The Plant NF-Y DNA Matrix In Vitro and In Vivo
Abstract
1. Introduction
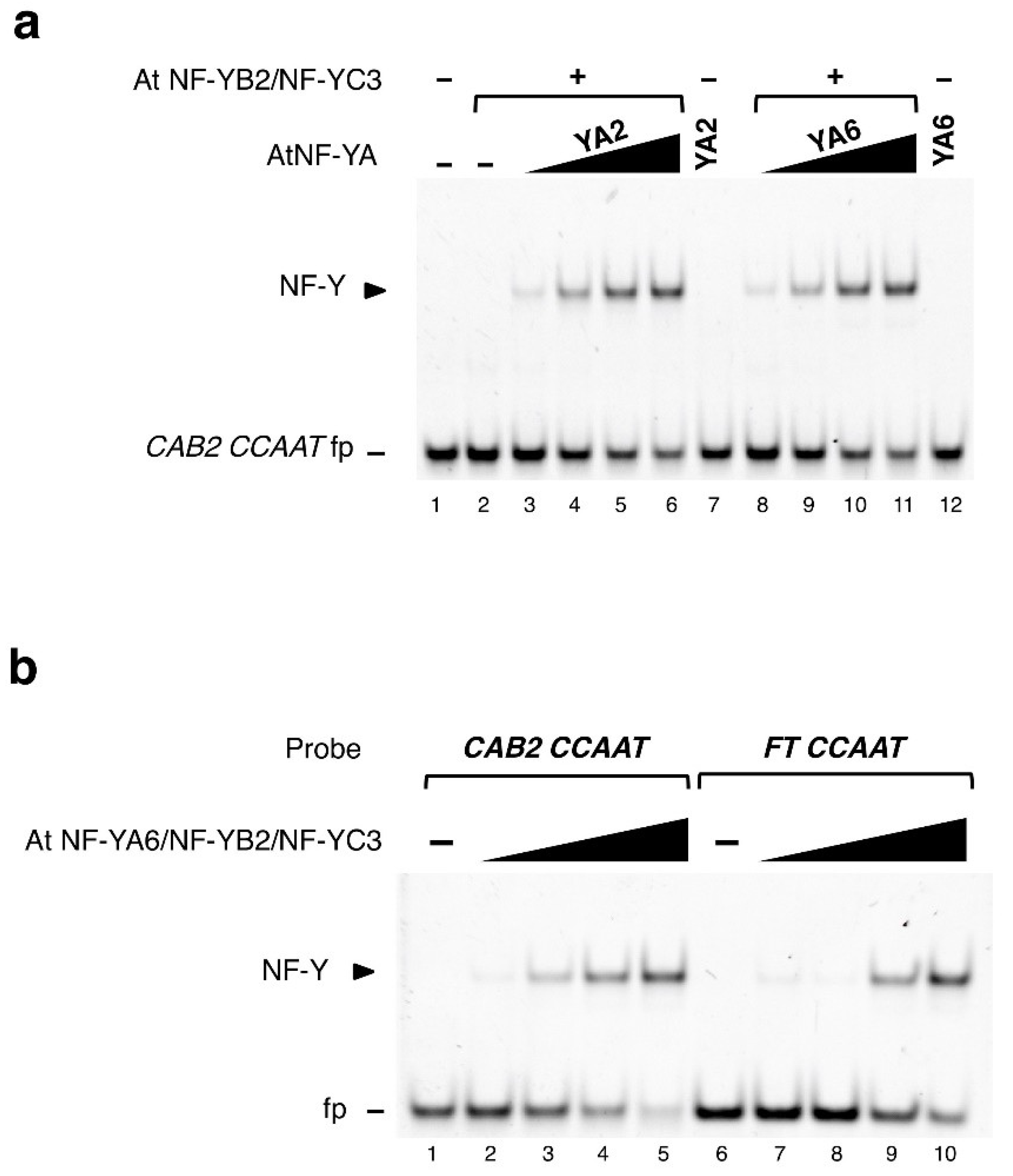
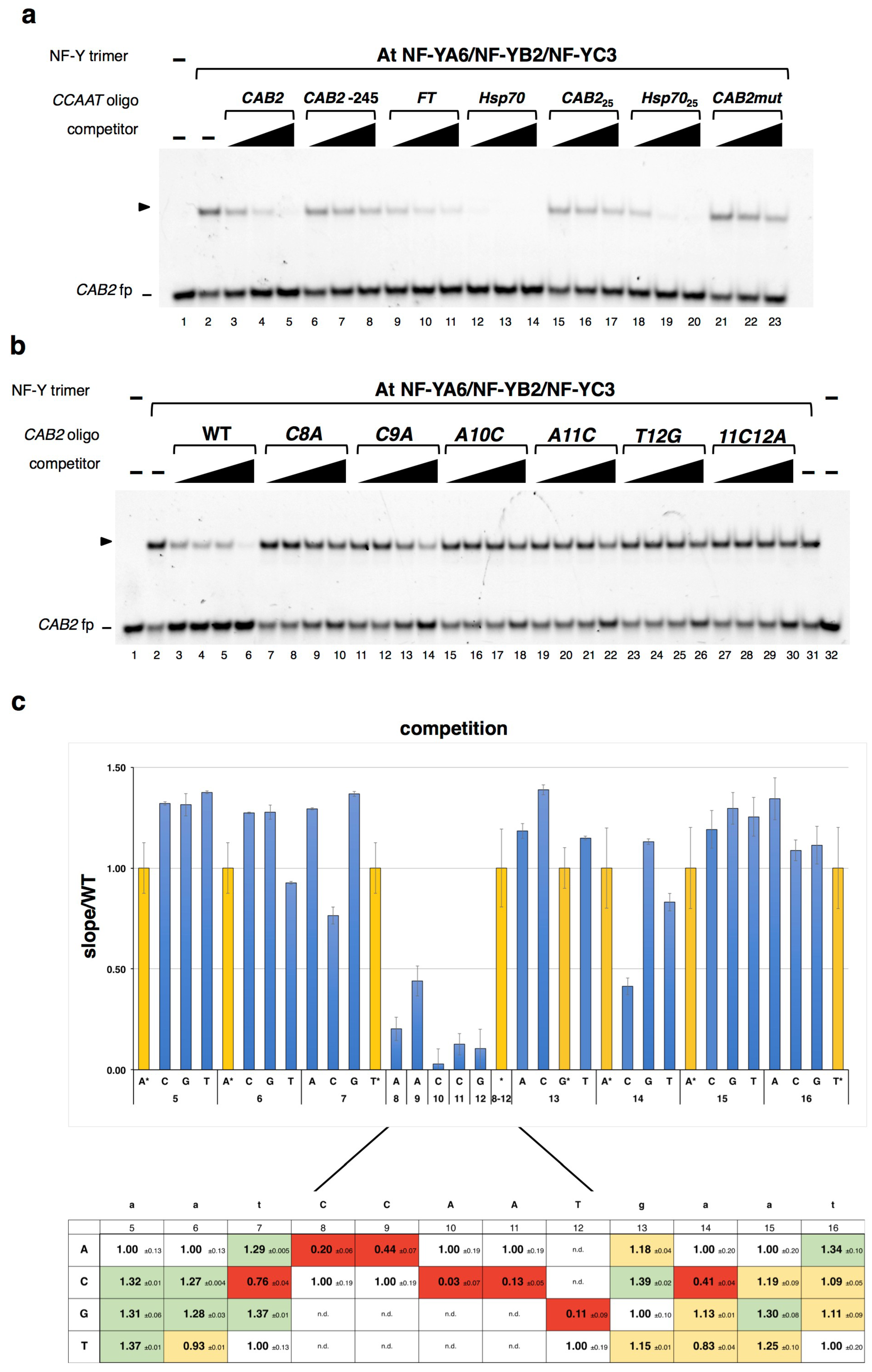
2. Results
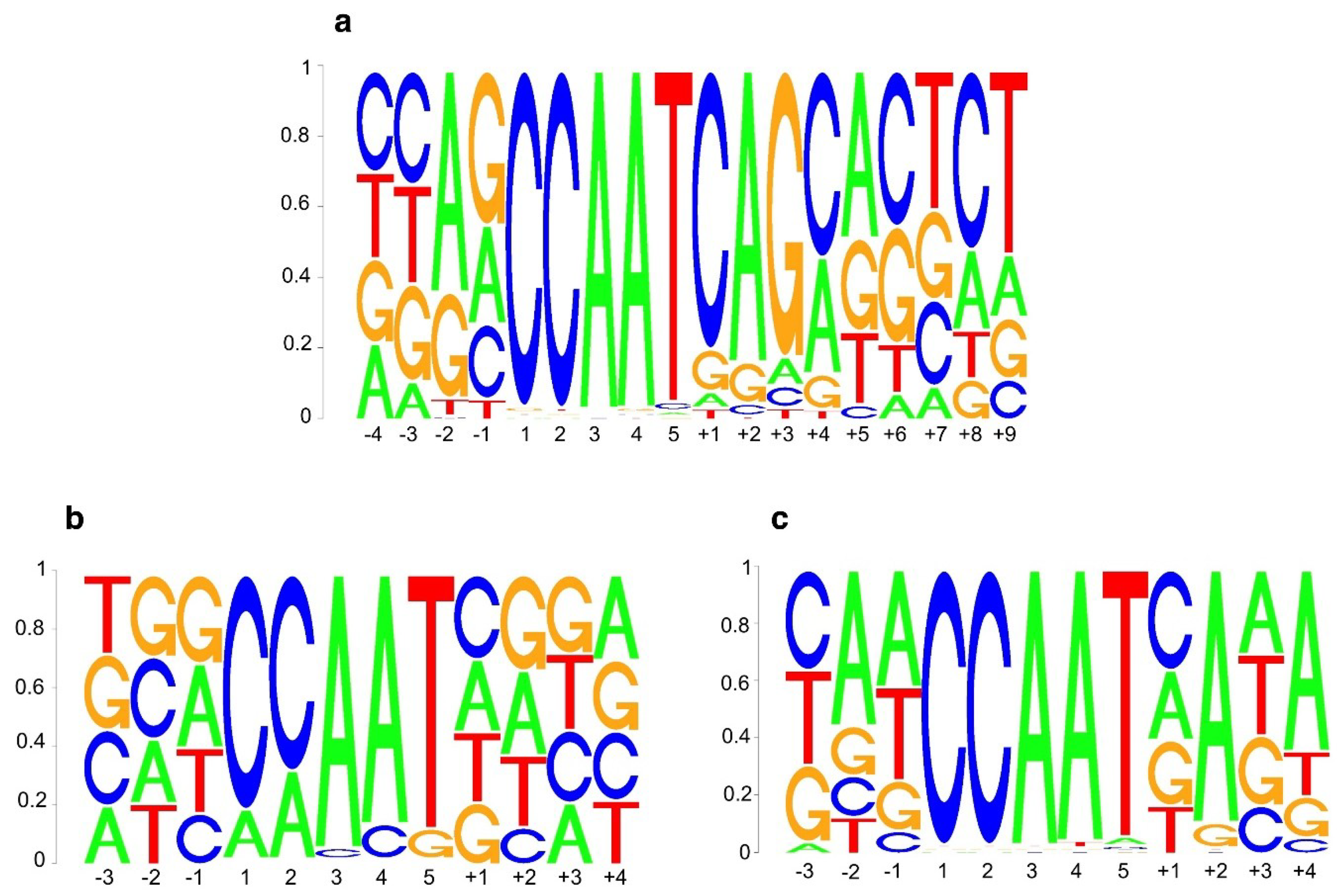
2.1. DNA Sequence-Specificity of the NF-Y Trimer
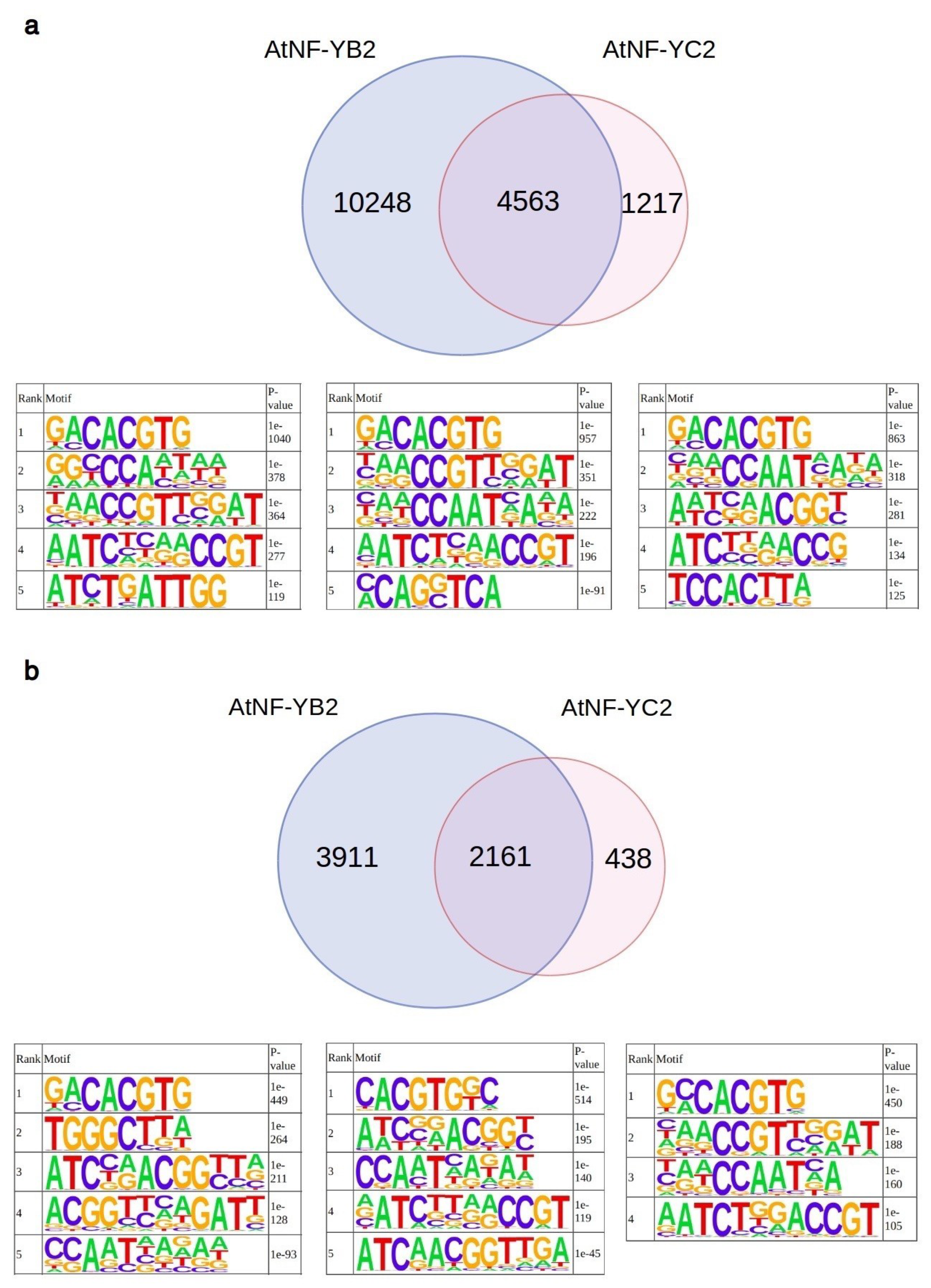
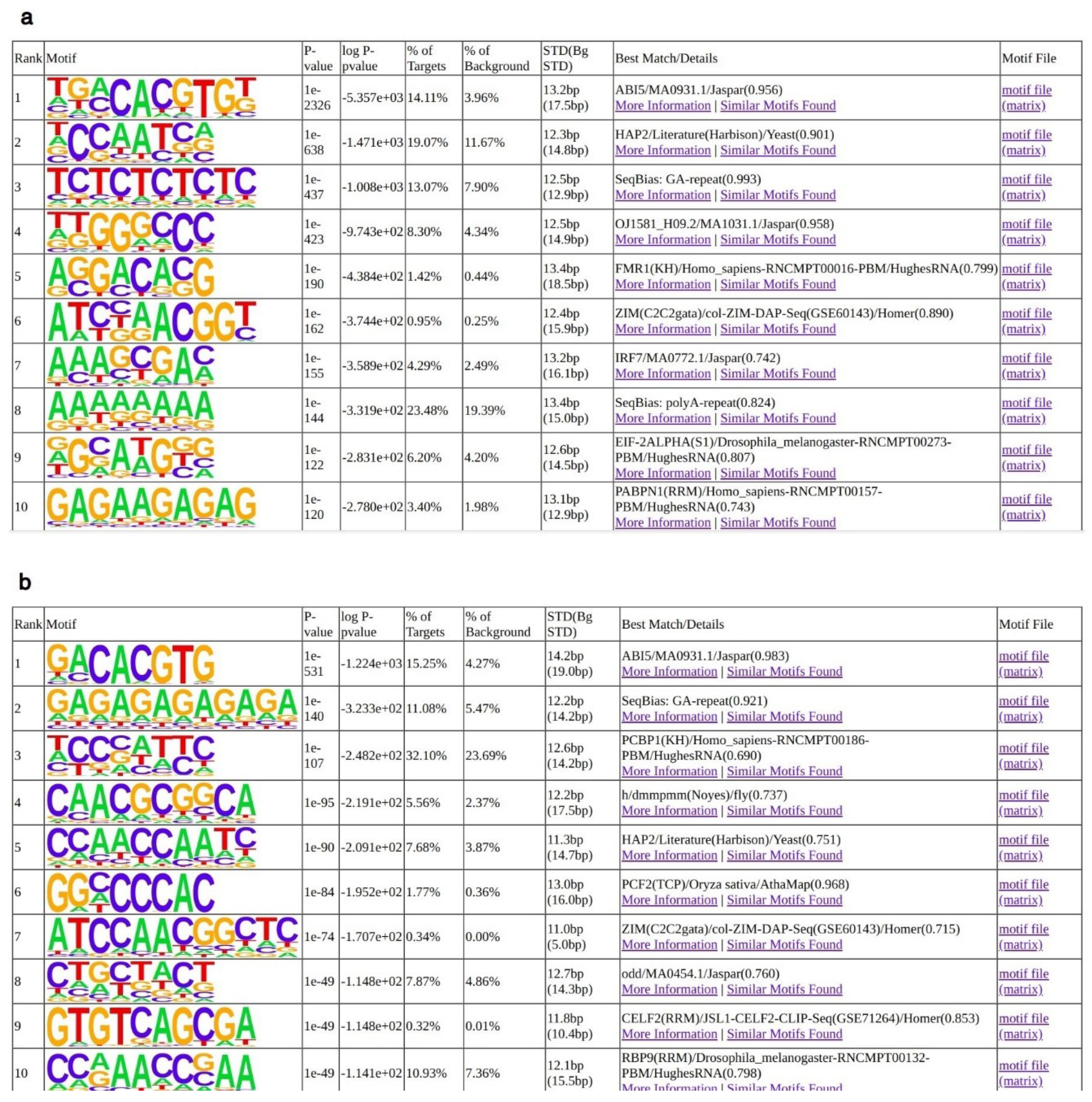
2.2. Analyses of AtNF-YB2, AtNF-YC2 and LEC1 Binding Sites In Vivo
3. Discussion
4. Materials and Methods
4.1. Protein Expression, Purification
4.2. EMSAs and In Vitro Competition Analyses
4.3. Analysis of AtNF-YB2, AtNF-YC2 and LEC1 Binding Sites In Vivo
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.; Cattoglio, C.; Tjian, R. Looping back to leap forward: Transcription enters a new era. Cell 2014, 157, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.D.; Pavesi, G.; Benatti, P.; Imbriano, C.; Mantovani, R.; Struhl, K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013, 23, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, A.J.; Yang, P.; Conway, A.E.; Cinghu, S.; Freudenberg, J.M.; Yellaboina, S.; Jothi, R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 2014, 55, 708–722. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Shen, L.; Gu, X.; Wang, Y.; Yu, H.; He, Y. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 2017, 551, 124–128. [Google Scholar] [CrossRef]
- Oldfield, A.J.; Henriques, T.; Burkholder, A.B.; Paulet, D.; Cinghu, S.; Yang, P.; Scruggs, B.S.; Lavender, C.A.; Kumar, D.; Bennett, B.; et al. NF-Y Controls Fidelity of Transcription Initiation at Gene Promoters Through Maintenance of the Nucleosome-Depleted Region. Nat. Commun. 2019, 10, 3072. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Zambelli, F.; Pavesi, G.; Mantovani, R. A perspective of promoter architecture from the CCAAT box. Cell Cycle 2009, 8, 4127–4137. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Scharf, D.H.; Hortschansky, P.; Groll, M.; Brakhage, A.A. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 2012, 20, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 2002, 283, 41–48. [Google Scholar] [CrossRef]
- Swain, S.; Myers, Z.A.; Siriwardana, C.L.; Holt, B.F. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 636–644. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Rípodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 645–654. [Google Scholar] [CrossRef]
- Brambilla, V.; Fornara, F. Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 655–660. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Goretti, D.; Martignago, D.; Landini, M.; Brambilla, V.; Gómez-Ariza, J.; Gnesutta, N.; Galbiati, F.; Collani, S.; Takagi, H.; Terauchi, R.; et al. Transcriptional and Post-Transcriptional Mechanisms Limit Heading Date 1 (Hd1) Function to Adapt Rice to High Latitudes. PLoS Genet. 2017, 13, e1006530. [Google Scholar] [CrossRef]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.; Horner, D.S.; Holt, B.F.; Mantovani, R. CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef]
- Luo, X.; Gao, Z.; Wang, Y.; Chen, Z.; Zhang, W.; Huang, J.; Yu, H.; He, Y. The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 2018, 95, 17–29. [Google Scholar] [CrossRef]
- Gatta, R.; Ceribelli, M.; Dolfini, D.; Vigano, A.M.; Pavesi, G.; Mantovani, R.; Merico, D. The Histone-Like NF-Y Is a Bifunctional Transcription Factor. Mol. Cell. Biol. 2008, 28, 2047–2058. [Google Scholar]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.Y.; Greven, M.C.; Kim, B.H.; Moore, J.; Pierce, B.G.; Dong, X.; Virgil, D.; et al. Factorbook.org: A Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013, 41, D171–D176. [Google Scholar] [CrossRef] [PubMed]
- Haubrock, M.; Hartmann, F.; Wingender, E. NF-Y Binding Site Architecture Defines a C-Fos Targeted Promoter Class. PLoS ONE 2016, 11, e0160803. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Adrian, J.; Farrona, S.; Reimer, J.J.; Albani, M.C.; Coupland, G.; Turck, F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 2010, 22, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Siriwardana, C.L.; Gnesutta, N.; Kumimoto, R.W.; Jones, D.S.; Myers, Z.A.; Mantovani, R.; Holt, B.F. NUCLEAR FACTOR Y, Subunit A (NF-YA) Proteins Positively Regulate Flowering and Act through FLOWERING LOCUS T. PLoS Genet. 2016, 12, e1006496. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, D.M.; Degenhardt, J.; Winicov, I.; Tobin, E.M. Two 10-bp Regions Are Critical for Phytochrome Regulation of a Lemna gibba Lhcb Gene Promoter. Plant Cell 1994, 6, 1123–1134. [Google Scholar] [PubMed]
- Folta, K.M.; Kaufman, L.S. Regions of the Pea Lhcb 1*4 Promoter Necessary for Blue-Light Regulation in Transgenic Arabidopsis. Plant Physiol. 1999, 120, 747–756. [Google Scholar] [CrossRef]
- Warpeha, K.M.; Upadhyay, S.; Yeh, J.; Adamiak, J.; Hawkins, S.I.; Lapik, Y.R.; Anderson, M.B.; Kaufman, L.S. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007, 143, 1590–1600. [Google Scholar] [CrossRef]
- Carre, I.A.; Kay, S.A. Multiple DNA-Protein Complexes at a Circadian-Regulated Promoter Element. Plant Cell 1995, 7, 2039–2051. [Google Scholar] [CrossRef]
- Song, L.; Huang, S.-S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, aag1550. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef]
- Gnesutta, N.; Saad, D.; Chaves-Sanjuan, A.; Mantovani, R.; Nardini, M. Crystal Structure of the Arabidopsis thaliana L1L/NF-YC3 Histone-Fold Dimer Reveals Specificities of the LEC1 Family of NF-Y Subunits in Plants. Mol. Plant 2017, 10, 645–648. [Google Scholar] [CrossRef]
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 2019, 61, 564–580. [Google Scholar] [CrossRef]
- Calvenzani, V.; Testoni, B.; Gusmaroli, G.; Lorenzo, M.; Gnesutta, N.; Petroni, K.; Mantovani, R.; Tonelli, C. Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 2012, 7, e42902. [Google Scholar] [CrossRef]
- Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef]
- Trigg, S.A.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.-S.C.; et al. CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 2017, 14, 819–825. [Google Scholar] [CrossRef]
- Dorn, A.; Bollekens, J.; Staub, A.; Benoist, C.; Mathis, D. A multiplicity of CCAAT box-binding proteins. Cell 1987, 50, 863–872. [Google Scholar] [CrossRef]
- Liberati, C.; di Silvio, A.; Ottolenghi, S.; Mantovani, R. NF-Y binding to twin CCAAT boxes: Role of Q-rich domains and histone fold helices 1 1Edited by M. Yaniv. J. Mol. Biol. 1999, 285, 1441–1455. [Google Scholar] [CrossRef]
- Bi, W.; Wu, L.; Coustry, F.; de Crombrugghe, B.; Maity, S.N. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 1997, 272, 26562–26572. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.-S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnesutta, N.; Chiara, M.; Bernardini, A.; Balestra, M.; Horner, D.S.; Mantovani, R. The Plant NF-Y DNA Matrix In Vitro and In Vivo. Plants 2019, 8, 406. https://doi.org/10.3390/plants8100406
Gnesutta N, Chiara M, Bernardini A, Balestra M, Horner DS, Mantovani R. The Plant NF-Y DNA Matrix In Vitro and In Vivo. Plants. 2019; 8(10):406. https://doi.org/10.3390/plants8100406
Chicago/Turabian StyleGnesutta, Nerina, Matteo Chiara, Andrea Bernardini, Matteo Balestra, David S. Horner, and Roberto Mantovani. 2019. "The Plant NF-Y DNA Matrix In Vitro and In Vivo" Plants 8, no. 10: 406. https://doi.org/10.3390/plants8100406
APA StyleGnesutta, N., Chiara, M., Bernardini, A., Balestra, M., Horner, D. S., & Mantovani, R. (2019). The Plant NF-Y DNA Matrix In Vitro and In Vivo. Plants, 8(10), 406. https://doi.org/10.3390/plants8100406





