Abstract
The photoperiod-insensitivity allele e1 is known to be essential for the extremely low photoperiod sensitivity of rice, and thereby enabled rice cultivation in high latitudes (42–53° north (N)). The E1 locus regulating photoperiod-sensitivity was identified on chromosome 7 using a cross between T65 and its near-isogenic line T65w. Sequence analyses confirmed that the E1 and the Ghd7 are the same locus, and haplotype analysis showed that the e1/ghd7-0a is a pioneer allele that enabled rice production in Hokkaido (42–45° N). Further, we detected two novel alleles, e1-ret/ghd7-0ret and E1-r/Ghd7-r, each harboring mutations in the promoter region. These mutant alleles alter the respective expression profiles, leading to marked alteration of flowering time. Moreover, e1-ret/ghd7-0ret, as well as e1/ghd7-0a, was found to have contributed to the establishment of Hokkaido varieties through the marked reduction effect on photoperiod sensitivity, whereas E1-r/Ghd7-r showed a higher expression than the E1/Ghd7 due to the nucleotide substitutions in the cis elements. The haplotype analysis showed that two photoperiod-insensitivity alleles e1/ghd7-0a and e1-ret/ghd7-0ret, originated independently from two sources. These results indicate that naturally occurring allelic variation at the E1/Ghd7 locus allowed expansion of the rice cultivation area through diversification and fine-tuning of flowering time.
1. Introduction
Rice is a major cereal extensively cultivated in a wide range of latitudes from 55° N to 35° S. Because rice is formerly a facilitative short-day (SD) plant well adapted to warm climate, photoperiodic control of flowering time is a key factor in the regional and seasonal adaptability of rice varieties [1]. In high latitudes (>ca. 40° N), rice cultivation had been impracticable due to the short summer and long-day (LD) more than 15 h during the summer, until early flowering varieties with extremely weak photoperiod sensitivity were raised [2,3,4]. It was during 1900 to 1930 that such varieties were first released and planted in the northernmost rice cultivation area, Hokkaido, in Japan (42–45° N) [5]. The varieties raised for Hokkaido also enabled rice cultivation even in Hei Long Jiang province (43–53° N) of China [6].
Also in low latitudes (ca. 20° S–20° N), a recent rice breeding program aims to produce varieties with weak photoperiod sensitivity (PS), though a long basic vegetative growth period is necessary at the same time, because such a combination of the two traits for heading will permit almost constant and adequate vegetative growth periods under SD (less than 13.5 h) [7,8,9]. In addition, in middle latitudes (30–40° N), there is a close relation between the photoperiod sensitivity of varieties and the latitude of their cultivation area [3,4]. Thus, understanding of the genetic factors responsible for photoperiod sensitivity, as well as basic vegetative growth, will be essential for not only guaranteeing stable rice production but also allowing further expansion of rice cultivation area.
Genetic studies on rice flowering (heading) time started in 1915 [10]. Since then, many flowering time loci were reported: among them, E1 [11,12,13,14], Photosensitivity 1 (Se1) [15], and Earliness 1 (Ef1) [16], have been intensively studied about their genetic characteristics, such as allelic variation, response to photoperiod, geographical distribution, and interaction with other loci. The geographical studies showed that these three loci play especially important roles in regional adaptabilities of Japanese and Taiwanese japonica rice varieties and japonica/indica cross varieties in Korea [4,5,6,15,17,18,19,20,21,22,23].
The Committee on Gene Symbolization, Nomenclature and Linkage Groups of the Rice Genetics Cooperative made a rule that the gene symbols which have been commonly used by many workers in the past should be retained [24], and recommended to categorize flowering time genes into three types, earliness and lateness (gene symbol: E), photoperiod sensitivity (gene symbol: Se), and basic vegetative growth (gene symbol: Ef). With the advance of quantitative trait locus (QTL) analysis, however, it has become difficult to categorize newly found QTLs into three types because they are detected only from flowering time data. Since then, the gene symbols, E, Se, and Ef, did not come to be retained. Recent molecular genetic analyses identified three key flowering time loci, Heading date 1 (Hd1) [25], Early heading date 1 (Ehd1) [26], and Grain number, plant height, and heading date 7 (Ghd7) [27], all of which were named regardless of the rule, and subsequent studies on these three loci provided new numerous molecular-based knowledges of rice flowering. Similarly, about the E1, Se1 and Ef1 loci, the information useful for rice breeding has been accumulated with enormous numbers until now. Therefore, it is significant to clarify the relationships of the three loci, E1, Se1 and Ef1, to the loci named regardless of the rule. To date, the Se1 and Ef1 loci proved to be identical with the Hd1 [21,28] and the Ehd1 loci [7], respectively.
The E1 was first identified as a late flowering time locus: the functional allele E1 is completely dominant over the nonfunctional allele e1 [11,12]. This locus was also involved in plant height. Later, this locus proved to control PS [13], and its functional allele E1 was shown essentially important in rice varieties for temperate areas in Japan (30–40° N) [17,18] because of firmly inhibiting the panicle primordial differentiation under LD until it becomes SD conditions, and thereby ensuring normal vegetative growth and stable yields. In contrast, a photoperiod-insensitivity allele e1 was found to be essential for the varieties commercially cultivated in Hokkaido (42–45° N), because of its marked reducing effect on photoperiod sensitivity: use of e1 enabled rice cultivation in high latitudes where LD conditions continue during the summer [5,21]. The E1 locus was found to be located on chromosome 7, linked to the rfs (rolled fine strip) and slg (slender glume) loci with recombination values of 16.3% and 9.1%, respectively [29]. This locus has been well investigated for its effects on photoperiod sensitivity and regional adaptabilities of rice plants [5,11,12,13,14,15,16,17,18,19], but little is known about the relationship with the loci which were identified by molecular genetic analysis and named regardless of the rule. Recently, the Ghd7 locus was precisely mapped on chromosome 7, and this locus exert major effects on not only heading date but also number of grains per panicle and plant height [27]. In addition, subsequent molecular analyses of the Ghd7 locus demonstrated that a loss-of-function allele of Ghd7 is essential for the extremely early flowering of Hokkaido varieties [30,31,32]. These reports make a conjecture that the E1 and Ghd7 are the same locus.
In the present study, we first analyzed the effects of three alleles at the E1 locus on photoperiod sensitivity using the Taiwanese japonica rice variety “Taichung 65 (T65)” harboring E1 [33], its isogenic line T65m harboring e1 [16,33,34,35], and T65w that harbors a chromosome segment of O. rufipogon Griff. including the E1 locus in the genetic background of T65 [36]. Subsequently, we attempted to determine the precise chromosomal location of the E1 locus using the progenies from T65 × T65w, and then conducted sequence analysis to learn the sequences of the three alleles, also to investigate the relationship between the two loci, E1 and Ghd7. We finally applied a haplotype analysis of the chromosomal region surrounding the E1/Ghd7 locus to 44 Hokkaido and 50 Japanese-core-collection varieties in order to prove correctness of the findings by Okumoto et al. (1996) [5] and Ichitani et al. (1998) [21] that e1 is the key allele for establishing the varieties for the northernmost rice cultivation area, and its history and origin.
2. Results
2.1. Photoperiod Sensitivities of T65, T65w, and T65m
Days to heading (DH) of T65, T65m, and T65w under a SD were 84.7, 81.4, and 82.0 respectively, while those under a LD were 95.6, 90.4, and 118.0, respectively (Figure 1). Thus, the photoperiod sensitivities of T65, T65m, and T65w were estimated at 11.1, 9.0, and 36.0, respectively. Since T65m is an isogenic line of T65 for the E1 locus, the weaker PS of T65m was attributable to the photoperiod-insensitivity allele e1 at the E1 locus. T65w showed far stronger photoperiod sensitivity than T65 and T65m. The genotypic difference between T65w and T65 is only in the chromosome region including the E1 locus, where only T65w harbors a chromosome segment induced from O. rufipogon Griff. Since any other photoperiod sensitivity genes have not yet been reported in this region, we conclude that the chromosome segment introduced from O. rufipogon Griff. in T65w certainly harbors a strong photoperiod-sensitivity allele, probably at the E1 locus.
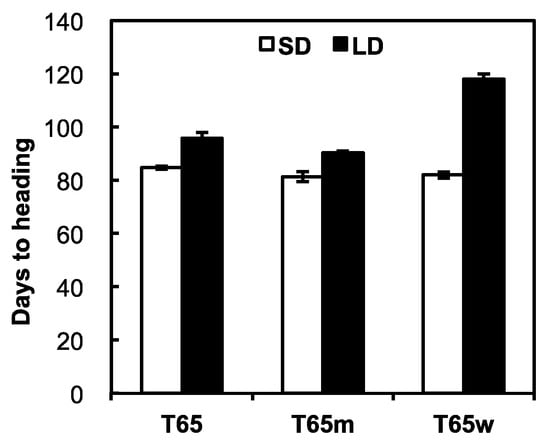
Figure 1.
Days to heading of T65, T65m, and T65w under short-day (SD, white bar) and long-day (LD, black bar) conditions.
2.2. Chromosomal Location of the E1 Locus
The F2 population from the cross between T65 and T65w, comprising 205 plants, showed a continuous distribution of DH within the parental ranges (Figure 2a). We conducted a progeny test using 38 F3 lines, which were derived from randomly selected F2 plants. In the test, all the F3 lines were clearly classified into three groups. The ratio of [T65-type]:[segregating-type]:[T65w-type] lines was 13:14:11, which fitted the 1:2:1 ratio expected for one-locus segregation (χ2 = 8.904, P > 0.05) (Table S1). In contrast, the F2 population from the cross between T65w and T65m showed a bimodal distribution of DH within the parental ranges, with a clear breakpoint dividing the population into early (T65m-type) and late (T65w-type) groups (Figure 2b). The ratio of early type (34 plants): late type (91 plants) fitted the 1:3 ratio expected for one-locus segregation (χ2 = 0.570, P > 0.05). In the progeny test, all the 40 F3 lines were clearly classified into three groups. The ratio of [T65m type]:[segregating type]:[T65w type] lines fitted the 1:2:1 ratio expected for one-locus segregation (χ2 = 0.150, P > 0.05) (Table S2). T65m is an isogenic line of T65 harboring a recessive allele e1 at the E1 locus. We accordingly inferred that T65w harbors a novel allele at the E1 locus, whose heading-date delaying effect was stronger than E1 in T65. We designated this allele E1-r (a novel photoperiod-sensitivity allele at the E1 locus).
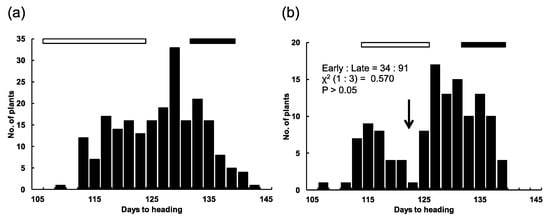
Figure 2.
Distributions of days to heading in two F2 populations from crosses between (a) T65 × T65w and (b) T65w × T65m. The black bar indicates the range of days to heading of T65w. The white bar indicates the ranges of days to heading of (a) T65 and (b) T65m. The arrow indicates the breakpoint between early and late heading groups.
Using 546 F3 plants from the cross between T65 and T65w, we tried to identify the chromosomal location of the E1 locus. The result showed that the E1 locus was present in the region with a physical distance of 4.11 Mb between two simple sequence repeat (SSR) markers, RM1253 and RM3635, on chromosome 7 (Figure 3). Subsequently, we attempted to narrow down the candidate region of the E1 locus, using 1263 F4 progenies derived from several F3 recombinants between RM1253 and RM3635; consequently, the chromosomal location of the E1 locus was narrowed down to the region with a physical length of approximately 228.1-kb between RM5436 and RM21341 (Figure 3). In this region, 11 genes are reported in Rice Annotation Program Database [37]. Among them, we proposed that Os07g0261200, which was reported as Ghd7, a repressor of flowering time under LD conditions [27], was likely to be a candidate of E1.
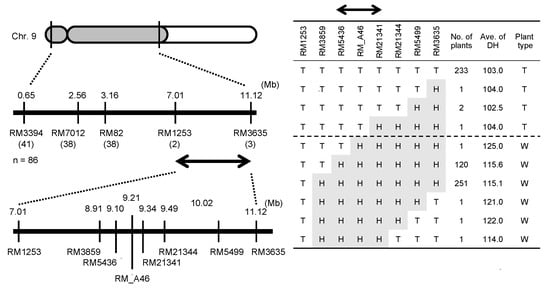
Figure 3.
Map-based cloning and graphical genotypes of the candidate region of the E1 locus. “T” and “H” at each marker indicate T65 homozygous and heterozygous, respectively. “T” and “W” at plant type indicate T65-type (early heading) and T65w-type (late heading), respectively.
Sequence analyses showed that the sequences of the alleles at the Ghd7 locus in T65 and T65m were completely consistent with a functional allele Ghd7-2 [38] and a nonfunctional allele ghd7-0a [27], respectively (Figure 4a,b). Since the genotypic difference in flowering time between T65 and T65m is only at the E1 locus, this suggests that E1 and Ghd7 are the same locus (hereafter we tentatively designate E1 (=Ghd7) as E1/Ghd7), and that T65m flowered earlier than T65 because the former harbors a loss-of-function allele e1/ghd7-0a. In contrast, the allele of T65w at the E1 locus harbored four nonsynonymous substitutions and two nucleotide substitutions in the promoter region (Figure 4a,b). Among the substitutions, two in the promoter region were in the transcriptional signal motifs (cis elements): low temperature response element (LTRE) core actor (located at −284) and the TATA box (located at −564). Thus, the two nucleotide substitutions were considered to modify the expression of E1/Ghd7. Subsequent expression analysis of E1/Ghd7 showed that the expression of T65w was higher than that of T65 (Figure 4c). This suggests that the late flowering of T65w is caused by high expression of E1/Ghd7 due to the nucleotide substitutions in the cis elements.
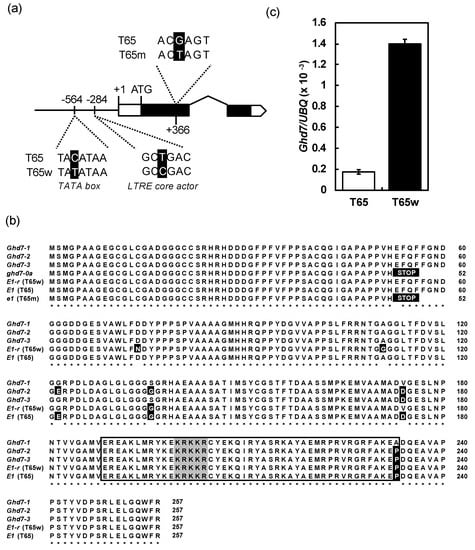
Figure 4.
(a) Schematic diagrams of the alleles at the E1/Ghd7 focused on nucleotide substitutions among T65 T65m, and T65w. (b) Alignments of amino acid sequences of the alleles at the E1/Ghd7 locus. The white and black characters with black and gray cells indicate amino acid substitutions and CONSTANS, CO-like, and TOC1 (CCT)-motif, respectively. The box indicates the CCT-motif region. Ghd7-1, Ghd7-2 and Ghd7-3 were functional alleles [29]. (c) Comparison of the expression level of the allele at the E1/Ghd7 locus between T65 and T65w.
2.3. A Novel Nonfunctional Allele at the E1/Ghd7 Locus
Okumoto et al. (1996) [5] showed that nine Hokkaido varieties tested all harbored a nonfunctional (photoperiod-insensitivity) allele e1 at the E1 locus thorough a conventional genetic analysis, and assumed that this allele has played an essential role in the establishment of rice varieties for the Hokkaido district. To confirm this assertion, we analyzed the presence of the nucleotide substitution from GAG (Glu) to TAG (stop codon) in exon 1 at the E1/Ghd7 locus (Figure 4a) of 44 Hokkaido varieties using a cleaved amplified polymorphic sequence (CAPS) marker. The result showed that 37 varieties harbored the e1/ghd7-0a allele, and 7 varieties did not (Table S3). This single nucleotide substitution was not observed in EG5 (Aikoku), which is one of the tester lines for the E1, E2 and E3 loci involved in the flowering time, and which harbors e1 allele at the E1 locus [11,12,13]. Sequence analysis for the EG5 revealed that a Ty1-copia like retrotransposon (TE) was inserted in the promoter region of the E1/Ghd7 allele (Figure 5a). The seven varieties, which did not harbor the e1/ghd7-0a allele, also harbored the same TE insertion. We named this novel nonfunctional allele e1-ret/ghd7-0ret. The expression of the e1-ret/ghd7-0ret allele was far lower than the E1/Ghd7-2 allele in the Japanese variety “Nipponbare” with the reference genome (Figure 5b), indicating that e1-ret/ghd7-0ret confers extremely weak photoperiod sensitivity by losing the normal function of the promoter.
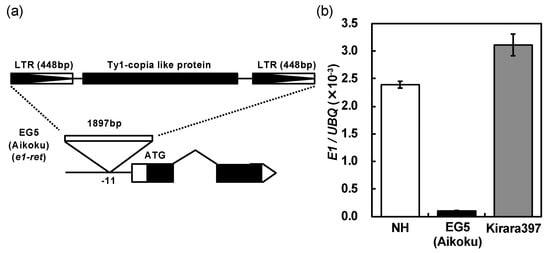
Figure 5.
(a) Schematic diagram of the allele at the E1/Ghd7 locus in “EG5 (Aikoku)”. (b) Comparison of the expression level of the allele at the E1/Ghd7 locus among three Japanes varieties “Nipponbare” (NH), “EG5 (Aikoku)” and “Kirara397”. “EG5 (Aikoku)” and “Kirara397” are a tester line for the E1, E2 and E3 locus and an elite Hokkaido variety, respectively.
2.4. Haplotype Patterns of the Chromosomal Region Surrounding E1/Ghd7 Locus
We surveyed DNA polymorphisms between EG5 (Aikoku) (e1-ret/ghd7-0ret) and Kirara397 (e1/ghd7-0a) around E1/Ghd7 locus. Subsequently, we found three polymorphisms (two SNPs and a 20-bp deletion) other than the nucleotide substitution from GAG (Glu) to TAG (stop codon) and the TE insertion. To know the origins of two nonfunctional alleles e1/ghd7-0a and e1-ret/ghd7-0ret, we investigated the haplotypes of Hokkaido and Japanese-core collection varieties using five markers surrounding the E1/Ghd7 locus (three single nucleotide polymorphisms (SNPs), a 20-bp deletion, and a TE insertion). The Japanese-core-collection varieties were classified into at least four haplotypes, Hap2, Hap3, Hap4, and Hap5 (Figure 6 and Table S4). In contrast, Hokkaido varieties were classified into two distinct haplotypes, Hap1 and Hap3. This suggests that Hap1 was derived from Hap2 via nucleotide substitution from GAG (Glu) to TAG (stop codon) in exon 1, whereas Hap3 was derived from Hap4 via the TE insertion in the promoter region. These results indicate that two independent mutational events contributed to the occurrence of the two nonfunctional alleles e1/ghd7-0a and e1-ret/ghd7-0ret. Interestingly, although Hap1 was found only in Hokkaido varieties, Hap3 was found not only in Hokkaido varieties but also in some Japanese varieties, particularly in the Aikoku-related varieties (Figure 6 and Table S4). Further, the varieties of the Hap3 group, except for Hokkaido varieties, flowered about 20 days later than the Hokkaido varieties, implying that such varieties do not adapt to Hokkaido where autumn comes early (Figure 6 and Table S4). These findings indicate that other genetic factor (s) were involved in the early flowering of Hokkaido varieties belonging to the Hap3 group. We accordingly investigated allelic variations in the Se1/Hd1 and another major photoperiod-sensitivity gene, Hd5, which is known to be involved in the PS in the Hokkaido varieties [30,31,32]. The results showed that varieties with e1/ghd7-0a flowered early regardless of harboring a functional allele(s) (photoperiod-sensitivity allele) at the Se1/Hd1 and/or Hd5 locus, whereas varieties with e1-ret/ghd7-0ret flowered early only when harboring a nonfunctional allele at either of the Se1/Hd1 or Hd5 locus (Figure 7). These results indicate that coexistence of e1-ret/ghd7-0ret with a photoperiod-insensitivity allele either at the Se1/Hd1 or at the Hd5 locus is necessary to promote flowering under LD conditions.
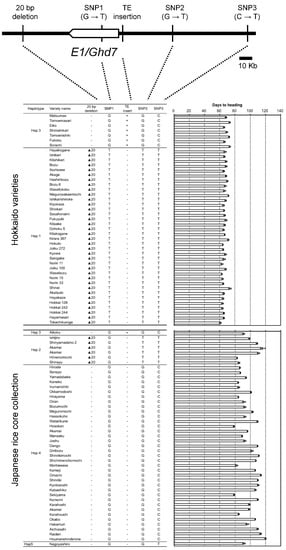
Figure 6.
Haplotypes around the E1/Ghd7 locus and days to heading of Hokkaido and Japanese core collection varieties.
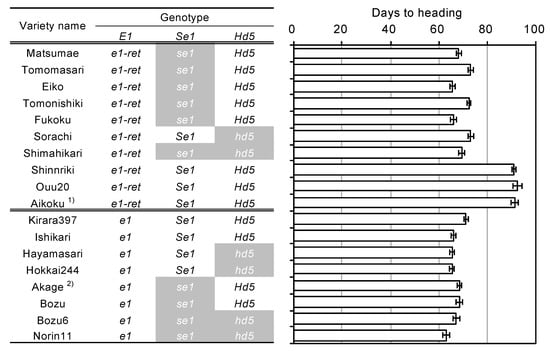
Figure 7.
Gene combinations for the E1, Se1, and Hd5 loci and days to heading. 1) This “Aikoku” variety belongs to the Japanese core collections. 2) This “Akage” variety belongs to the Hokkaido varieties.
3. Discussion
Since Hoshino (1915) [10], many genes (loci) controlling flowering time have been reported (reviewed in [39,40,41]). Among them, the E1 is an important locus closely associated with the regional adaptability of rice varieties: its photoperiod insensitivity allele e1 enabled rice cultivation even in Hokkaido, one of the northernmost rice cultivation area (42–45° N) [5,13,21]. A dominant photoperiod-sensitivity allele E1 at the E1 locus widely deployed among Japanese varieties for all regions other than Hokkaido in Japan [17,18,21]. Ichitani et al. (1998) [21] reported that the E1 locus was identical to the Heading date 4 (Hd4) locus, which was identified by Quantitative Trait Locus (QTL) analysis of flowering time using progenies from the cross of the indica variety Kasalath and the japonica variety Nipponbare [25]. Fujino and Sekiguchi (2005) [30] identified two QTLs, qDTH-7-1 and qDTH-7-2, for flowering time using progenies from the cross between two Hokkaido varieties, Hoshinoyume and Nipponbare. They concluded that qDTH-7-1 is the same locus as the E1 (Hd4). Later, Xue et al. (2008) [27] isolated the grain number, plant height, and heading date 7 (Ghd7) locus on chromosome 7, whose functional allele Ghd7 encodes a CONSTANS, CO-like, and TOC1 (CCT) domain-containing protein that delays flowering under LD conditions. They also reported that a nonfunctional allele ghd7-0a, harboring a premature termination in the predicted coding region, deployed among varieties commercially cultivated in Hei Long Jiang province, China (43–53° N). In the present study, we successfully determined the chromosomal location of the E1 locus within a 228.1-kb physical region on chromosome 7 (Figure 3). According to the rice public database RAP-DB [38], 11 loci (genes) exist in this region. Among the genes, only Os07g0261200 (=Ghd7) showed a SNP in exon 1 of a photoperiod insensitive allele e1 in T65m (Figure 4). This substitution was the same as that of the nonfunctional allele ghd7-0a [27]. From these results, we concluded that the E1, Hd4, qDTH-7-1 and Ghd7 are the same locus. Then we finally designate this locus as E1/Ghd7. It is noteworthy that we identified a novel photoperiod-insensitivity allele e1-ret/ghd7-0ret that harbored a TE insertion in the promoter region of E1/Ghd7. We conclude that this TE-insertional mutation causes the loss of function of the E1/Ghd7 allele.
In addition, we identified a novel strong photoperiod-sensitivity allele E1-r/Ghd7-r, which harbors three nonsynonymous substitutions in the coding sequence (CDS) and two SNPs in the promoter region: one is in the LTRE core actor (CCGAC), and the other is in the TATA-box (Figure 5a). The LTRE core actor was identified in the regulatory regions of all cold-induced genes in Arabidopsis [42]. The CCGAC core motif, also known as C-repeat / drought response element (CRT/DRE), is essential for transcriptional activation in response to cold, drought, and/or high-salt treatments [43]. Kim et al. (2002) [44] showed that light signaling mediated by phytochrome activates cold-induced gene expression through CRT/DRE. The expression of E1-r/Ghd7-r was higher than that of a PS allele of E1/Ghd7-2 in Nipponbare (Figure 5b). The expression level of E1/Ghd7 is regulated by red light signal and correlated well with its LD specific activity [45,46]. These suggest that the mutations in the promoter region modify the E1-r/Ghd7-r expression, which delays flowering under LD conditions. Elucidation of the influences of the three amino acid substitutions in the CDS of E1-r/Ghd7-r awaits further study.
Haplotype analysis showed that two photoperiod-insensitive alleles, e1/ghd7-0a and e1-ret/ghd7-0ret, originated independently from two sources (Figure 6). e1/ghd7-0a widely deployed among improved and landrace varieties in Hokkaido, including “Akage”, which was one of the pioneers of the Hokkaido rice varieties in the late 1800’s [47]. This indicates that e1/ghd7-0a is a pioneer allele, leading to raising extremely early heading varieties with extremely weak photoperiod sensitivity during 1900–1930 (see Introduction). In contrast, e1-ret/ghd7-0ret was detected in “Fukoku”, one of the past leading varieties in Hokkaido. “Fukoku” was bred from the cross between the Japanese warm region variety “Nakate-Aikoku”, and the Hokkaido variety “Bozu 6”. “Nakate-Aikoku” harbors e1-ret/ghd7-0ret, whereas “Bozu 6” does not. This indicates that e1-ret/ghd7-0ret of “Fukoku” was derived from “Nakate-Aikoku” (Figure S1). Interestingly, although most of the Aikoku-related varieties harbor e1-ret/ghd7-0ret (Table S4), none of them flowered as early as “Fukoku”. This indicates that some genetic factor(s) other than e1-ret/ghd7-0ret is also responsible for the early flowering of “Fukoku”. The varieties with e1-ret/ghd7-0ret flowered extremely early when harboring a nonfunctional allele either at the Se1/Hd1 locus or at the Hd5 locus (Figure 7). Therefore, we conclude that a nonfunctional allele either at the Se1/Hd1 locus or at the Hd5 locus is necessary for early flowering of the Hokkaido varieties with e1-ret/ghd7-0ret. Fujino et al. (2013) [32] reported that the nonfunctional allele at the Hd5 locus was a spontaneous mutant gene that occurred in the Hokkaido local landrace “Bozu,” and that this allele contributed to the expansion of rice cultivation to the northern area of Hokkaido. In contrast, the nonfunctional allele at the Se1/Hd1 locus widely deployed among the varieties for the Tohoku-Hokuriku region (37–40° N) [15,18]. In the early rice breeding in Hokkaido, many Aikoku-related varieties, which were chiefly cultivated in the Tohoku-Hokuriku region, Japan, were frequently used as cross-parents to increase the genetic diversity and improve grain quality [47]. This suggests that e1-ret/ghd7-0ret was introduced from the Aikoku-related varieties, and the combination with the nonfunctional allele at the Hd1 or Hd5 locus made e1-ret/ghd7-0ret available for rice breeding programs in Hokkaido. It is also suggested that mutations in the promoter region often make functional differentiations of alleles at the E1/Ghd7 locus, bringing about flowering time diversification in rice.
Recent molecular genetic studies revealed that there are large natural allelic variations in key loci controlling flowering time, such as Ghd7, Hd1, DTH2, and DTH7, which contribute to the diversity of flowering time, and to the regional adaptability by adjusting flowering time to distinct environmental conditions [27,32,48,49,50,51]. Zhang et al. (2015) [52] and Zheng et al. (2016) [53] classified Ghd7 alleles into three (strong function, weak function and non-function) and two (function and non-function) groups based on the sequence polymorphisms in the coding region, respectively. On the other hand, Lu et al. (2012) [38] analyzed 104 varieties (O. sativa) and three wild rice accessions (O. rufipogon) and found that 76 SNPs and six insertions and deletions within a 3932-bp DNA fragment of Ghd7. Among them, the functional C/T mutation in the promoter region was related to plant height probably by altering gene expression. In this study, we identified two novel alleles with functional differentiations in the promoter region of the E1/Ghd7 allele. The interaction of these alleles with other flowering time genes except Hd1 and Hd5 have not yet been elucidated, additional detailed analysis of their effects on flowering time will contribute to fine-tuning of the flowering time well adapting to the climatic conditions in each area.
Photoperiod sensitivity is an important trait responsible for regional and seasonal adaptability of rice varieties. In this study, we detected two novel alleles at the E1/Ghd7 locus. It is known that many other loci are involved in the photoperiod sensitivity pathway of flowering in rice. Recent studies showed that photoperiod sensitivity loci, Se1/Hd1, OsPRR37 and Ghd8, each have a large allelic variation [49,54,55,56,57]. Therefore, analyzing the functional and inter-locus interactions should be advanced, which will lead to practice the fine-tuning of flowering time in rice breeding programs. In addition, we elucidated that the E1 and Ghd7 are the same locus. Until now, numerous genetic information has been accumulated for each locus (E1, Ghd7). For further genetic analyses of the E1/Ghd7 locus, however, all information about E1 and Ghd7 should be available.
4. Materials and Methods
4.1. Photoperiod Sensitivities of T65, T65m, and T65
The Taiwanese japonica rice variety “Taichung 65 (T65)”, and its isogenic line T65m and its near-isogenic line T65w were used. T65 harbors a photoperiod sensitivity allele E1 at the E1 locus, while T65m harbors a photoperiod-insensitivity allele e1, which is derived from the cross with Bozu5 [16,19,34,35]. T65w is a chromosome substitution line which was developed by introducing the E1 region of O. rufipogon (W107) into the genetic background of T65 [36]. Five seeds were sown on field soil in a 3.6 L pot and covered with granulated soils. Seedlings were thinned to one plant per pot 14 days after sowing, and were grown under two photoperiod conditions, SD (12-h light/12-h dark) and LD (14-h light/10-h dark). Photoperiod treatments were conducted using two growth cabinets without temperature control. In addition to natural daylight (8:00–18:00), supplementary artificial light was used for the 12-h and 14-h light conditions. The degree of photoperiod sensitivity of each line was expressed as a difference of DH between SD and LD. The experiment was conducted by using five plants per line with three replications from 1 May to October 16. Heading date was recorded for each plant when the first panicle emerged from the sheath of the flag leaf.
4.2. Chromosomal Location of the E1 Locus
Two F2 populations from crosses of T65 × T65w, comprising 205 plants, and T65w × T65m, comprising 125 plants, were subjected to genetic analysis of heading date. They were grown in a paddy field of Kyoto University, Kyoto, Japan (35°01′ N). Seeds were sown on April 25 in 2007, and seedlings were transplanted on May 16 in 2007. The progeny test was conducted with 40 F3 lines (sowing on April 30 in 2008 and transplanting on May 21 in 2008). Each F3 line with approximately 25 plants was the progeny of an F2 plant randomly selected from the F2 population. To narrow down the candidate region of the E1 locus, the F4 progenies derived from several F3 recombinants between RM1253 and RM3635 were cultivated (sowing on May 1 in 2009 and transplanting on May 22 in 2009).
4.3. Expression Analysis of E1/Ghd7
Plants of T65, T65m, and T65w were grown in a growth cabinet with a temperature controller under a LD condition (14.5-h light, 30 °C/9.5-h dark, 25 °C) at 70% relative humidity. Seedlings were grown on sand in 3.6 L pots (two plants/pot) with additional liquid fertilizer (Kimura’s B Culture Solution, Nippon Medical and Chemical Instruments Co., Ltd., Osaka, Japan). Thirty days before flowering, leaves were collected at 4-h intervals during that day. Total RNAs were extracted with Trizol reagent (Life Technologies Inc., Gaithersburg, Maryland, USA) according to the manufacturer’s protocols. Total RNA was subjected to DNA digestion by treatment with RNase-free DNase I (Takara Bio Inc.). The transcriptor first-strand cDNA synthesis kit (Roche Applied Science, Indianapolis, Indiana, USA) was used to reverse-transcribe cDNA from 1 µg of RNA using anchored-oligo (dT)18 primers. Real-time PCR analysis was performed by TaqMan PCR using a LightCycler 1.5 (Roche Applied Science) according to the manufacturer’s instructions. The primer sets of Ghd7 and UBQ genes and Universal Probe Library probes of each gene were designed with ProbeFinder version 2.45 (Roche; https://www.roche-applied-science.com/). Primer and probe sequences are shown in Table S5. Expression analysis using the standard curve method was performed to determine the expression level of each gene. The relative expression level of each gene was calculated using UBQ gene. The RNA gene standards for the seven genes were applied to their plasmids prepared by the pGEM-T Easy Vector System (Promega Corp., Madison, Wisconsin, USA) using PCR amplicons from the total RNA of T65.
4.4. Identification of the Genotype at the E1/Ghd7 Locus
To identify the E1/Ghd7 genotype, we developed two DNA markers based on an SNP and a TE insertion. CAPS marker analysis was based on a nucleotide substitution from GAG (Glu) to TAG (stop codon) in exon 1, producing an additional restriction enzyme (Spe I) site. A pair of PCR primers (Ghd7_CAPS_F: 5′-CCAACTTGCCCTGTCTTCTT-3′, Ghd7_CAPS_R: 5′-AGCTGCTGCAAGCCAGTAAT-3′) was designed to amplify the 950 bp. PCR was performed with a 20 μL reaction mixture containing 2 μL template DNA, 10× PCR buffer, 25 mM MgCl2, 2 mM of each deoxyriboside-triphosphate (dNTP), 0.2 μL Taq DNA polymerase (5 U/μL), 4 μL of a 2.5 mM solution of each primer, and 3.2 μL H2O. PCR conditions were as follows: 94 °C for 5 min, followed by 35 cycles (1 min at 94 °C, 1 min at 60 °C, and 2 min at 72 °C) with a final extension of 7 min at 72 °C. The amplified products were digested with Spe Ι at 37 °C for 6 h. After digestion, the nonfunctional allele produced three fragments (81, 287, and 582 bp), whereas the functional allele produced two fragments (81 and 869 bp). Amplicons and digested amplicons were separated on 1% agarose gel. After electrophoresis, the gel was stained with ethidium bromide, and the DNA fragments were visualized under UV light. Insertion and deletion (INDEL) marker analysis was based on an 1897-bp copia-like TE insertion. A pair of PCR primers (Ghd7_INDEL_F: 5′-CGTTTCAGCAATAGCATTATGG-3′, Ghd7_INDEL_R: 5′-GCGGGTAGTCATCGAACAG-3′) was designed to amplify the 824 bp in the wild type and the 2721 bp in the insertion type. PCR was performed with a 20 μL reaction mixture containing 2 μL template DNA, 10× PCR buffer, 25 mM MgCl2, 2 mM of each dNTP, 0.2 μL Taq DNA polymerase (5 U/μL), 4 μL of a 2.5 mM solution of each primer, and 3.2 μL H2O. PCR conditions were as follows: 94 °C for 5 min, followed by 35 cycles (1 min at 94 °C, 1 min at 60 °C, and 2 min at 72 °C) with a final extension of 7 min at 72 °C. Amplicons were separated on 1% agarose gel. After electrophoresis, the gel was stained with ethidium bromide, and the DNA fragments were visualized under UV light.
4.5. Haplotype Patterns of the Chromosomal Region around the E1/Ghd7 Locus
Three DNA polymorphisms (two single-nucleotide substitutions and a 20 bp deletion) within the 100-kb chromosomal region surrounding Ghd7 existed among 44 Hokkaido, 50 Japanese-core-collection [58] and 71 Aikoku-related varieties, which are considered to the derivatives of “Aikoku” variety based on their names. The Japanese rice core collection is a limited set of accessions representing, with a minimum set repetitiveness, the genetic diversity among Japanese rice varieties [58]. The two substitutions (Hap_SNP1 and Hap_SNP2) were detected by CAPS marker analyses. Hap_SNP1 harbors a G-to-T substitution, resulting in changing the recognition site of the restriction enzyme Hpy188 I. Hap_SNP2 harbors a C-to-T substitution, resulting in changing the recognition site of Hpy188 I. Primer pairs for Hap_SNP1 and Hap_SNP2 were designed to amplify 623 and 295 bp, respectively (Table S6). In addition, two polymorphism surveys of the copia-like TE insertion and the SNP in the first exon of Ghd7 were performed. The CAPS and INDEL marker analyses to detect the insertion and SNP were described above.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/12/550/s1, Figure S1: Pedigree of the varieties, which harbor e1-ret/ghd7-0ret allele. Bold with underline indicates that the varieties harbor e1-ret/ghd7-0ret allele. Gray indicates varieties whose allele is unknown. Other indicates that the varieties harbor e1/ghd7-0a allele., Table S1: Frequency distributions of days to heading in F3 lines (T65 × T65w), Table S2: Frequency distributions of days to heading in F3 lines (T65m × T65w), Table S3: Lists of Hokkaido and Japanese core collection varieties used in this study, Table S4: Lists of Aikoku-related varieties used in this study, Table S5: Primer and probe sequences for expression analysis, Table S6 Primer sequences for haplotype analysis.
Author Contributions
Conceptualization, H.S., Y.O. and T.T. (Takatoshi Tanisaka); Methodology, H.S., Y.O. and T.T. (Takatoshi Tanisaka); Formal analysis, H.S., T.T. (Takuji Tsukiyama) and M.T.; Investigation, H.S. and C.X.; Writing—original draft preparation, H.S.; Writing—review and editing, H.S., Y.O. and T.T. (Takatoshi Tanisaka); Project administration, H.S. and T.T. (Takatoshi Tanisaka).
Acknowledgments
We thank the genetic resources center, NARO (National Agriculture and Food Research Organization) for providing the seeds of Hokkaido, Japanese-core-collection and Aikoku-related varieties. T65m was kindly provided from Shigetoshi Sato in Ryukyu University and Kuo Hai Tsai in National Chung-Hsing University. T65w was kindly provided from Yoshio Sano in Hokkaido University. We also would like to thank Syo Asano and Shinsuke Hamada for their supports of our experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Wada, E. Studies on the response of heading to daylength and temperature in rice plants. : II. Response of varieties and the relation to their geographical distribution in Japan. Jpn. J. Breed. 1952, 2, 55–62. [Google Scholar] [CrossRef]
- Hosoi, T. Studies on meteorological fluctuation in the growth of rice plants. V. Regional references of thermo-sensitivity, photo-sensitivity, basic vegetative growth and factors determining the growth duration of Japanese varieties. Jpn. J. Breed. 1981, 31, 239–250. [Google Scholar] [CrossRef]
- Tanisaka, T.; Inoue, H.; Uozu, S.; Yamagata, H. Basic vegetative growth and photoperiod sensitivity of heading-time mutants induced in rice. Jpn. J. Breed. 1992, 42, 657–668. [Google Scholar] [CrossRef]
- Okumoto, Y.; Ichitani, K.; Inoue, H.; Tanisaka, T. Photoperiod insensitivity gene essential to the varieties grown in the northern limit region of paddy rice (Oryza sativa L.) cultivation. Euphytica. 1996, 92, 63–66. [Google Scholar] [CrossRef]
- Zhang, Y. An Economic Analysis on Growth and Technological Progress of Rice Production in Three Northeastern Provinces of China. J. Rural Problem 2002, 38, 1–12. [Google Scholar] [CrossRef][Green Version]
- Saito, H.; Yuan, Q.; Okumoto, Y.; Doi, K.; Yoshimura, A.; Inoue, H.; Teraishi, M.; Tsukiyama, T.; Tanisaka, T. Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 119, 315–323. [Google Scholar] [CrossRef]
- Saito, H.; Ogiso-Tanaka, E.; Okumoto, Y.; Yoshitake, Y.; Izumi, H.; Yokoo, T.; Matsubara, K.; Hori, K.; Yano, M.; Inoue, H.; et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short-and long-day conditions. Plant Cell Physiol. 2012, 53, 717–728. [Google Scholar] [CrossRef]
- Yuan, Q.; Saito, H.; Okumoto, Y.; Inoue, H.; Nishida, H.; Tsukiyama, T.; Teraishi, M.; Tanisaka, T. Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor. Appl. Genet. 2009, 119, 675–684. [Google Scholar] [CrossRef]
- Hoshino, Y. On the inheritance of the flowering time in peas and rice. J. Col. Agric. 1915, 6, 229–288. [Google Scholar]
- Syakudo, K.; Kawase, T. Studies on the quantitative inheritance (11): A Rice (Oryza sativa L.) (d) Inheritance of the heading duration and the quantitative function of the causal genes in its determination. : (1) On the quantitative function of the genes E1, E2 and D1. Jpn. J. Breed. 1953, 3, 6–12. [Google Scholar] [CrossRef]
- Syakudo, K.; Kawase, T.; Yoshino, K. Studies on the quantitative inheritance (13): A Rice (Oryza sativa L.) (d) Inheritance of the heading duration and the quantitative function of the causal genes in its determination. : (2) On the quantitative function of the genes E3, E4 and E5. Jpn. J. Breed. 1954, 4, 83–91. [Google Scholar] [CrossRef]
- Yamagata, H.; Okumoto, Y.; Tanisaka, T. Analysis of genes controlling heading time in Japanese rice. Rice Genet. 1986, 1, 351–359. [Google Scholar]
- Okumoto, Y.; Tanisaka, T.; Yamagata, H. Genotypic difference in response to light interruption in Japanese rice varieties. Rice Genet. 1991, 2, 778–780. [Google Scholar]
- Yokoo, M.; Kikuchi, F.; Nakane, A.; Fujimaki, H. Genetical analysis of heading time by aid of close linkage with blast resistance in rice. Bull. Nat. Inst. Agric. Sci. Ser. 1980, D31, 95–126. [Google Scholar]
- Tsai, K.H. Studies on earliness genes in rice, with special reference to analysis of isoalleles at the E locus. Jpn. J. Genet. 1976, 51, 115–128. [Google Scholar] [CrossRef]
- Okumoto, Y.; Tanisaka, T.; Yamagata, H. Heading-time genes of the rice varieties grown in south-west-warm region in Japan. Jpn. J. Breed. 1991, 41, 135–152. [Google Scholar] [CrossRef]
- Okumoto, T.; Tanisaka, T.; Yamagata, H. Heading-time genes of the rice varieties grown in the Tohoku-Hokuriku region in Japan. Jpn. J. Breed. 1992, 42, 121–135. [Google Scholar] [CrossRef]
- Okumoto, Y.; Yoshimura, A.; Tanisaka, T.; Yamagata, H. Analysis of a rice variety Taichung 65 and its isogenic early-heading lines for late-heading genes E1, E2 and E3. Jpn. J. Breed. 1992, 42, 415–429. [Google Scholar] [CrossRef]
- Ichitani, K.; Okumoto, Y.; Tanisaka, T. Photoperiod sensitivity gene of Se-1 locus found in photoperiod insensitive rice cultivars of the northern limit region of rice cultivation. Breed. Sci. 1997, 47, 145–152. [Google Scholar]
- Ichitani, K.; Okumoto, Y.; Tanisaka, T. Genetic analysis of low photoperiod sensitivity of rice cultivars for the northernmost regions of Japan. Plant Breed. 1998, 117, 543–547. [Google Scholar] [CrossRef]
- Ichitani, K.; Inoue, H.; Nishida, H.; Okumoto, Y.; Tanisaka, T. Interactive effects of two heading-time loci, Se1 and Ef1, on pre-flowering developmental phases in rice (Oryza sativa L.). Euphytica. 2002, 12, 227–234. [Google Scholar] [CrossRef]
- Saito, H.; Okumoto, Y.; Teranishi, T.; Yuan, Q.; Nakazaki, T.; Tanisaka, T. Heading time genes responsible for the regional adaptability of ‘Tongil-type short-culmed rice cultivars’ developed in Korea. Breed. Sci. 2007, 57, 135–143. [Google Scholar] [CrossRef][Green Version]
- Kinoshita, T. Report of the committee on gene symbolization, nomenclature and linkage groups. Rice Genet. Let. 1986, 3, 3. [Google Scholar]
- Yano, M.; Sasaki, T. Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 1997, 35, 145–153. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Gene Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Matsuoka, M. Genome-wide association study using whole genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Okumoto, Y.; Tanisaka, T. Trisomic analysis of a strong photoperiod-sensitivity gene E1 in rice (Oryza sativa L.). Euphytica 1997, 95, 301–307. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H. Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.). Theor. Appl. Genet. 2005, 111, 393–398. [Google Scholar] [CrossRef]
- Nonoue, Y.; Fujino, K.; Hirayama, Y.; Yamanouchi, U.; Lin, S.Y.; Yano, M. Detection of quantitative trait loci controlling extremely early heading in rice. Theor. Appl. Genet. 2008, 116, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Yamanouchi, U.; Yano, M. Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 2013, 126, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nishida, H.; Okumoto, Y.; Tanisaka, T. Identification of an early heading time gene found in the Taiwanese rice cultivar Taichung 65. Breed. Sci. 1998, 48, 103–108. [Google Scholar] [CrossRef]
- Tsai, K.H.; Oka, H. Genetic studies of yielding capacity and adaptability in crop plants. 4. Effects on an earliness gene, mb in the genetic background of a rice variety, Taichung 65. Bot. Bull. Acad. Sinica. 1970, 11, 16–26. [Google Scholar]
- Itoh, Y.; Sano, Y. Phyllochron dynamics under controlled environments in rice (Oryza sativa L.). Euphytica 2006, 150, 87–95. [Google Scholar] [CrossRef]
- Eiguchi, M.; Sano, Y. Evolutionary significance of chromosome 7 in an annual type of wild rice. Rice Genet. Newsl. 1995, 12, 187. [Google Scholar]
- The Rice Annotation Project Database. Available online: http://rapdb.dna.affrc.go.jp (accessed on 29 August 2019).
- Lu, L.; Yan, W.H.; Shao, D.; Xing, Y.Z. Evolution and association analysis of Ghd7 in rice. PLoS ONE 2012, 7, e34021. [Google Scholar] [CrossRef]
- Vergara, B.S.; Chang, T.T. The Flowering Response of the Rice Plant to Photoperiod: A Review of the Literature, 2nd ed.; IRRI: Manila, Philippine, 1985; pp. 1–65. [Google Scholar]
- Izawa, T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2011, 58, 3091–3097. [Google Scholar] [CrossRef]
- Tsuji, H.; Taoka, K.; Shimamoto, K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 2011, 14, 45–52. [Google Scholar] [CrossRef]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Kim, H.J.; Kim, Y.K.; Park, J.Y.; Kim, J. Light signaling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002, 29, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Izawa, T. The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 2013, 6, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Osugi, A.; Itoh, H.; Ikeda-Kawakatsu, K.; Takano, M.; Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011, 157, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Shinada, H.; Yamamoto, T.; Yamamoto, E.; Hori, K.; Yonemaru, J.; Matsuba, S.; Fujino, K. Historical changes in population structure during rice breeding programs in the northern limits of rice cultivation. Theor. Appl. Genet. 2014, 127, 995–1004. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, T.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2001, 12, 2473–2484. [Google Scholar] [CrossRef]
- Fujino, K.; Wu, J.; Sekiguchi, H.; Ito, T.; Izawa, T.; Matsumoto, T. Multiple introgression events surrounding the Hd1 flowering-time gene in cultivated rice, Oryza sativa L. Mol. Gen. Genom. 2010, 284, 137–146. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, X.M.; Lu, G.; Zhong, Z.; Gao, H.; Chen, L.; Wu, C.; Wang, H.J.; Wang, Q.; Zhou, K.; et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 2775–2780. [Google Scholar] [CrossRef]
- Gao, H.; Jin, M.; Zheng, X.M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Yan, W.; Zhang, Z.; Lu, L.; Han, Z.; Zhao, H.; Liu, H.; Song, P.; Hu, Y.; et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015, 208, 1056–1066. [Google Scholar] [CrossRef]
- Zheng, X.M.; Feng, L.; Wang, J.; Qiao, W.; Zhang, L.; Cheng, Y.; Yang, Q. Nonfunctional alleles of long-day suppressor genes independently regulate flowering time. J. Int. Plant Biol. 2016, 58, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Teshima, K.M.; Yokoi, S.; Innan, H.; Shimamoto, K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 2009, 106, 4555–4560. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Ebana, K.; Shibaya, T.; Wu, J.; Matsubara, K.; Kanamori, H.; Yamane, H.; Yamanouchi, U.; Mizubayashi, T.; Kono, I.; Shomura, A.; et al. Uncovering of major genetic factors generating naturally occurring variation in heading date among Asian rice cultivars. Theor. Appl. Genet. 2011, 122, 1199–1210. [Google Scholar] [CrossRef]
- Koo, B.H.; Yoo, S.C.; Park, J.W.; Kwon, C.T.; Lee, B.D.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 2013, 6, 1877–1888. [Google Scholar] [CrossRef]
- Ebana, K.; Kojima, K.; Fukuoka, S.; Nagamine, T.; Kawase, M. Development of mini core collection of Japanese rice landrace. Breed Sci. 2008, 58, 281–291. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).