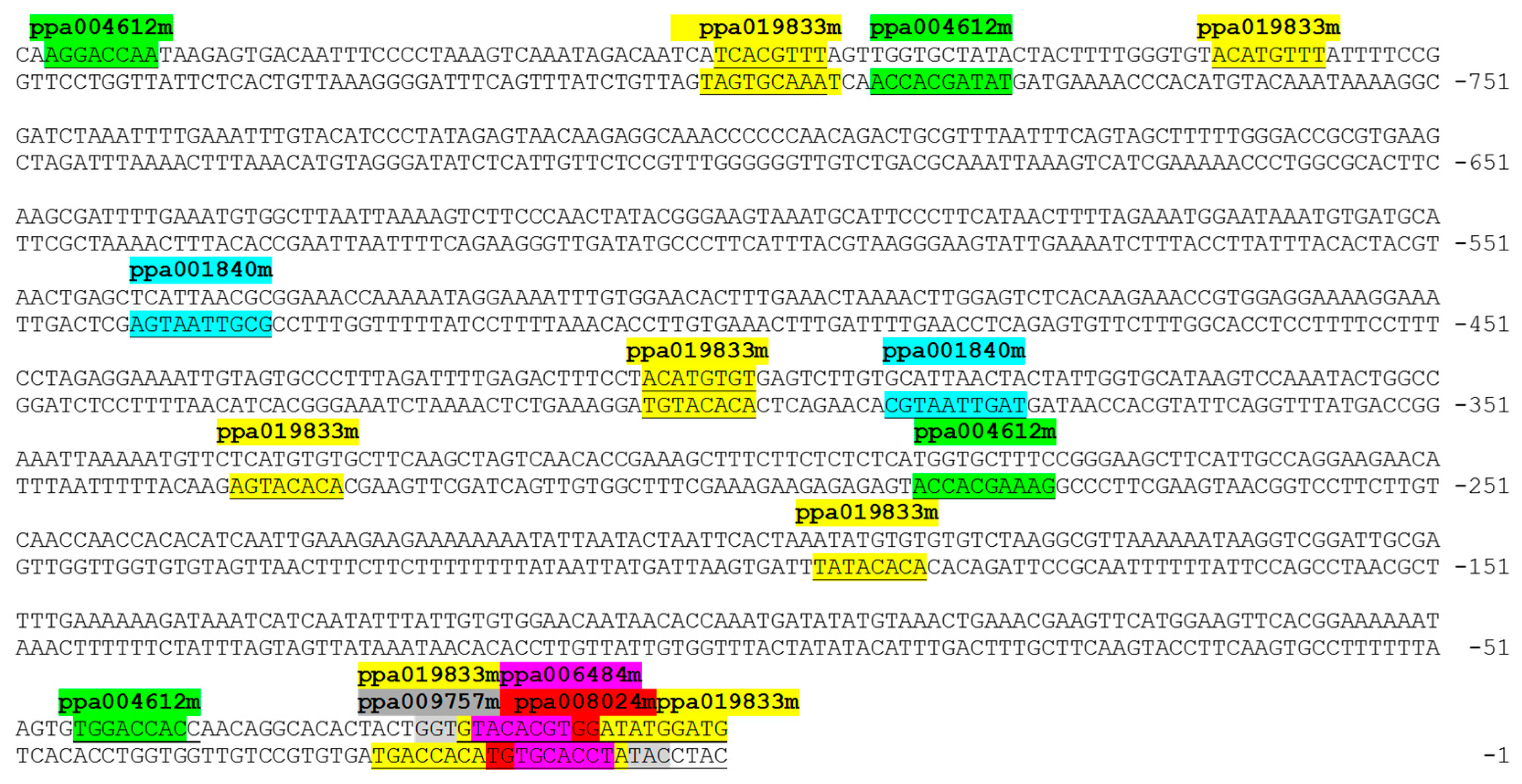
2.2. Isolation of ACS1 Promoter and Bioinformatics Analysis
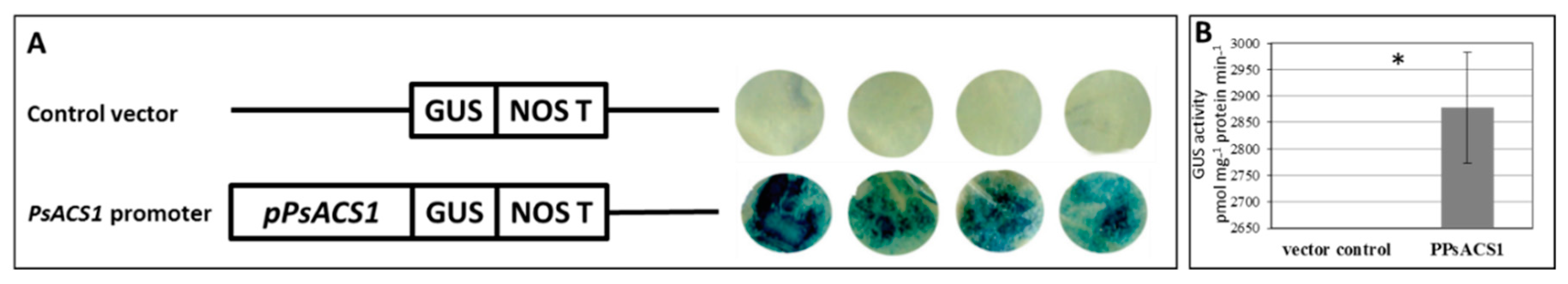
2.3. Promoter Activity Assays
2.4. Screening with Yeast One-Hybrid Assay
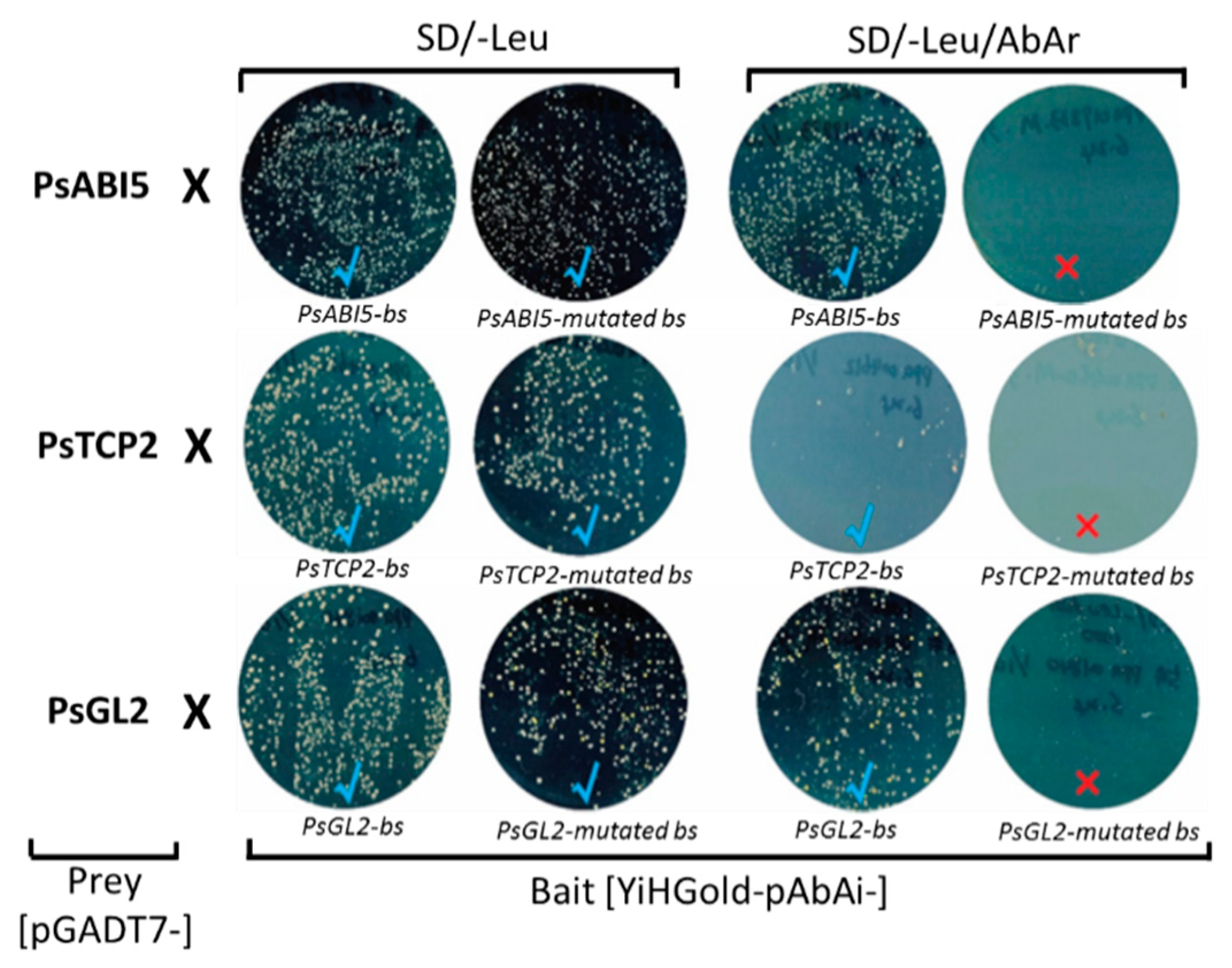
2.5. Verification of Positive Interactions using Yeast One-Hybrid Assay
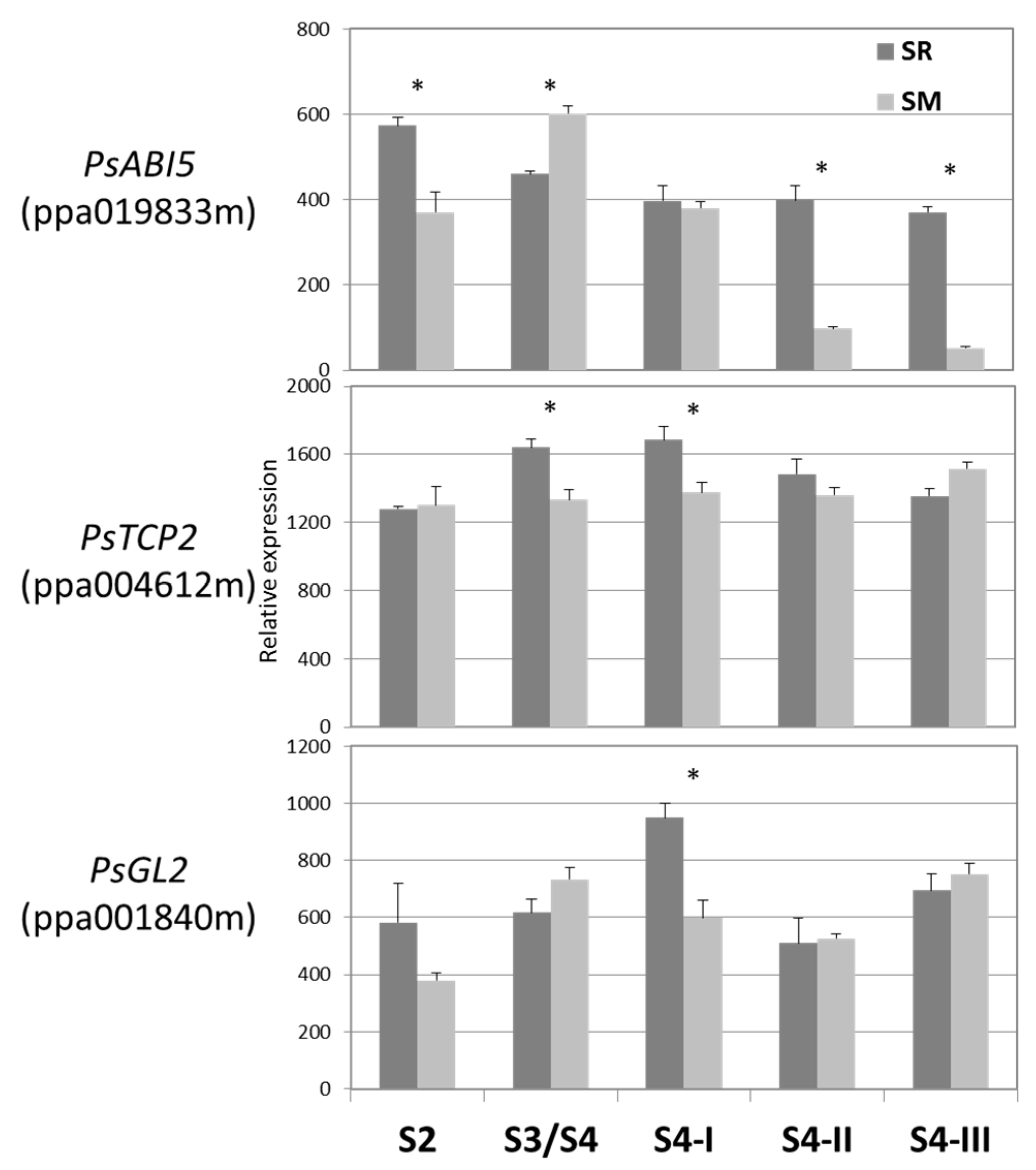
2.6. Real-Time RT-PCR and Phylogenic Analysis of Transcription Factors
2.7. Statistical Analyses
3. Results
3.1. Isolation and Prediction of ACS1 Promoter Regulatory Elements
3.2. pPsACS1 Basal Activity
3.3. Isolation of Transcription Factors Interacting with pPsACS1
3.4. Interaction between PsAB15, PsTCP2, and PsGL2 and their Corresponding Cis-Acting Elements
3.5. Relative Expression of the TFs during Japanese Plum Fruit Development
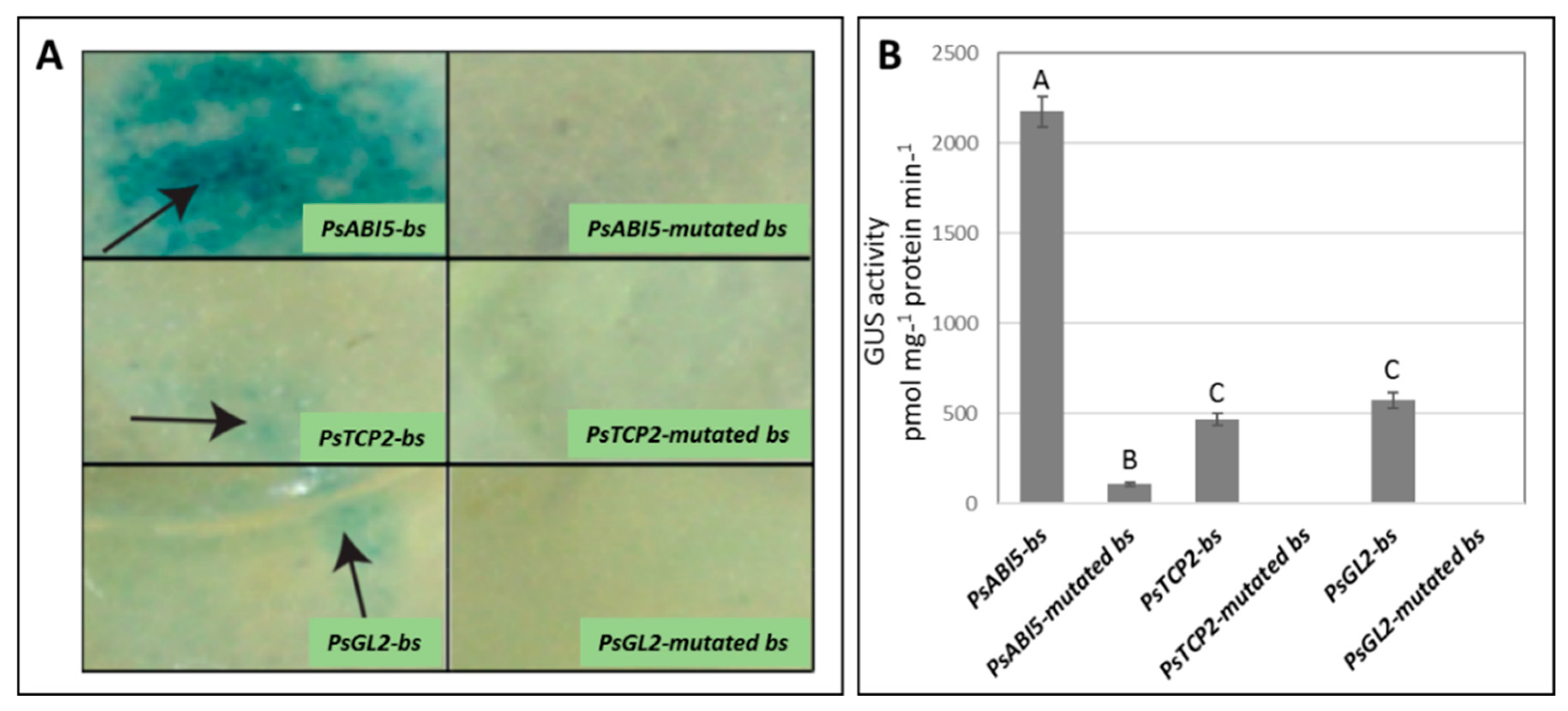
3.6. Basal in vivo Activity of pPsAB15, pPsTCP2, and pPsGL2 and their Corresponding Mutated Forms
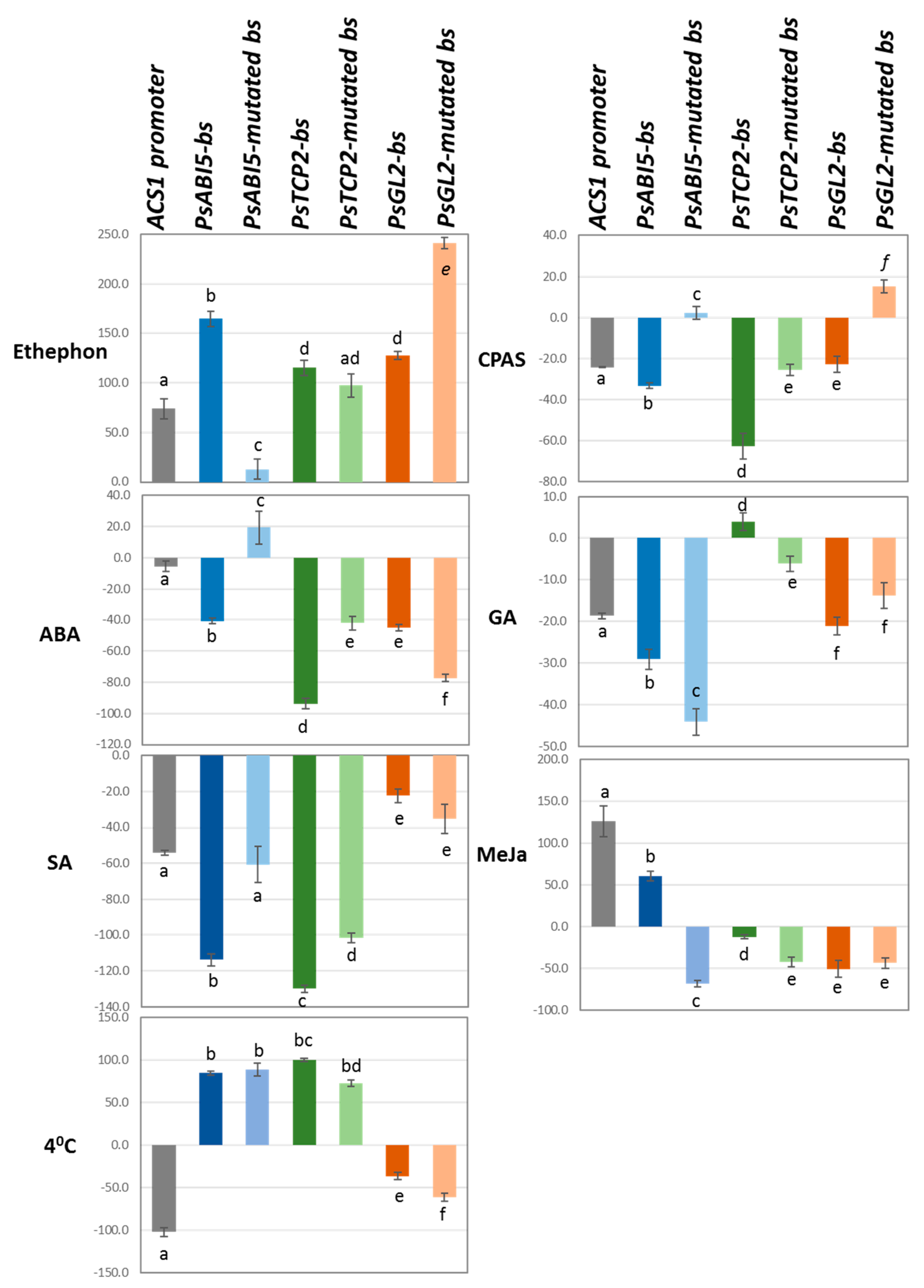
3.7. Effect of Hormone and other Treatments on GUS Activities Driven by pACS1, Synthetic Promoter Fragments, and their Mutated Forms
4. Discussion
4.1. ACS1 Promoter Activity in Response to Various Treatments
4.2. Activity of Synthetic Promoter Fragments and their Mutated Forms
4.3. Interactions among ABI5, Ethylene, and ABA
4.4. Binding of GL2 and TCP2 to the ACS1 Promoter
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seymour, G.B.; Ostergaard, L.; Chapman, N.H.; Knapp, S.; Martin, C. Fruit development and ripening. Ann. Rev. Plant Biol. 2013, 64, 219–241. [Google Scholar] [CrossRef] [PubMed]
- Biale, J.B. Growth, Maturation, and Senescence in Fruits: Recent knowledge on growth regulation and on biological oxidations has been applied to studies with fruits. Science 1964, 146, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.S.; Kieber, J.J. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 2005, 10, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [PubMed]
- Osorio, S.; Scossa, F.; Fernie, A.R. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef]
- Li, L.; Zhu, B.; Yang, P.; Fu, D.; Zhu, Y.; Luo, Y. The regulation mode of RIN transcription factor involved in ethylene biosynthesis in tomato fruit. J. Sci. Food Agric. 2011, 91, 1822–1828. [Google Scholar] [CrossRef]
- Li, L.; Zhu, B.; Fu, D.; Luo, Y. RIN transcription factor plays an important role in ethylene biosynthesis of tomato fruit ripening. J. Sci. Food Agric. 2011, 91, 2308–2314. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Han, B.Y.; Foo, C.S.; Wu, S.; Cyster, J.G. The C2H2-ZF transcription factor Zfp335 recognizes two consensus motifs using separate zinc finger arrays. Genes Dev. 2016, 30, 1509–1514. [Google Scholar] [CrossRef]
- Li, T.; Tan, D.; Liu, Z.; Jiang, Z.; Wei, Y.; Zhang, L.; Li, X.; Yuan, H.; Wang, A. Apple MdACS6 Regulates Ethylene Biosynthesis During Fruit Development Involving Ethylene-Responsive Factor. Plant Cell Physiol. 2015, 56, 1909–1917. [Google Scholar] [CrossRef]
- Yoon, G.M. New Insights into the Protein Turnover Regulation in Ethylene Biosynthesis. Mol. Cells 2015, 38, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Stone, S.L. Regulation of ethylene biosynthesis through protein degradation. Plant Signal. Behav. 2012, 7, 1438–1442. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Meng, X.; Wang, R.; Mao, G.; Han, L.; Liu, Y.; Zhang, S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. Plos Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. Comparative Analysis of DNA Methylation Reveals Specific Regulations on Ethylene Pathway in Tomato Fruit. Genes 2018, 9, 266. [Google Scholar] [CrossRef]
- Barry, C.S.; Llop-Tous, M.I.; Grierson, D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000, 123, 979–986. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Cavallaro, A.S.; Botella, J.R. Cloning and characterisation of ripening-induced ethylene biosynthetic genes from non-climacteric pineapple (Ananas comosus) fruits. Funct. Plant Biol. 1998, 25, 513–518. [Google Scholar] [CrossRef]
- Trebitsh, T.; Staub, J.E.; O’Neill, S.D. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiol. 1997, 113, 987–995. [Google Scholar] [CrossRef]
- Yamagami, T.; Tsuchisaka, A.; Yamada, K.; Haddon, W.F.; Harden, L.A.; Theologis, A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 2003, 278, 49102–49112. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
- Costa, F.; Stella, S.; Van de Weg, W.E.; Guerra, W.; Cecchinel, M.; Dallavia, J.; Koller, B.; Sansavini, S. Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica 2005, 141, 181–190. [Google Scholar] [CrossRef]
- Harada, T.; Sunako, T.; Wakasa, Y.; Soejima, J.; Satoh, T.; Niizeki, M. An allele of the 1-aminocyclopropane-1-carboxylate synthase gene (Md-ACS1) accounts for the low level of ethylene production in climacteric fruits of some apple cultivars. Theor. Appl. Genet. 2000, 101, 742–746. [Google Scholar] [CrossRef]
- Wakasa, Y.; Kudo, H.; Ishikawa, R.; Akada, S.; Senda, M.; Niizeki, M.; Harada, T. Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol. Technol. 2006, 39, 193–198. [Google Scholar] [CrossRef]
- Oraguzie, N.C.; Iwanami, H.; Soejima, J.; Harada, T.; Hall, A. Inheritance of the Md-ACS1 gene and its relationship to fruit softening in apple (Malus x domestica Borkh.). Theor. Appl. Genet. 2004, 108, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Sunako, T.; Sakuraba, W.; Senda, M.; Akada, S.; Ishikawa, R.; Niizeki, M.; Harada, T. An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol. 1999, 119, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Mathooko, F.M.; Tsunashima, Y.; Owino, W.Z.O.; Kubo, Y.; Inaba, A. Regulation of genes encoding ethylene biosynthetic enzymes in peach (Prunus persica L.) fruit by carbon dioxide and 1-methylcyclopropene. Postharvest Biol. Technol. 2001, 21, 265–281. [Google Scholar] [CrossRef]
- Tatsuki, M.; Haji, T.; Yamaguchi, M. The involvement of 1-aminocyclopropane-1-carboxylic acid synthase isogene, Pp-ACS1, in peach fruit softening. J. Exp. Bot. 2006, 57, 1281–1289. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Kim, W.S.; Jayasankar, S.; Svircev, A.M.; Brown, D.C. Differential regulation of four members of the ACC synthase gene family in plum. J. Exp. Bot. 2008, 59, 2009–2027. [Google Scholar] [CrossRef]
- Farcuh, M.; Li, B.; Rivero, R.M.; Shlizerman, L.; Sadka, A.; Blumwald, E. Sugar metabolism reprogramming in a non-climacteric bud mutant of a climacteric plum fruit during development on the tree. J. Exp. Bot. 2017, 68, 5813–5828. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Sherif, S.; Mila, I.; Bouzayen, M.; Jayasankar, S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J. Exp. Bot. 2009, 60, 907–922. [Google Scholar] [CrossRef]
- Kim, H.Y.; Farcuh, M.; Cohen, Y.; Crisosto, C.; Sadka, A.; Blumwald, E. Non-climacteric ripening and sorbitol homeostasis in plum fruits. Plant Sci. 2015, 231, 30–39. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Saha, P.; Farcuh, M.; Li, B.; Sadka, A.; Blumwald, E. RNA-Seq Analysis of Spatiotemporal Gene Expression Patterns During Fruit Development Revealed Reference Genes for Transcript Normalization in Plums. Plant Mol. Biol. Rep. 2015, 33, 1634–1649. [Google Scholar] [CrossRef]
- Minas, I.S.; Font, I.F.C.; Dangl, G.S.; Gradziel, T.M.; Dandekar, A.M.; Crisosto, C.H. Discovery of non-climacteric and suppressed climacteric bud sport mutations originating from a climacteric Japanese plum cultivar (Prunus salicina Lindl.). Front. Plant Sci. 2015, 6, 316. [Google Scholar] [CrossRef] [PubMed]
- Farcuh, M.; Rivero, R.M.; Sadka, A.; Blumwald, E. Ethylene regulation of sugar metabolism in climacteric and non-climacteric plums. Postharvest Biol. Technol. 2018, 139, 20–30. [Google Scholar] [CrossRef]
- Farcuh, M.; Toubiana, D.; Sade, N.; Rivero, R.M.; Doron-Faigenboim, A.; Nambara, E.; Sadka, A.; Blumwald, E. Hormone balance in a climacteric plum fruit and its non-climacteric bud mutant during ripening. Plant Sci. 2019, 280, 51–65. [Google Scholar] [CrossRef]
- Fernandez i Marti, A.; Saski, C.A.; Manganaris, G.A.; Gasic, K.; Crisosto, C.H. Genomic Sequencing of Japanese Plum (Prunus salicina Lindl.) Mutants Provides a New Model for Rosaceae Fruit Ripening Studies. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.; Ye, G.-N.; Weeden, N.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Qi, M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000, 22, 543–551. [Google Scholar] [CrossRef]
- Zhang, M.; Yuan, B.; Leng, P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 2009, 60, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Dubos, C.; Kelemen, Z.; Sebastian, A.; Bülow, L.; Huep, G.; Xu, W.; Grain, D.; Salsac, F.; Brousse, C.; Lepiniec, L.; et al. Integrating bioinformatic resources to predict transcription factors interacting with cis-sequences conserved in co-regulated genes. BMC Genom. 2014, 15, 317. [Google Scholar] [CrossRef] [PubMed]
- Huberman, M.; Riov, J.; Goldschmidt, E.E.; Apelbaum, A.; Goren, R. The novel ethylene antagonist, 3-cyclopropyl-1-enyl-propanoic acid sodium salt (CPAS), increases grain yield in wheat by delaying leaf senescence. Plant Growth Regul. 2014, 73, 249–255. [Google Scholar] [CrossRef]
- Wang, N.; Chen, H.; Nonaka, S.; Sato-Izawa, K.; Kusano, M.; Ezura, H. Ethylene biosynthesis controlled by NON-RIPENING: A regulatory conflict between wounding and ripening. Plant Physiol. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.N.; Chang, L.; Wu, S.A.; Li, P.L.; Liu, G.Q.; Wang, N.N. Auto-regulation of the promoter activities of Arabidopsis 1-aminocyclopropane-1-carboxylate synthase genes AtACS4, AtACS5, and AtACS7 in response to different plant hormones. Plant Sci. 2008, 175, 161–167. [Google Scholar] [CrossRef]
- Wang, N.N.; Shih, M.C.; Li, N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 2005, 56, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Alessio, V.M.; Cavacana, N.; Dantas, L.L.D.; Lee, N.; Hotta, C.T.; Imaizumi, T.; Menossi, M. The FBH family of bHLH transcription factors controls ACC synthase expression in sugarcane. J. Exp. Bot. 2018, 69, 2511–2525. [Google Scholar] [CrossRef]
- Yu, M.M.; Shen, L.; Fan, B.; Zhao, D.Y.; Zheng, Y.; Sheng, J.P. The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol. Technol. 2009, 54, 153–158. [Google Scholar] [CrossRef]
- Leng, P.; Yuan, B.; Guo, Y.D. The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 2014, 65, 4577–4588. [Google Scholar] [CrossRef]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2014, 65, 4561–4575. [Google Scholar] [CrossRef]
- Mou, W.; Li, D.; Bu, J.; Jiang, Y.; Khan, Z.U.; Luo, Z.; Mao, L.; Ying, T. Comprehensive Analysis of ABA Effects on Ethylene Biosynthesis and Signaling during Tomato Fruit Ripening. PloS ONE 2016, 11, e0154072. [Google Scholar] [CrossRef]
- Zaharah, S.S.; Singh, Z.; Symons, G.M.; Reid, J.B. Mode of action of abscisic acid in triggering ethylene biosynthesis and softening during ripening in mango fruit. Postharvest Biol. Technol. 2013, 75, 37–44. [Google Scholar] [CrossRef]
- Ampa, K.; Ikeura, H.; Saito, T.; Okawa, K.; Ohara, H.; Kondo, S. Effects of pre-harvest application of ethephon or abscisic acid on ‘Kohi’ kiwifruit (Actinidia chinensis) ripening on the vine. Sci. Hortic. 2016, 209, 255–260. [Google Scholar] [CrossRef]
- Zuzunaga, M.; Serrano, M.; Martinez-Romero, D.; Valero, D.; Riquelme, F. Comparative study of two plum (Prunus salicina Lindl.) cultivars during growth and ripening. Food Sci. Technol. Int. 2001, 7, 123–130. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant. Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- McCourt, P.; Creelman, R. The ABA receptors—We report you decide. Curr. Opin. Plant Biol. 2008, 11, 474–478. [Google Scholar] [CrossRef]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the Arabidopsis Abscisic Acid-Insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef]
- Bossi, F.; Cordoba, E.; Dupre, P.; Mendoza, M.S.; Roman, C.S.; Leon, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Tian, H.; Wang, S.; Chen, J.G. The Small Ethylene Response Factor ERF96 is Involved in the Regulation of the Abscisic Acid Response in Arabidopsis. Front. Plant Sci. 2015, 6, 1064. [Google Scholar] [CrossRef]
- Danisman, S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Lopez, J.A.; Sun, Y.; Blair, P.B.; Mukhtar, M.S. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 2015, 20, 238–245. [Google Scholar] [CrossRef]
- Song, C.B.; Shan, W.; Yang, Y.Y.; Tan, X.L.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J.; Kuang, J.F. Heterodimerization of MaTCP proteins modulates the transcription of MaXTH10/11 genes during banana fruit ripening. Bba-Gene Regul. Mech. 2018, 1861, 613–622. [Google Scholar] [CrossRef]
- Guo, Z.H.; Shu, W.S.; Cheng, H.Y.; Wang, G.M.; Qi, K.J.; Zhang, S.L.; Gu, C. Expression Analysis of TCP Genes in Peach Reveals an Involvement of PpTCP.A2 in Ethylene Biosynthesis During Fruit Ripening. Plant Mol. Biol. Rep. 2018, 36, 588–595. [Google Scholar] [CrossRef]
- Lin, Q.; Ohashi, Y.; Kato, M.; Tsuge, T.; Gu, H.Y.; Qu, L.J.; Aoyama, T. GLABRA2 Directly Suppresses Basic Helix-Loop-Helix Transcription Factor Genes with Diverse Functions in Root Hair Development. Plant Cell 2015, 27, 2894–2906. [Google Scholar] [CrossRef]
- Qing, L.; Aoyama, T. Pathways for Epidermal Cell Differentiation via the Homeobox Gene GLABRA2: Update on the Roles of the Classic Regulator. J. Integr. Plant Biol. 2012, 54, 729–737. [Google Scholar] [CrossRef]
| Expression library | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SR-S2 | SR-S4 | SM-S2 | SM-S4 | ||||||
| Number of Clones | % | Number of Clones | % | Number of Clones | % | Number of Clones | % | ||
| Total screened clones | 4,800,000 | 8,600,000 | 4,800,000 | 6,800,000 | |||||
| Positive clones | 151 | 196 | 160 | 165 | |||||
| Annotation of positive clones | Unknown | 41 | 27.2% | 46 | 23.5% | 39 | 24.4% | 53 | 32.1% |
| Known proteins of no apparent relevance | 64 | 42.4% | 117 | 59.7% | 92 | 57.5% | 86 | 52.1% | |
| Transcription factors | 33 | 21.9% | 17 | 8.7% | 11 | 6.9% | 8 | 4.8% | |
| DNA/RNA binding | 13 | 8.6% | 16 | 8.2% | 18 | 11.3% | 16 | 9.7% | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadka, A.; Qin, Q.; Feng, J.; Farcuh, M.; Shlizerman, L.; Zhang, Y.; Toubiana, D.; Blumwald, E. Ethylene Response of Plum ACC Synthase 1 (ACS1) Promoter is Mediated through the Binding Site of Abscisic Acid Insensitive 5 (ABI5) . Plants 2019, 8, 117. https://doi.org/10.3390/plants8050117
Sadka A, Qin Q, Feng J, Farcuh M, Shlizerman L, Zhang Y, Toubiana D, Blumwald E. Ethylene Response of Plum ACC Synthase 1 (ACS1) Promoter is Mediated through the Binding Site of Abscisic Acid Insensitive 5 (ABI5) . Plants. 2019; 8(5):117. https://doi.org/10.3390/plants8050117
Chicago/Turabian StyleSadka, Avi, Qiaoping Qin, Jianrong Feng, Macarena Farcuh, Lyudmila Shlizerman, Yunting Zhang, David Toubiana, and Eduardo Blumwald. 2019. "Ethylene Response of Plum ACC Synthase 1 (ACS1) Promoter is Mediated through the Binding Site of Abscisic Acid Insensitive 5 (ABI5) " Plants 8, no. 5: 117. https://doi.org/10.3390/plants8050117
APA StyleSadka, A., Qin, Q., Feng, J., Farcuh, M., Shlizerman, L., Zhang, Y., Toubiana, D., & Blumwald, E. (2019). Ethylene Response of Plum ACC Synthase 1 (ACS1) Promoter is Mediated through the Binding Site of Abscisic Acid Insensitive 5 (ABI5) . Plants, 8(5), 117. https://doi.org/10.3390/plants8050117






