Abstract
In the last decade, several studies have relied on a small number of plastid genomes to deduce deep phylogenetic relationships in the species-rich Myrtaceae. Nevertheless, the plastome of Rhodomyrtus tomentosa, an important representative plant of the Rhodomyrtus (DC.) genera, has not yet been reported yet. Here, we sequenced and analyzed the complete chloroplast (CP) genome of R. tomentosa, which is a 156,129-bp-long circular molecule with 37.1% GC content. This CP genome displays a typical quadripartite structure with two inverted repeats (IRa and IRb), of 25,824 bp each, that are separated by a small single copy region (SSC, 18,183 bp) and one large single copy region (LSC, 86,298 bp). The CP genome encodes 129 genes, including 84 protein-coding genes, 37 tRNA genes, eight rRNA genes and three pseudogenes (ycf1, rps19, ndhF). A considerable number of protein-coding genes have a universal ATG start codon, except for psbL and ndhD. Premature termination codons (PTCs) were found in one protein-coding gene, namely atpE, which is rarely reported in the CP genome of plants. Phylogenetic analysis revealed that R. tomentosa has a sister relationship with Eugenia uniflora and Psidium guajava. In conclusion, this study identified unique characteristics of the R. tomentosa CP genome providing valuable information for further investigations on species identification and the phylogenetic evolution between R. tomentosa and related species.
1. Introduction
The family Myrtaceae has over 3000 species distributed predominantly in tropical and subtropical regions of Australia and America [1]. Within this family, Rhodomyrtus tomentosa, an evergreen shrub of genera Rhodomyrtus, is commonly found in east and southeast Asia, including southern China, Japan, and Thailand [2]. R. tomentosa is an important plant used in traditional Chinese medicine and has a long history of clinical application. Its leaves, fruits and roots have all been used as alternative medicines, with different medicinal efficacies [3]. In addition, its fruits are one of the most popular foods in the wild. The vital medicinal and nutritional properties of R. tomentosa have drawn the attention of researchers in recent years [4,5]. The major chemical components of R. tomentosa include hydrolytic tannins, phloroglucin, flavonoids, and triterpenes [6], which possess antioxidant, anti-inflammatory, anti-tumor, antibacterial and other biological activities [7,8].
Eukaryotic cells possess large amounts of nuclear DNA, among which there are two organelles that carry independent genetic material, namely mitochondria and chloroplasts (CPs). CPs contain the enzymatic machinery necessary for photosynthesis and other important metabolism, and generally have small, highly conserved genomes [9]. The CP genome, also known as CP DNA, can be duplicated, transcribed and involved in expression [10]. Furthermore, compared to nuclear genomes and mitochondrial genomes, CP genomes are much smaller, and often have a conserved tetrad structure, which facilitates rapid and efficient sequencing and assembly. In addition, the nucleotide evolution rate of the chloroplast genome is moderate. This has allowed chloroplast genomes to be used for phylogenetic studies at different taxonomic levels [11]. The first chloroplast genome was successfully obtained from Nicotiana tabacum [12]. Subsequently, the CP genomes of several species, including Arabidopsis thaliana [13] and Panax ginseng [14], were determined using the first generation of sequencing technology, also called the Sanger method. With significant advances in sequencing technologies and bioinformatics, elucidation and characterization of CP genome has become more rapid and efficient and smoother than ever before [15].
CP genomes are circular DNA molecules generally ranging from 115 to 165 kb in length [16]. A pair of inverted repeat (IR) regions are among the largest components of the CP genome, which are reverse complement to another and are separated by a large-single-copy (LSC) region and a small-single-copy (SSC) region [17]. Two of the most important factors linked to size changes in the CP genome are the expansion and contraction of the IR regions [18].
Overall, literatures reports on complete CP genome sequences in the Myrtaceae are still very scarce, hindering phylogenetic analyses based on large-scale genomes. Therefore, it is particularly important to supplement the chloroplast genome information in the Myrtaceae for the phylogenetic purpose. Here, a comprehensive analysis of the complete CP genome of R. tomentosa was reported. A detailed map of the CP genome was constructed to help identify characteristics of the R. tomentosa CP genome, codon usage and RNA editing sites in R. tomentosa CP genome were analyzed to facilitate a better understanding of R. tomentosa CP genome. For the purpose of identifying conserved and unique features of CP genomes in different species, four species closely related to R. tomentosa were selected for comparison. Interestingly, one protein-coding gene, atpE was found to a have a premature termination codon (PTC). Although there are few studies related to PTCs in plant CP genomes, the discovery of PTC in atpE in R. tomentosa may provide a basis for further studies at the protein level through cloning and expression.
Phylogenetic trees were constructed based on the CP genomes of R. tomentosa along with several species within the same family and several unrelated species for the purpose of establishing the evolutionary position of R. tomentosa. Our results establish a better understanding of evolutionary history of the Myrtaceae clade and may accelerate phylogenic, population, and genetic engineering research on R. tomentosa.
2. Results and Discussion
2.1. Analysis and Discussion
2.1.1. Characters of Rhodomyrtus tomentosa Chloroplast (CP) Genome
The CP genome of R. tomentosa displays a typical circular double-chain structure, which is 156,129 bp in length. As shown in Figure 1 and Table 1, it is composed of four parts. A pair of inverted repeats (IRa and IRb) with lengths of 25,824 bp separates the large single-copy (LSC) region from the small single-copy (SSC) region, which have lengths of 86,298 bp and 18,183 bp, respectively. The overall length of the protein-coding sequence (CDS) is 76,113 bp. The total GC and AT contents of the R. tomentosa CP genome are 37.1% and 62.8%, respectively. Furthermore, the GC contents of the IR regions, the SSC region and the LSC region are 42.9%, 30.8% and 35.1%, respectively. Generally, the GC content of CP genomes ranges from 34% to 40%, but GC content is not evenly distributed in various regions of the genome. GC content in the IR region was significantly higher than that in the LSC and SSC region. The high GC content in the IR regions was due to the presence of four rRNA genes (rrn16, rrn23, rrn4.5 and rrn5). The content of GC in CDS (37.6%) is slightly higher than that of total GC content. In greater detail, the GC content for the first, second, and third codon positions are 44.9%, 37.3%, and 30.6%, respectively. Obviously, there is a bias toward a higher AT representation at the third codon position, a feature that was also found in other plant CP genomes [19,20].
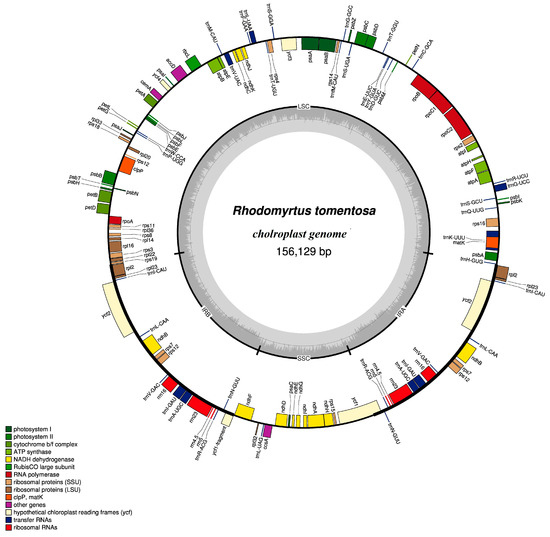
Figure 1.
Genome scheme of the Rhodomyrtus tomentosa chloroplast genome. Genes inside the circle are transcribed clockwise, while those outside are transcribed counterclockwise Filled colors represent different functional groups that specific genes fall into according to the legend on the bottom. Gray arrow represents gene direction. The darker gray color in the inner circle corresponds to GC (guanine and cytosine) content, whereas the lighter gray corresponds to AT (adenine and uracil) content.

Table 1.
Base composition in the chloroplast genome of R. tomentosa.
The CP genome of R. tomentosa encodes a total of 114 different genes (Table 2), of which 15 genes are duplicated in the IR regions. These 129 genes are comprised of 84 protein-coding genes, 37 tRNA genes and eight rRNA genes. Three pseudogenes (ycf1, rps19 and ndhF) are located around the IR-SSC, IR-LSC and SSC-IR boundaries, respectively. Four protein-coding genes, seven tRNA genes and four rRNA genes are duplicated in the IR regions. The coding regions constitute 56.7% of the genome, while the rest of the genome contains non-coding regions including introns, pseudogenes, and intergenic spacers.

Table 2.
Base composition in the chloroplast genome of R. tomentosa.
In the R.tomentosa CP genome, there are 18 genes containing introns (Table 3) that may participate in regulating gene expression and enhancing the expression of exogenous genes at specific sites and specific times in the plant [15]. Among those, six are tRNA genes and 12 are protein-coding genes. Most genes contain only one intron, while ycf3 and clpP contain two introns. The rps12 gene is unusual, containing one 5’ exon and two 3’ exons. The 5’ exon is located in the LSC region, while the 3’ exon is located in the IR regions, which is consistent with the CP genomes of Psidium guajava, Eugenia uniflora and Eucalyptus grandis [21]. The three pseudogenes which contain ycf1, rps19 and ndhF are located between IRB/SSC, IRA /LSC and SSC/IRA, respectively. Due to the inverse repeating nature of the IR regions, these three genes cannot be fully duplicated and lose the ability to encode complete proteins, which leads to their classification as pseudogene.

Table 3.
Base composition in the chloroplast genome of R. tomentosa.
2.1.2. Analysis of Premature Termination Codons in the R. tomentosa CP Genome
One protein-coding gene (atpE) with a premature termination codon (PTC) was identified during annotation. In order to validate this finding, raw (data) reads were used to conduct mapping on the spliced R. tomentosa sequences, followed by Integrative Genomics Viewer (IGV) visual processing to examine variable loci. The mapping rate of the aptE locus was found to be higher than 99%, suggesting that this locus was indeed variable and resulted in a PTC. PTCs lead to changes in protein coding. Because CP genomes are relatively conserved, especially within the same family, these plants from the Myrtaceae family were selected as control groups: Psidium guajava, Eugenia uniflora and Eucalyptus grandis. The atpE genes from these species were extracted using CLC Sequence Viewer (version 8) and then compared with that of R. tomentosa. The comparison results are shown in Figure 2. It can be seen that the premature termination of the atpE gene in R. tomentosa resulted in the absence of an amino acid compared to the three closely control species.
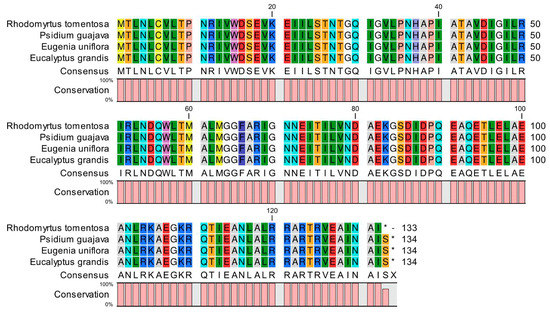
Figure 2.
Comparison of amino acid sequence of atpE in the Rhodomyrtus tomentosa chloroplast genome with those of three closely related species. * Indicates termination codons.
The atpE gene encodes a subunit of the chloroplast ATP synthase complex, which participates in photosynthetic phosphorylation necessary for plant growth [22]. As such, this gene is a critical component of the CP genome. Although literatures reports on PTC in genetics are common, few studies have identified PTCs in plant CP genomes. In genetics, nonsense point mutations often result in the production of nonfunctional proteins, assuming these proteins are properly transcribed and translated [23]. To be more exact, the effect of a nonsense mutation point relies on the proximity of the mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. Some genetic disorders such as thalassemia result from point-nonsense mutations [24,25,26].
With this in mind, the discovery of PTC in the R. tomentosa atpE gene may establish a foundation for further studies at the protein level through cloning and expression. Future work on CP transcription and translation are needed to verify the presence and functions of PTCs in atpE and potentially other genes.
2.1.3. Identification of Long Repeats (LRs) and Simple Sequence Repeats (SSRs)
Repetitive sequences in CP genome have been a major focus of research. There are an abundance of repeated sequences in the CP genome, which are distributed in intergenetic spacer and intron sequences [27]. Long repeats with length greater than 30 bp, might have functions in promoting chloroplast genome rearrangement and increasing population genetic diversity [28]. In order to verify the above-mentioned functions and obtain a comprehensive understanding of the long repeats within the R. tomentosa CP genome, the long repeats in CP genomes from four other species, Psidium guajava, Eugenia uniflora, Eucalyptus grandis and Melastoma candidum were selected for comparison according to the ties of consanguinity between species. These three species were used to compare and analyze the conserved and unique characteristics of chloroplast CP genomes between different genera of the same family. M. candidum, which belongs to another family within the Myrtiflorae order, is the most closely related among other species whose CP genome sequences are available from NCBI except for the three species of Myrtaceae family. Similarly, M. candidum was used to analyze differences between species in different families. The resulting data revealed the repeat structure of these four species, demonstrating that there are 38 (14 forward, 24 palindromic), 31 (14 forward, 15 palindromic, 2 reverse), 33 (16 forward, 16 palindromic, 1 reserve), 30 (16 forward, 14 palindromic) and 49 (22 forward, 19 palindromic, 4 reverse, 4 complement) large repeats (LRs) in R. tomentosa, Psidium guajava, Eugenia uniflora, Eucalyptus grandis and Melastoma candidum, respectively. In detail, there is no reverse or complement repeats in R. tomentosa, similar to Eucalyptus grandis. At the same time, complement repeats exists in M. candidum, which is not a member of the Myrtaceae family unlike the other four species. Thus, population genetic diversity is revealed by LRs differences, which is consistent with LRs functional analysis (Figure 3).
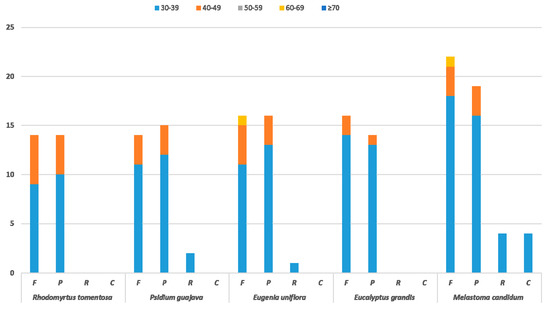
Figure 3.
Repeat sequences in four chloroplast genomes. F, P, R, and C indicates the repeat types: F (forward), P (palindrome), R (reverse), and C (complement). Repeats with different lengths are indicated in different colors.
Simple sequence repeats (SSRs) are composed of small repeated sequences of 1 to 6 bp, which are extensively distributed in intergenic regions, intron regions, and even protein-coding regions. High mutation rates in these regions also reflect the genetic diversity [29]. CP SSRs, which are widely used in phylogenetic and population genetic analyses [30], are important sources for developing molecular markers. A total of 282 SSRs were identified in the R. tomentosa CP genome and were summarized in Table 4, including 173 mononucleotide, 37 dinucleotide, 63 trinucleotide and nine tetranucleotide repeat units. In addition, 98.8% of the mononucleotide SSRs belongs to the A/T type, which is consistent with previous studies where proportions of polyadenine (polyA) and polythymine (polyT) were higher than those of polycytosine (polyC) and polyguanine (polyG) within CP SSRs in many plants [31].

Table 4.
Base composition in the chloroplast genome of R. tomentosa.
2.1.4. Codon Usage and RNA Editing Sites
In different organisms, synonymous codons occur at different frequencies—this is called preference [30,32]. As for highly expressed genes, the preference of codons is closely related to the abundance of tRNA. An improved understanding of preference of codons will facilitate further studies on the preference of base composition of DNA sequences, finding optimal codons, and designing expression vectors accordingly to improve the efficiency of protein synthesis [33].
In the R. tomentosa CP Genome, all the protein-coding genes were composed of 23,939 codons in sum, among which 2,724 codons (accounting for 11.38%) encode leucine and 286 (1.19%) encode cysteine, respectively. These represent the most and least universal amino acids, respectively, out of the 20 amino acids that can be used for protein biosynthesis by tRNA found in the R. tomentosa CP genome. The relative synonymous codon usage (RSCU) value (Figure 4 and Table S1) increases with the quantity of codons that encode for a specific amino acid. As illustrated, most of the amino acid codons, except for methionine and tryptophan, have preferences. This phenomenon was also found in the CP genomes of other species [10,34].
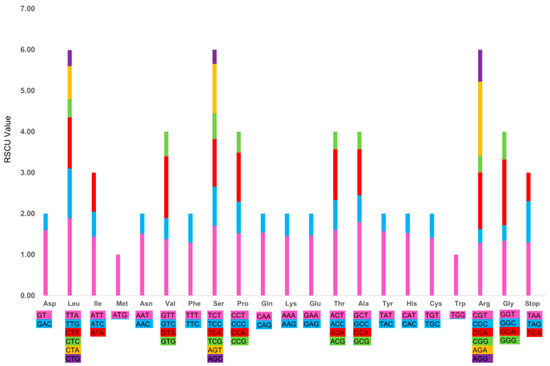
Figure 4.
Codon content of 20 amino acid and stop codons in all protein-coding genes of the Rhodomyrtus tomentosa chloroplast genome.
In addition, RNA editing is a very common phenomenon that occurs in plant CP genomes. The core functions of RNA editing include modifying mutations, correcting and regulating translation [35]. RNA editing sites in the R. tomentosa CP genome were predicted based on 35 genes by the predictive RNA editor for plants (PREP) program, among which, a total of 20 genes were analyzed and summarized in Table S2. In sum, 64 RNA editing sites were identified in the R. tomentosa CP genome, in which amino acid conversion from serine to leucine occurred most frequently, while threonine to methionine occurred least often.
2.1.5. Contraction and Expansion of IRs in the R. tomentosa CP Genome
As mentioned above, the typical quadripartite structure of the CP genome includes two different single-copy regions and two IR regions [36]. Although the inverted repeat regions (IRa and IRb) are the most conserved regions of the CP genome, contraction and expansion at the borders of the IR regions are hypothesized to explain size differences between CP genomes [37,38]. A comparison between R. tomentosa and four other closely related species may explain size differences between their respective CP genomes.
As presented in Figure 5, the IR/SSC and IR/LSC boundaries of R. tomentosa (MK_044696) were compared to those in Psidium guajava (NC_033355), Eugenia uniflora, (NC_027744), Eucalyptus grandis (HM_347959) and Melastoma candidum (NC_034716). The length of the IR regions in the five CP genomes showed a modest expansion, ranging from 25,824 to 26,390 bp. The IR regions expanded to partially include rps19, ycf1 and ndhF, correspondingly creating truncated ψrps19, ψycf1 and ψndhF copies at the junction of IRa/LSC and IRb/SSC and IRa/LSC, respectively. Long ycf1 pseudogene exists in all species, which has been used to analyze CP genome variation in plants [28,38]. Moreover, it has been reported that the rps19 gene is one of the most abundant transcripts in the CP genome, which exists in most species except for Eugenia uniflora and Eucalyptus grandis. The ndhF gene, related to photosynthesis, was found to be 67 bp, 112 bp, 209 bp away from the IRb/SSC border in R. tomentosa, P. guajava, Eugenia. uniflora, and Eucalyptus grandis, respectively. The trnH gene is present at the longest distance (32 bp) from the LSC edge in the R. tomentosa CP genome.

Figure 5.
Comparison of the borders of the LSC, SSC, and IR regions among five chloroplast genomes. Ψ: pseudogenes, /: distance from the edge.
2.1.6. Comparative CP Genomic Analysis
The whole CP genome sequence of R. tomentosa (MK_044696) was compared to those of Psidium guajava (NC_033355), Eugenia uniflora, (NC_027744), Eucalyptus grandis (HM_347959), and Melastoma candidum (NC_034716) using the mVISTA program (Figure 6). By comparison, the two IR regions were less divergent than the LSC and SSC regions, which also occurred in most plants [6,39]. Moreover, it was found that the non-coding region was more variable than the coding region, and the different regions may provide candidate DNA barcodes for future studies. In the coding region, most genes were relatively conserved except for matK, accD, ndhF, ycf1 and ycf2. These divergence hotspot regions of the four plant CP genome sequences provided abundant information for developing molecular markers for phylogenetic analyses and plant identification of Myrtaceae species.
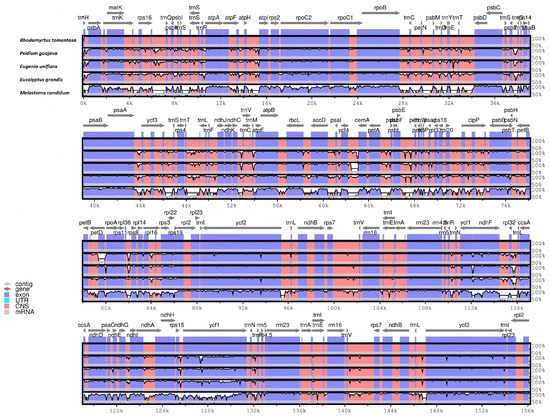
Figure 6.
Sequence identity plot comparison of the chloroplast genome of Rhodomyrtus. tomentosa with three others using mVISTA. Gray arrows and thick black lines above the alignment indicate genes with their orientation and the position of the IRs, respectively. A cut-off of 70% identity was used for the plots, and the Y-scale represents the percentage identity ranging from 50 to 100%.
2.1.7. Phylogenetic Analysis of the R. tomentosa CP Genome
The availability of a complete CP genome provides us with abundant sequence information that can be used to study the molecular evolution and phylogeny of plants [8,40]. To identify the evolutionary position of R. tomentosa, the whole CP genomes of 17 species were used to reconstruct a phylogenetic tree using the maximum likelihood (ML) method, in which four species from Myrtaceae along with 13 species from other families were chosen. Figure 7 shows that most nodes were strongly supported by 100 % bootstrap values (BP). Furthermore, R. tomentosa exhibited a sister relationship with two species of Eugenia uniflora and Psidium guajava and then grouped with Eucalyptus grandis. These four species all belong to the Myrtaceae family and were clustered distinctly from other families, which could help reveal the relationship between different families and orders. Nevertheless, node branching of this phylogenetic tree showed high consistency with the angiosperm phylogeny group (APG) IV classification system, which is a modern classification system of angiosperms based on the research of molecular system development. This classification situation differs from that of Flora of China, a series of books that summarize the systematic classification of vascular plants (ferns and seed plants) in China.
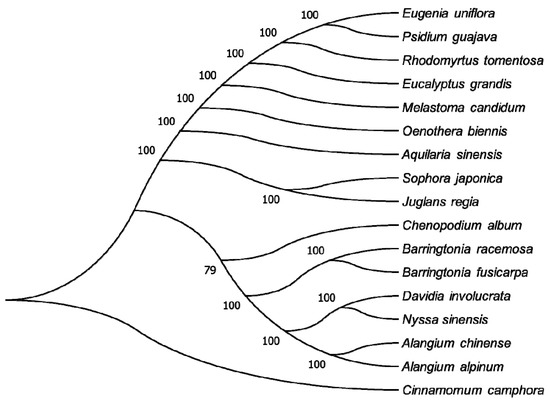
Figure 7.
Phylogenetic relationship of the 17 species inferred from maximum likelihood analyses based on the complete chloroplast genome excluding the IRA region. Numbers at nodes represent bootstrap support values.
Due to the limited availability of CP genome sequences from Myrtaceae deposited in databases, phylogenetic relationships among Myrtaceae plants based on CP genome sequence can be difficult to determine. Therefore, more data is needed to evaluate phylogenetic relationships of Myrtaceae plants in the future.
3. Materials and Methods
3.1. Plant Material, DNA Extraction and Sequencing
Fresh leaves of Rhodomyrtus tomentosa were collected from the Medicinal Botanical Garden of Guangzhou University of Chinese Medicine. Total genomic DNA was extracted from clean leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The extracted genomic DNA was measured in terms of purity and integrity by ultraviolet spectrophotometry and gel electrophoresis. DNA samples with good integrity and purity were submitted for library construction and sequencing using an Illumina Hiseq 2000 Sequencing platform (Illumina Inc., San Diego, CA, USA).
3.2. Chloroplast Genome Assembly and Annotation
Trimmomatic (v0.36, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) was used to filter and trim low-quality reads. The complete sequence of Psidium guajava chloroplast genome was downloaded from NCBI and served as a reference. Based on their coverage and similarity, CP-like reads were extracted and assembled using the Abyss2.0 program to form a complete chloroplast genome sequence. BLASTn was used to conduct self-alignment for locating the precise position of the quadripartite structure. In order to verify the assembly, four regions between the IR regions and the LSC/SSC region were confirmed through PCR amplification.
The preliminarily gene annotation of the R. tomentosa CP genome was performed using the GeSeq online tool (https://chlorobox.mpimp-golm.mpg.de/geseq.Html) with default parameters [41]. The annotation information was further examined and revised manually using the CLC Sequence Viewer (version 8), which was used to compare the CP genome of R. tomentosa and the related species, Psidium guajava. Since sequences at both ends of the exon are relatively conserved if genes contain introns, the chloroplast introns can be predicted according to the revised annotation file. The Organellar Genome DRAW (OGDRAW) (v1.2, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) [42] was used to construct a detailed map of the CP genome. Finally, the whole CP genome of R. tomentosa was deposited into GenBank, with an accession number of MK_044696.2.1.
3.3. Genome Structure and Genome Comparison
Distribution of codon usage and GC content were analyzed using the Molecular Evolutionary Genetics Analysis (MEGA 6.06, Tokyo Metropolitan University, Tokyo, Japan) [43]. Thirty-five protein-coding genes of the chloroplast genome of R. tomentosa were used to predict potential RNA editing sites using the online program Predictive RNA Editor for Plants (PREP) suite [44], with a cutoff value of 0.8. MISA and REPuter (https://bibiserv.cebitec.uni-Bielefeld.de/session) were used to identify SSRs and LRs in the R. tomentosa CP genome [45]. For the purpose of comparison among genomes, the mVISTA program (http://genome. Lbl.gov/vista/index.shtml) was used to align the CP genome of R. tomentosa with the CP genomes of Psidium guajava, Eugenia uniflora and Eucalyptus grandis [46].
3.4. Phylogenetic Analysis
A total of 17 complete CP genome sequences were downloaded from the GenBank (NCBI) database. Nucleotide alignments were subjected to phylogenetic analyses with maximum likelihood (ML) using the GTR + G substitution model, which was selected based on model screening. Bootstrap analysis was conducted with 1000 replicates and TBR branch swapping. In addition, Cinnamomum camphora was set as the out-group.
4. Conclusions
In conclusion, the complete CP genome of Rhodomyrtus Tomentosa was obtained using high throughput sequencing, which is 156,129 bp in length and encodes 129 genes. Further analysis on genome structure and genome characteristics revealed that gene structure and gene content of the R. tomentosa CP genome are conserved. The phylogenetic analysis indicated that R. tomentosa has a sister relationship with Eugenia uniflora and Psidium guajava. These results provide valuable information for further investigations on species identification and the evolution of R. tomentosa and its related species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/4/89/s1, Table S1: Codon usage of the Rhodomyrtus tomentosa chloroplast genome, Table S2: Predicted RNA editing sites in the chloroplast genomes of Rhodomyrtus tomentosa by the PREP program.
Author Contributions
Conceptualization, Z.Y.; Data curation, Y.H., Z.Y., W.A. and J.L.; Formal analysis, S.H.; Resources, S.H. and J.L.; Writing—original draft, Y.H.; Writing—review & editing, X.Z.
Funding
This research received no external funding.
Acknowledgments
We sincerely appreciate Professor Ming Li from School of Foreign Studies, Guangzhou University of Chinese Medicine for her kind help in reviewing and editing of this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Freire, C.G.; Giachini, A.J.; Gardin, J.P.P.; Rodrigues, A.C.; Vieira, R.L.; Baratto, C.M.; Werner, S.S.; Abreu, B.H. First record of in vitro formation of ectomycorrhizae in Psidium cattleianum Sabine, a native Myrtaceae of the Brazilian Atlantic Forest. PLoS ONE 2018, 13, e0196984. [Google Scholar] [CrossRef] [PubMed]
- He, S.M.; Wang, X.; Yang, S.C.; Dong, Y.; Zhao, Q.M.; Yang, J.L.; Cong, K.; Zhang, J.J.; Zhang, G.H.; Wang, Y.; et al. De novo Transcriptome Characterization of Rhodomyrtus tomentosa Leaves and Identification of Genes Involved in alpha/beta-Pinene and beta-Caryophyllene Biosynthesis. Front. Plant Sci. 2018, 9, 1231. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Ma, G.; Li, N.; Deng, Q.; Yin, Y.; Huang, R. Investigation of in vitro and in vivo antioxidant activities of flavonoids rich extract from the berries of Rhodomyrtus tomentosa(Ait.) Hassk. Food Chem. 2015, 173, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, J.G.; Su, J.C.; Huang, X.J.; Ye, W.C.; Wang, Y. Tomentodione E, a new sec-pentyl syncarpic acid-based meroterpenoid from the leaves of Rhodomyrtus tomentosa. J. Asian Nat. Prod. Res. 2018, 20, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Chen, C.; Wang, X.B.; Wu, L.; Yang, M.H.; Luo, J.; Zhang, C.; Sun, H.B.; Luo, J.G.; Kong, L.Y. Rhodomyrtials A and B, Two Meroterpenoids with a Triketone-Sesquiterpene-Triketone Skeleton from Rhodomyrtus tomentosa: Structural Elucidation and Biomimetic Synthesis. Org. Lett. 2016, 18, 4068–4071. [Google Scholar] [CrossRef] [PubMed]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Lavanya, G.; Voravuthikunchai, S.P.; Towatana, N.H. Acetone Extract from Rhodomyrtus tomentosa: A Potent Natural Antioxidant. Evid. Based Complement. Altern. Med. 2012, 2012, 535479. [Google Scholar] [CrossRef]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.Y.; Gao, L.Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Kaila, T.; Chaduvla, P.K.; Rawal, H.C.; Saxena, S.; Tyagi, A.; Mithra, S.V.A.; Solanke, A.U.; Kalia, P.; Sharma, T.R.; Singh, N.K.; et al. Chloroplast Genome Sequence of Clusterbean (Cyamopsis tetragonoloba L.): Genome Structure and Comparative Analysis. Genes 2017, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Cremen, M.C.M.; Leliaert, F.; West, J.; Lam, D.W.; Shimada, S.; Lopez-Bautista, J.M.; Verbruggen, H. Reassessment of the classification of Bryopsidales (Chlorophyta) based on chloroplast phylogenomic analyses. Mol. Phylogenetics Evol. 2019, 130, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Tabata, S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999, 6, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Feng, D.; Song, G.; Wei, X.; Chen, L.; Wu, X.; Li, X.; Zhu, Z. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Ser. Life Sci. 2003, 46, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Kim, K.; Lee, S.C.; Yang, T.J.; et al. The Complete Chloroplast Genome Sequences of Five Epimedium Species: Lights into Phylogenetic and Taxonomic Analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K. Chloroplast and mitochondrial genomes from a liverwort, Marchantia polymorpha--gene organization and molecular evolution. Biosci. Biotechnol. Biochem. 1996, 60, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Shao, J.; Zhang, H.; Jiang, M.; Huang, L.; Zhang, Z.; Yang, D.; He, M.; Ronaghi, M.; Luo, X.; et al. Sequencing and Analysis of Strobilanthes cusia (Nees) Kuntze Chloroplast Genome Revealed the Rare Simultaneous Contraction and Expansion of the Inverted Repeat Region in Angiosperm. Front. Plant Sci. 2018, 9, 324. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Kwak, M.; Lee, B.; Park, S. Complete chloroplast genome of Tetragonia tetragonioides: Molecular phylogenetic relationships and evolution in Caryophyllales. PLoS ONE 2018, 13, e0199626. [Google Scholar] [CrossRef]
- Trosch, R.; Barahimipour, R.; Gao, Y.; Badillo-Corona, J.A.; Gotsmann, V.L.; Zimmer, D.; Muhlhaus, T.; Zoschke, R.; Willmund, F. Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. plants 2018, 4, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Paiva, J.A.; Prat, E.; Vautrin, S.; Santos, M.D.; San-Clemente, H.; Brommonschenkel, S.; Fonseca, P.G.; Grattapaglia, D.; Song, X.; Ammiraju, J.S.; et al. Advancing Eucalyptus genomics: Identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries. BMC Genom. 2011, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, J.; Schairer, H.U.; Sebald, W. Identification of amino-acid substitutions in the proteolipid subunit of the ATP synthase from dicyclohexylcarbodiimide-resistant mutants of Escherichia coli. Eur. J. Biochem. 1980, 112, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Bukowy-Bieryllo, Z.; Dabrowski, M.; Witt, M.; Zietkiewicz, E. Aminoglycoside-stimulated readthrough of premature termination codons in selected genes involved in primary ciliary dyskinesia. RNA Biol. 2016, 13, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Baradaran-Heravi, A.; Balgi, A.D.; Zimmerman, C.; Choi, K.; Shidmoossavee, F.S.; Tan, J.S.; Bergeaud, C.; Krause, A.; Flibotte, S.; Shimizu, Y.; et al. Novel small molecules potentiate premature termination codon readthrough by aminoglycosides. Nucleic Acids Res. 2016, 44, 6583–6598. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, H.; Wang, L.; Zhu, C.; Sheng, K.; Du, Y.; Wang, K.; Dias, A.; Chen, S.; Whitman, M.; et al. Premature Termination Codons Are Recognized in the Nucleus in A Reading-Frame Dependent Manner. Cell Discov. 2015, 1, 15001. [Google Scholar] [CrossRef]
- Sui, T.; Song, Y.; Liu, Z.; Chen, M.; Deng, J.; Xu, Y.; Lai, L.; Li, Z. CRISPR-induced exon skipping is dependent on premature termination codon mutations. Genome Biol. 2018, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.H.; Jiang, X.M.; Yan, C.C.; Zhang, W.Q.; Xiao, G.S.; Yue, B.S.; Zhou, C.Q. Distribution patterns and variation analysis of simple sequence repeats in different genomic regions of bovid genomes. Sci. Rep. 2018, 8, 14407. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Shanker, A. Identification of Simple Sequence Repeats in Chloroplast Genomes of Magnoliids Through Bioinformatics Approach. Interdiscip. Sci. 2016, 8, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, X.; Cui, Y.; Sun, W.; Li, Y.; Wang, Y.; Song, J.; Yao, H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Taylor, D.R. Testing for selection on synonymous sites in plant mitochondrial DNA: The role of codon bias and RNA editing. J. Mol. Evol. 2010, 70, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xing, H.; Yuan, Y.; Wang, X.; Saeed, M.; Tao, J.; Feng, W.; Zhang, G.; Song, X.; Sun, X. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE 2018, 13, e0194372. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, F.; Huang, P.; Guo, W.; Zheng, Y. Complete Chloroplast Genome Sequence of Decaisnea insignis: Genome Organization, Genomic Resources and Comparative Analysis. Sci. Rep. 2017, 7, 10073. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Luo, C. Molecular and Functional Diversity of RNA Editing in Plant Mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef]
- Reginato, M.; Neubig, K.M.; Majure, L.C.; Michelangeli, F.A. The first complete plastid genomes of Melastomataceae are highly structurally conserved. Peer J. 2016, 4, e2715. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M.; Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhihai, H.; Jiang, X.; Shuiming, X.; Baosheng, L.; Yuan, G.; Chaochao, Z.; Xiaohui, Q.; Wen, X.; Shilin, C. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. GigaScience 2016, 5, 1–5. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).