Genome-Wide Identification of WRKY Transcription Factors in the Asteranae
Abstract
1. Introduction
2. Results
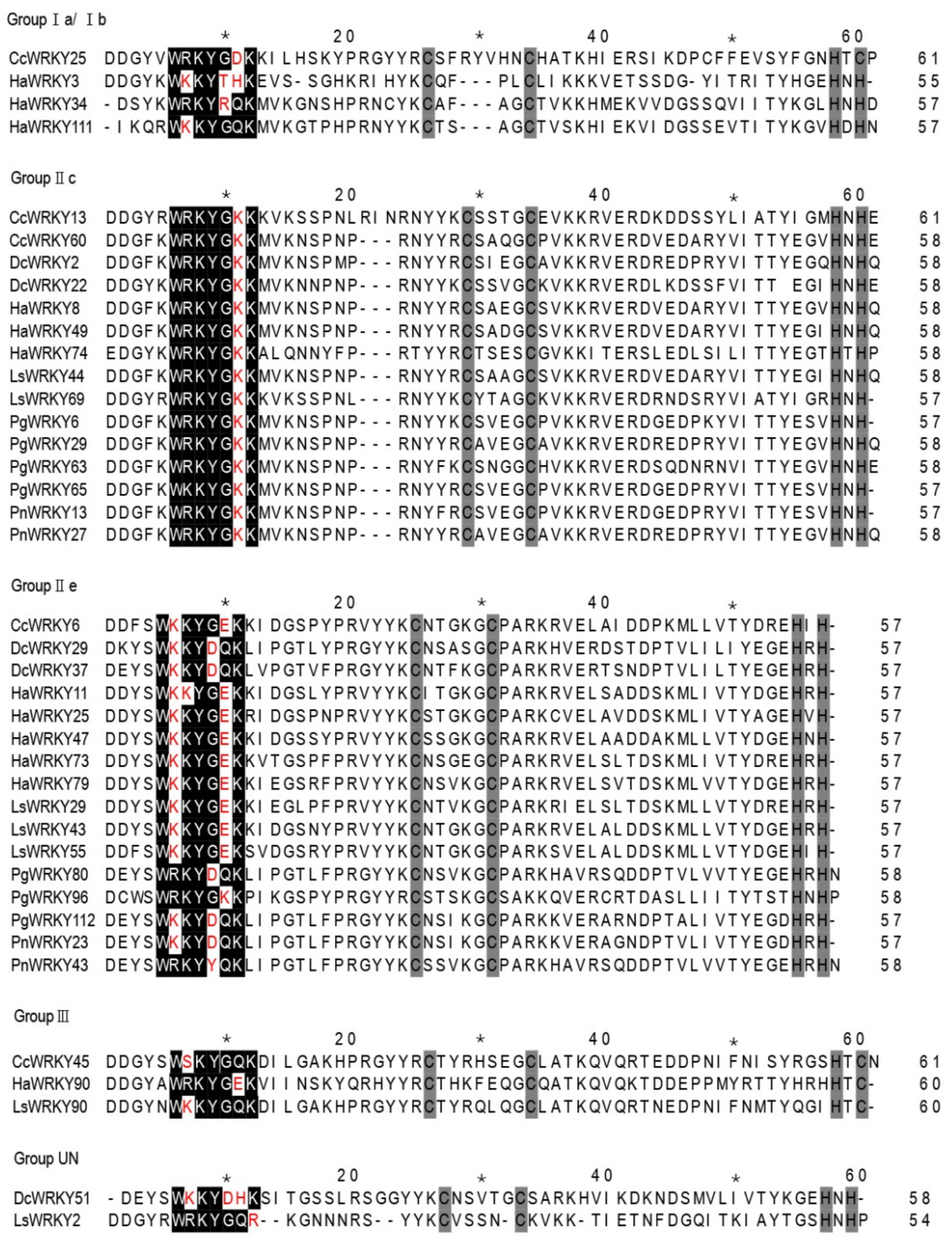
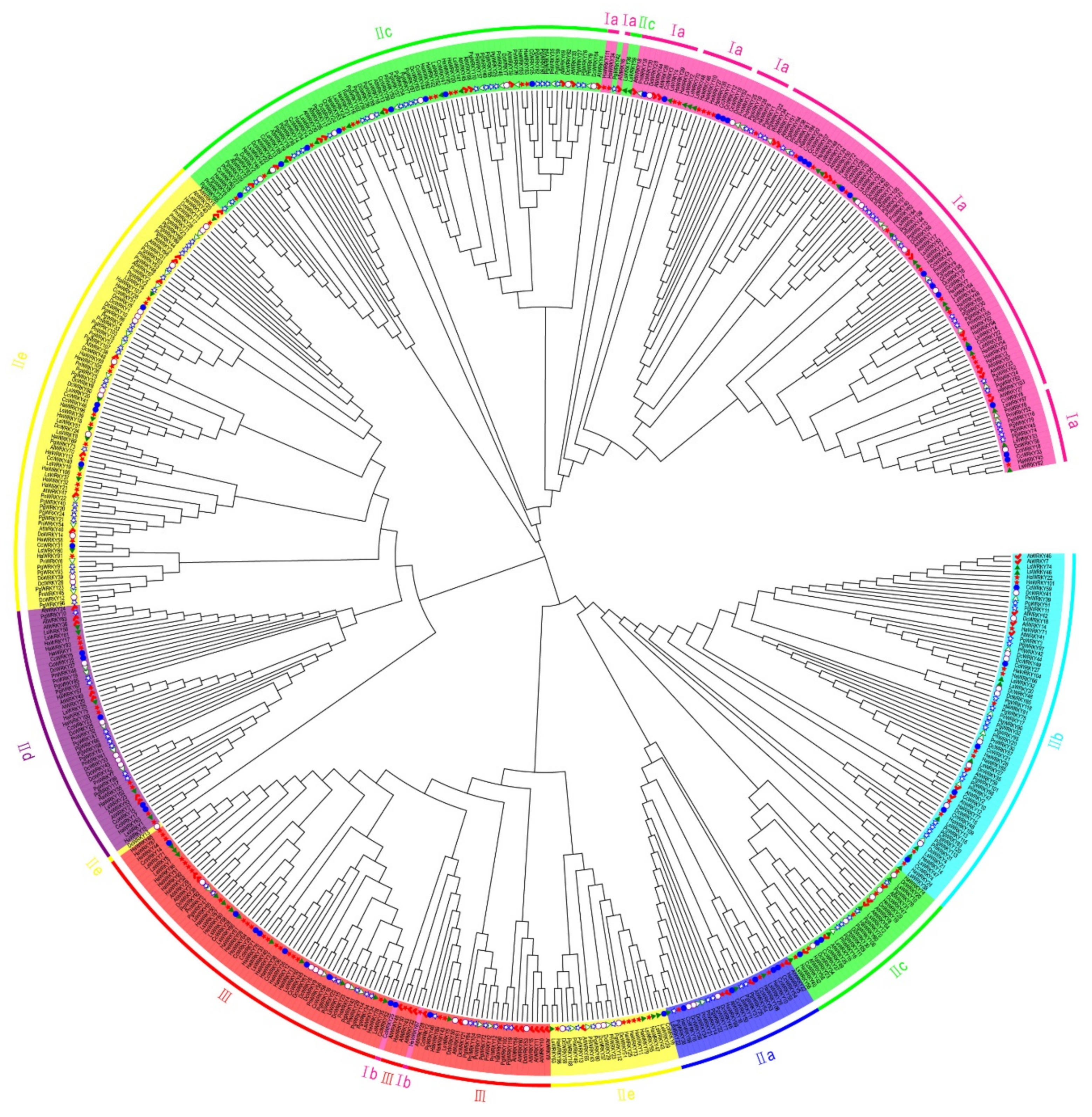
2.1. Phylogenetic Analysis and Classification of WRKY Genes
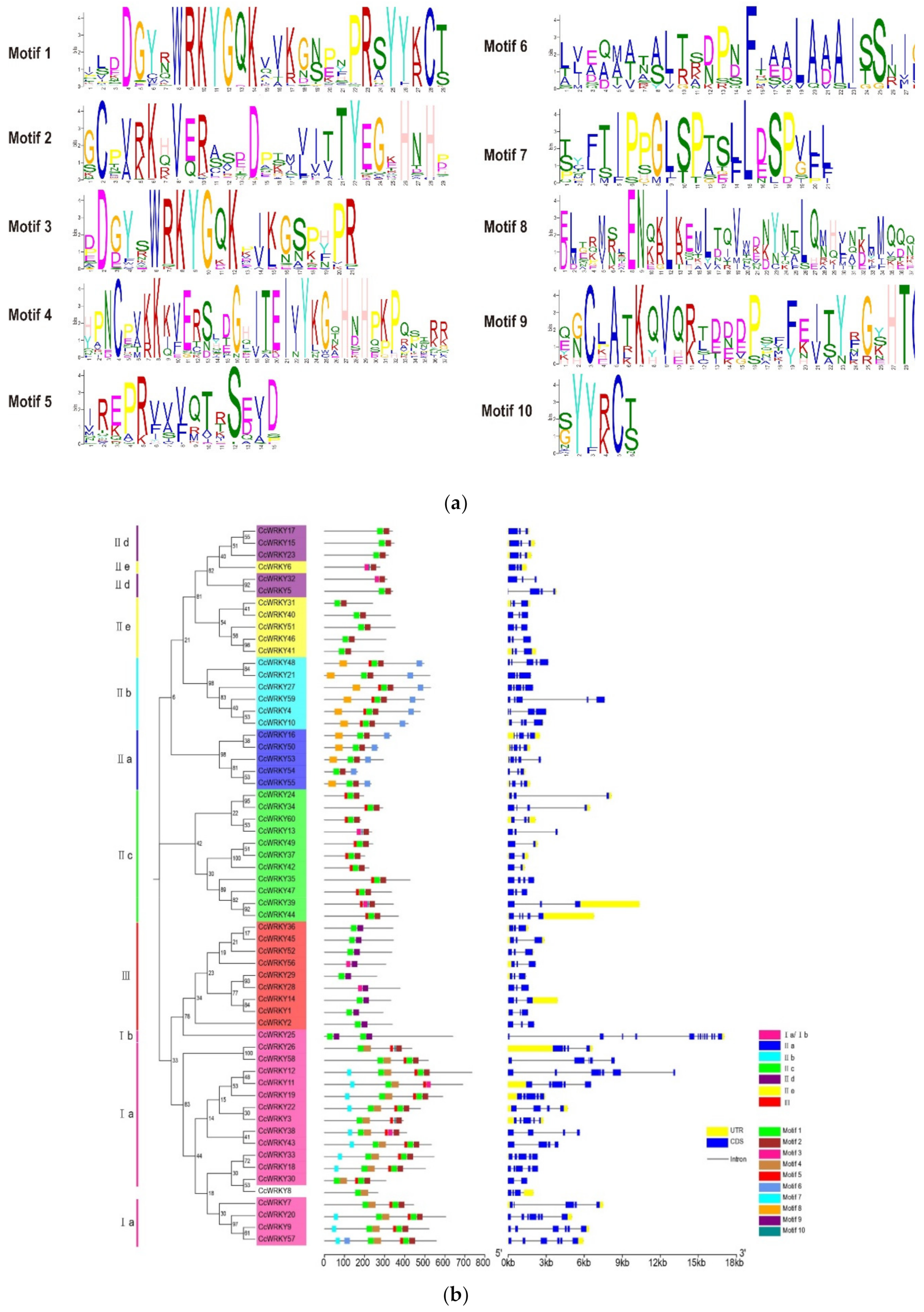
2.2. Conserved Motif and Gene Structure Analysis of WRKY Genes
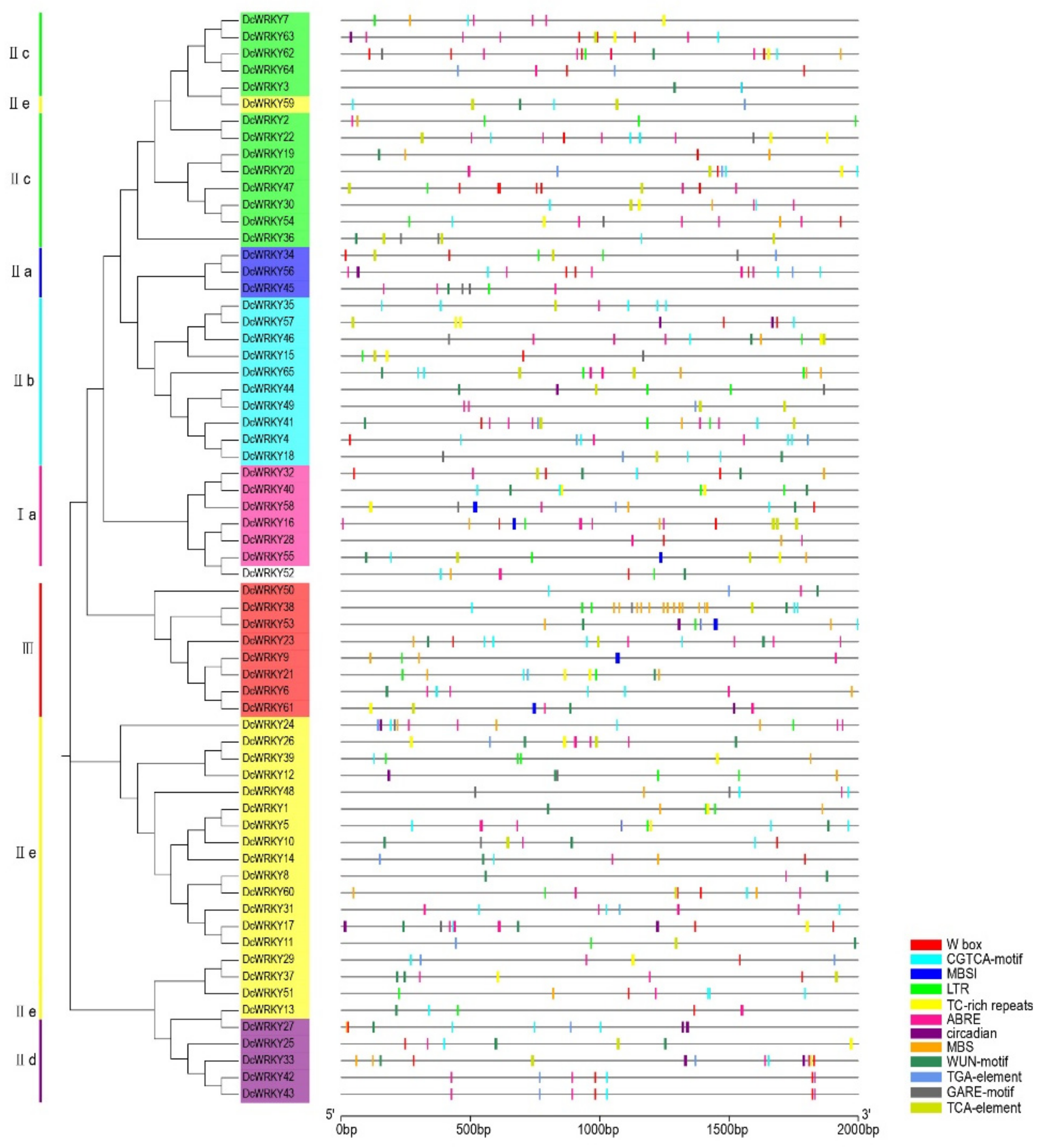
2.3. Analysis of WRKY Gene Promoters
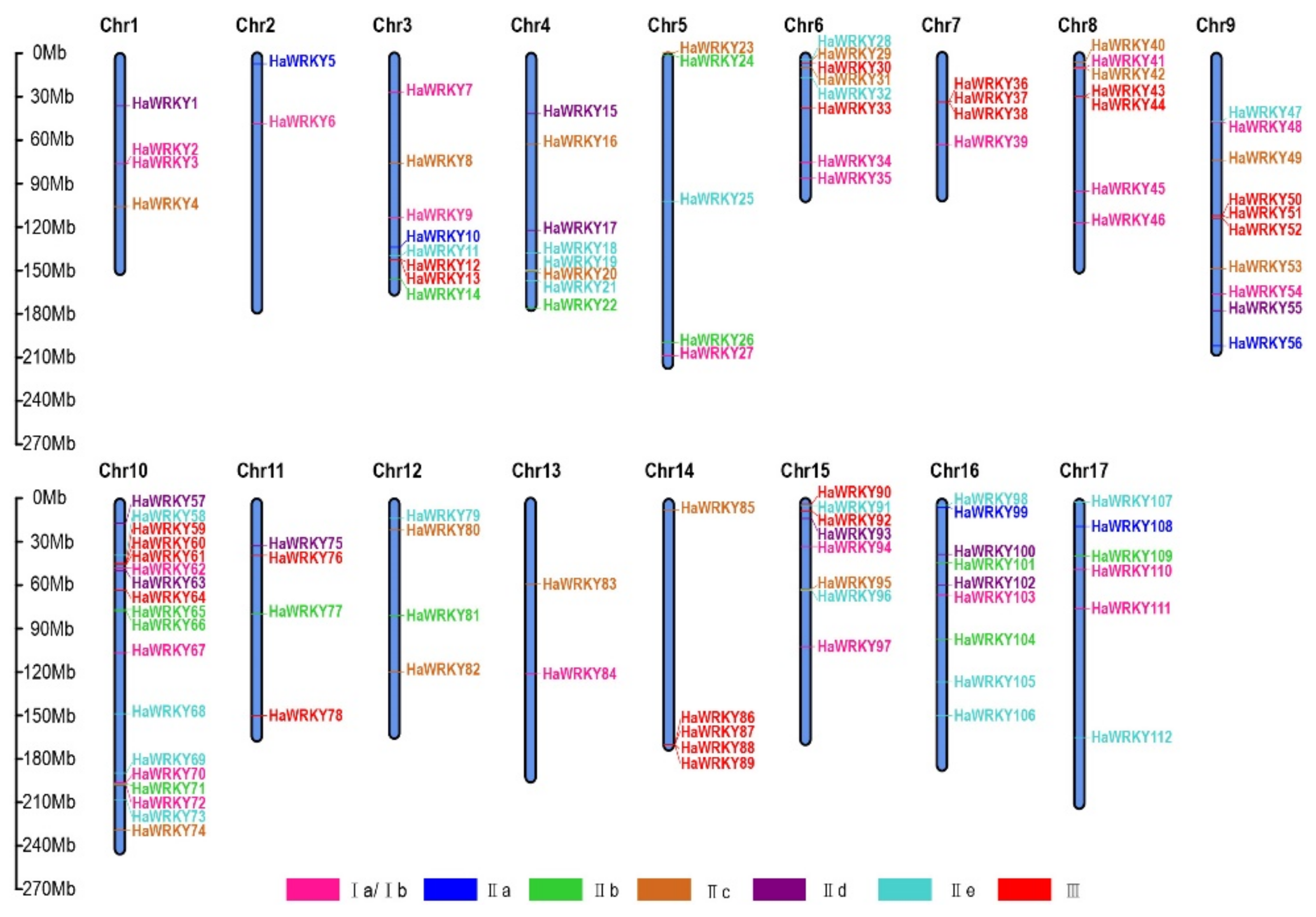
2.4. Chromosomal Distribution and Duplication of WRKY Genes
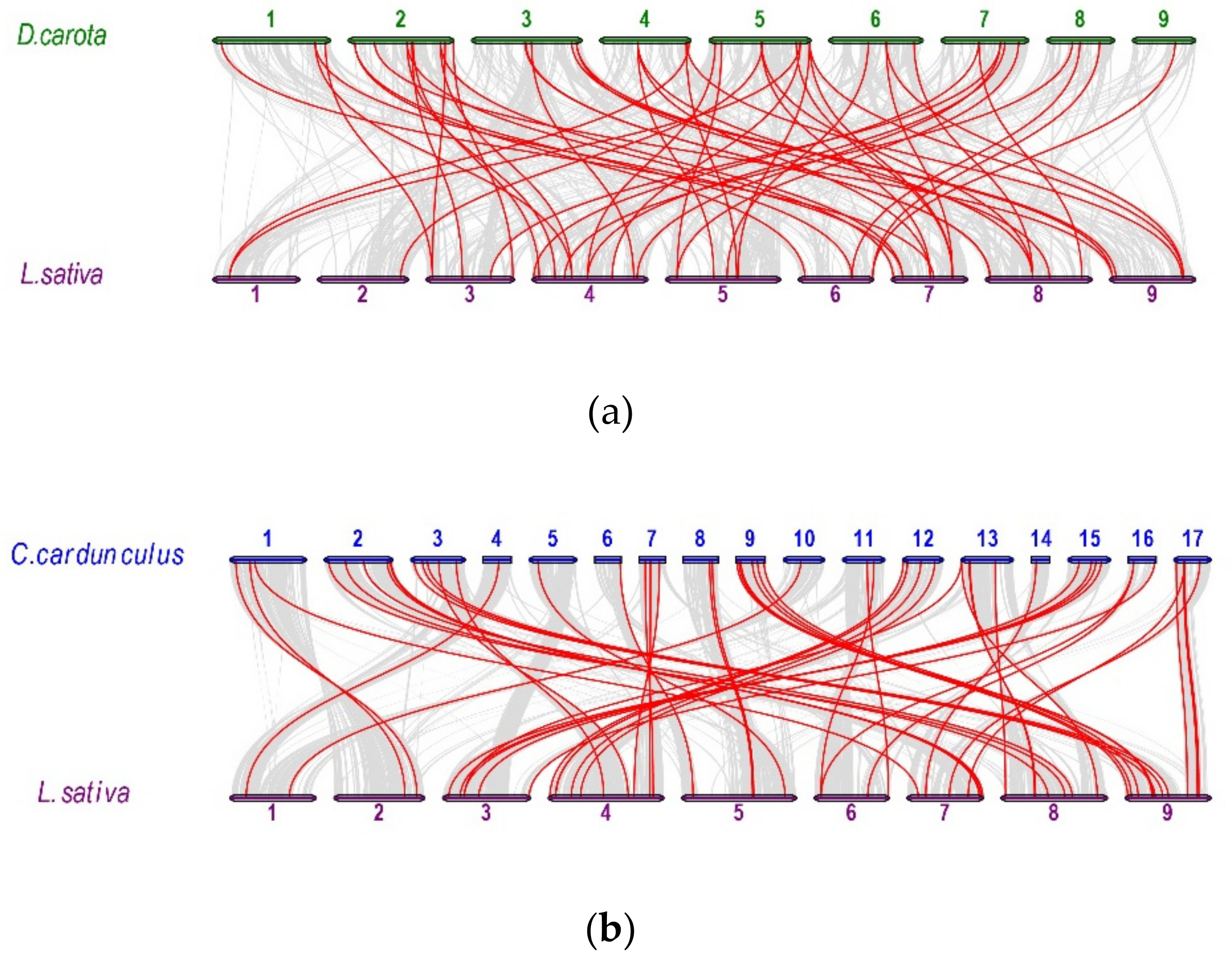
2.5. Analysis of Synteny Between Asterales and Apiales Genomes
3. Discussion
4. Materials and Methods
4.1. Identification of WRKY Genes
4.2. Classification and Phylogenetic Analysis of WRKY Genes
4.3. WRKY Protein Conserved Motif and Gene Structure Analysis
4.4. Chromosomal Localization of WRKY Genes and Cis-Acting Element Analysis
4.5. Analysis of WRKY Gene Duplication and Synteny Among Species
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, L.G.; Xiang, S.Y.; Chen, Y.L.; Li, D.B.; Yu, D.Q. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Xiu, H.; Nuruzzaman, M.; Guo, X.Q.; Cao, H.Z.; Huang, J.J.; Chen, X.H.; Wu, K.L.; Zhang, R.; Huang, Y.Z.; Luo, J.L.; et al. Molecular cloning and expression analysis of eight PgWRKY genes in Panax ginseng responsive to salt and hormones. Int. J. Mol. Sci. 2016, 17, 319. [Google Scholar] [CrossRef] [PubMed]
- Raineri, J.; Hartman, M.D.; Chan, R.L.; Iglesias, A.A.; Ribichich, K.F. A sunflower WRKY transcription factor stimulates the mobilization of seed-stored reserves during germination and post-germination growth. Plant Cell Rep. 2016, 35, 1875–1890. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Lv, S.S.; Liu, R.; Fan, S.X.; Liu, C.J.; Liu, R.Y.; Han, Y.Y. Transcriptomic analysis reveals the roles of gibberellin-regulated genes and transcription factors in regulating bolting in lettuce (Lactuca sativa L.). PLoS ONE 2018, 13, e0191518. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Li, M.; Zhao, D.; Yang, J.F.; Fu, J.D.; Xu, Z.S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.D.; Yu, D.Q. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [Google Scholar] [CrossRef]
- Chen, M.H.; Yan, T.X.; Shen, Q.; Lu, X.; Pan, Q.F.; Huang, Y.R.; Tang, Y.L.; Fu, X.Q.; Liu, M.; Jiang, W.M.; et al. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 2017, 214, 304–316. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.L.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Xu, W.Q.; Choi, H.K.; Huang, L.F. State of Panax ginseng research: A global analysis. Molecules 2017, 22, 1518. [Google Scholar] [CrossRef]
- Wang, T.; Guo, R.X.; Zhou, G.H.; Zhou, X.D.; Kou, Z.Z.; Sui, F.; Li, C.; Tang, L.Y.; Wang, Z.J. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: A review. J. Ethnopharmacol. 2016, 188, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Que, F.; Hou, X.L.; Wang, G.L.; Xu, Z.S.; Tan, G.F.; Li, T.; Wang, Y.H.; Khadr, A.; Xiong, A.S. Advances in research on the carrot, an important root vegetable in the Apiaceae. Hortic. Res. 2019, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, V.; Kroon, P.A.; Linsalata, V.; Cardinali, A. Globe artichoke: A functional food and source of nutraceutical ingredients. J. Funct. Foods 2009, 1, 131–144. [Google Scholar] [CrossRef]
- Guo, S.S.; Ge, Y.; Na Jom, K. A review of phytochemistry, metabolite changes, and medicinal uses of the common sunflower seed and sprouts (Helianthus annuus L.). Chem. Cent. J. 2017, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Moon, Y.; Tou, J.C.; Mou, B.; Waterland, N.L. Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.). J. Food Compos. Anal. 2016, 49, 19–34. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.N.; Wang, J.; Wong, G.K.; Li, S.G.; Liu, B.; Deng, Y.J.; Dai, L.; Zhou, Y.; Zhang, X.Q.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef]
- Rice WRKY Working Group. Nomenclature report on rice WRKY’s–Conflict regarding gene names and its solution. Rice 2012, 5, 3. [Google Scholar] [CrossRef]
- Dong, J.X.; Chen, C.H.; Chen, Z.X. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhou, L.X.; Lei, X.T.; Cao, H.X.; Wang, Y.; Dou, Y.J.; Tang, W.Q.; Xia, W. Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS ONE 2017, 12, e0189224. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mu, S.H.; Cheng, Z.C.; Cheng, Y.W.; Zhang, Y.; Miao, Y.; Hou, C.L.; Li, X.P.; Gao, J. Characterization and expression analysis of the WRKY gene family in moso bamboo. Sci. Rep. 2017, 7, 6675. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.L.; Guo, R.R.; Xu, X.Z.; Gao, M.; Li, X.Q.; Song, J.Y.; Zheng, Y.; Wang, X.P. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef]
- Tang, J.; Wang, F.; Hou, X.L.; Wang, Z.; Huang, Z.N. Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 2013, 32, 781–795. [Google Scholar]
- Jiang, Y.Z.; Duan, Y.J.; Yin, J.; Ye, S.L.; Zhu, J.R.; Zhang, F.Q.; Lu, W.X.; Fan, D.; Luo, K.M. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef]
- Song, H.; Wang, P.F.; Lin, J.Y.; Zhao, C.Z.; Bi, Y.P.; Wang, X.J. Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 2016, 7, 534. [Google Scholar] [CrossRef]
- Nan, H.; Gao, L.Z. Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Genet. 2019, 10, 363. [Google Scholar] [CrossRef]
- Li, M.Y.; Xu, Z.S.; Tian, C.; Huang, Y.; Wang, F.; Xiong, A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016, 6, 23101. [Google Scholar] [CrossRef]
- Jiang, C.M.; Shen, Q.J.; Wang, B.; He, B.; Xiao, S.Q.; Chen, L.; Yu, T.Q.; Ke, X.; Zhong, Q.F.; Fu, J.; et al. Transcriptome analysis of WRKY gene family in Oryza officinalis Wall ex Watt and WRKY genes involved in responses to Xanthomonas oryzae pv. oryzae stress. PLoS ONE 2017, 12, e0188742. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.D.A.D.; Oliveira, J.D.A.D.; Del-Bem, L.E.; Bronze, D.S.E.; Santana, S.R.J.; Peres, G.K.; Vincentz, M.; Micheli, F. Genome-wide identification and characterization of cacao WRKY transcription factors and analysis of their expression in response to witches’ broom disease. PLoS ONE 2017, 12, e0187346. [Google Scholar]
- Yang, Y.; Zhou, Y.; Chi, Y.J.; Fan, B.F.; Chen, Z.X. Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef]
- Sun, W.J.; Zhan, J.Y.; Zheng, T.R.; Sun, R.; Wang, T.; Tang, Z.Z.; Bu, T.L.; Li, C.L.; Wu, Q.; Chen, H. The jasmonate-responsive transcription factor CbWRKY24 regulates terpenoid biosynthetic genes to promote saponin biosynthesis in Conyza blinii H. Lev. J. Genet. 2018, 97, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. PlantCell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Ross, A.; Kramer, K.; Finkemeier, I.; Somssich, I.E. Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. 2018, 96, 487–502. [Google Scholar] [CrossRef]
- Chakraborty, J.; Ghosh, P.; Sen, S.; Das, S. Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci. 2018, 276, 250–267. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, J.; Lin, W.; Li, S.G.; Li, H.; Zhou, J.; Ni, P.X.; Dong, W.; Hu, S.N.; Zeng, C.Q.; et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; Li, H.; Yang, Y.C.; Wang, Y.Q.; Mo, Y.L.; Zhang, R.M.; Zhang, Y.; Ma, J.X.; Wei, C.H.; Zhang, X. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS ONE 2018, 13, e0191308. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.J.; Li, C.H.; Liu, J.R.; Liu, C.Y.; He, Y.H. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Karanja, B.K.; Fan, L.X.; Xu, L.; Wang, Y.; Zhu, X.W.; Tang, M.J.; Wang, R.H.; Zhang, F.; Muleke, E.M.; Liu, L.W. Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Rep. 2017, 36, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Yang, J.; Gu, X. Expression divergence between duplicate genes. Trends Genet. 2005, 21, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.G.; Sun, J.L.; Wang, C.Q.; Dong, Y.M.; Xiao, S.H.; Gao, X.L.; Cao, Q.W.; Li, L.B.; Li, W.D.; Gao, C. Genome-wide characterization, evolutionary analysis of WRKY genes in Cucurbitaceae species and assessment of its roles in resisting to powdery mildew disease. PLoS ONE 2018, 13, e0199851. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chu, Y.; Liao, B.S.; Xiao, S.M.; Yin, Q.G.; Bai, R.; Su, H.; Dong, L.L.; Li, X.W.; Qian, J.; et al. Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience 2017, 6, 1–15. [Google Scholar] [CrossRef]
- Chen, W.; Kui, L.; Zhang, G.H.; Zhu, S.S.; Zhang, J.; Wang, X.; Yang, M.; Huang, H.C.; Liu, Y.X.; Wang, Y.; et al. Whole-genome sequencing and analysis of the chinese herbal plant Panax notoginseng. Mol. Plant 2017, 10, 899–902. [Google Scholar] [CrossRef]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.Y.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Chin-Wo, S.; Wang, Z.W.; Yang, X.H.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.F.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv Bioinf. 2018, 289660. [Google Scholar] [CrossRef]
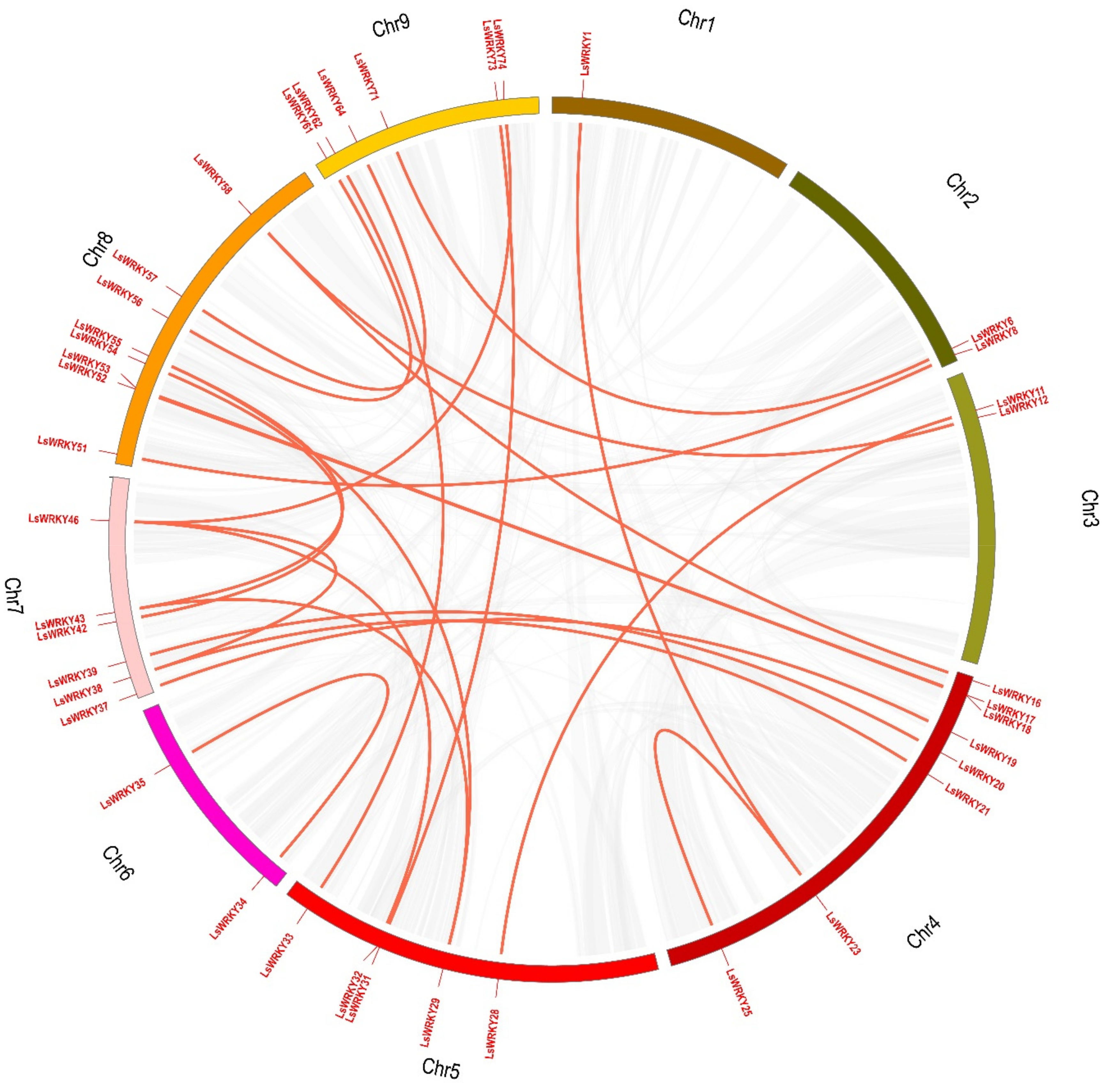
| Tandem Duplication Gene Pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| HaWRKY51–HaWRKY92 | 0.0678063 | 0.174569 | 0.388421 |
| HaWRKY51–HaWRKY52 | 0.0678063 | 0.174569 | 0.388421 |
| HaWRKY104–HaWRKY66 | 0.112547 | 0.468332 | 0.240314 |
| HaWRKY9–HaWRKY84 | 0.088504 | 0.399459 | 0.22156 |
| HaWRKY91–HaWRKY58 | 0.103471 | 0.483353 | 0.214069 |
| HaWRKY95–HaWRKY20 | 0.0970665 | 0.371965 | 0.260956 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Zhang, Y.; Wang, Z.; Lin, L.; Cui, M.; Long, Y.; Xing, Z. Genome-Wide Identification of WRKY Transcription Factors in the Asteranae. Plants 2019, 8, 393. https://doi.org/10.3390/plants8100393
Guo H, Zhang Y, Wang Z, Lin L, Cui M, Long Y, Xing Z. Genome-Wide Identification of WRKY Transcription Factors in the Asteranae. Plants. 2019; 8(10):393. https://doi.org/10.3390/plants8100393
Chicago/Turabian StyleGuo, Hongyu, Yantong Zhang, Zhuo Wang, Limei Lin, Minghui Cui, Yuehong Long, and Zhaobin Xing. 2019. "Genome-Wide Identification of WRKY Transcription Factors in the Asteranae" Plants 8, no. 10: 393. https://doi.org/10.3390/plants8100393
APA StyleGuo, H., Zhang, Y., Wang, Z., Lin, L., Cui, M., Long, Y., & Xing, Z. (2019). Genome-Wide Identification of WRKY Transcription Factors in the Asteranae. Plants, 8(10), 393. https://doi.org/10.3390/plants8100393




